Abstract
Purpose
To investigate the prognostic role of genomic gain for MET and epidermal growth factor receptor (EGFR) genes in surgically resected non–small-cell lung cancer (NSCLC).
Patients and Methods
This retrospective study included 447 NSCLC patients with available tumor tissue from primary lung tumor and survival data. EGFR and MET status was evaluated by fluorescent in situ hybridization (FISH) in tissue microarray sections.
Results
EGFR FISH results were obtained in 376 cases. EGFR gene amplification and high polysomy (EGFR FISH+) were observed in 10.4% and 32.4% of cases, respectively. EGFR FISH-positive patients had a nonsignificant shorter survival than EGFR FISH-negative patients (P = .4). Activating EGFR mutations were detected in 9.7% of 144 stage I-II disease with no impact on survival. MET FISH analysis was performed in 435 cases. High MET gene copy number (mean ≥ 5 copies/cell) was observed in 48 cases (MET+, 11.1%), including 18 cases with true gene amplification (4.1%). MET+ status was associated with advanced stage (P = .01), with grade 3 (P = .016) and with EGFR FISH+ result (P < .0001). No patient with activating EGFR mutation resulted MET+. In the whole population, MET-positive patients had shorter survival than MET-negative patients (P = .005). Multivariable model confirmed that MET-negative patients had a significant reduction in the risk of death than MET-positive patients (hazard ratio, 0.66; P = .04).
Conclusion
MET increased gene copy number is an independent negative prognostic factor in surgically resected NSCLC. EGFR gene gain does not impact survival after resection.
INTRODUCTION
Since its identification, the epidermal growth factor receptor (EGFR) has emerged as one of the most relevant targets for cancer treatment.1 During the past few years, anti-EGFR strategies offered new hopes to patients with metastatic non–small-cell lung cancer (NSCLC). Cetuximab (C225, Erbitux; ImClone System Inc, New York, NY), a monoclonal antibody against the extracellular domain of EGFR, modestly but significantly prolonged survival of chemotherapy-naive NSCLC patients when used in combination with cisplatin and vinorelbine.2 Erlotinib (OSI 774; Tarceva; Genentech, South San Francisco, CA), an orally available EGFR tyrosine kinase inhibitor (TKI), significantly prolonged survival when used as single agent in pretreated NSCLC.3 Studies in NSCLC with EGFR-TKIs or cetuximab showed that these agents are particularly effective in individuals with certain biologic characteristics.4–6 Increased EGFR gene copy number detected by fluorescent in situ hybridization (FISH) emerged as the strongest predictor for survival in retrospective analyses of large phase III trials comparing EGFR-TKI versus placebo.7–9 More recently, a retrospective analysis of NSCLC patients treated with chemotherapy plus cetuximab showed prolonged progression-free survival for individuals with increased EGFR gene copy number (EGFR FISH positive) when compared to EGFR FISH negative.10 Although these data indicated a predictive value of EGFR gene gain, other studies raised the possibility that this event may be associated with a better natural history.11–12 The impact of EGFR protein expression and gene copy number on survival of surgically resected NSCLC patients is not a resolved issue. Distinct studies conducted at the protein level did not reach similar conclusions regarding duration of survival and level of EGFR expression.13–22 Studies at the gene level using FISH have not shown significant survival difference between patients with high or low EGFR copy numbers; however, the scoring criteria were different than described in studies of patients treated with EGFR-TKIs.22–24
MET is the receptor for hepatocyte growth factor and frequently overexpresses in NSCLC.25–27 Previous studies described MET gene amplification in up to 10% of gastric cancers,28 in 4% of esophageal cancer,29 and in endometrial cancer.30 In addition to proliferative and antiapoptotic activities that are common to many growth factors, MET activation demonstrated to stimulate cell-cell detachment, migration, and invasiveness.31 Preclinical findings suggested that lung cancer cell lines harboring MET gene amplification are dependent on MET for growth and survival.32 Recent data showed that MET amplification is a rare event in NSCLC, occurring in up to 7% of cases.33–35 The rarity of MET amplification in NSCLC, particularly at the high level observed in TKI-resistant cell-line models,34,36 suggested that this event plays a limited role in primary resistance to EGFR-TKI. In contrast, MET gene amplification is one of the most relevant mechanisms involved in EGFR-TKI acquired resistance. Engelman et al36 reported that NSCLC overcomes inhibition of EGFR-TKIs by amplifying the MET oncogene to activate HER3, a member of the EGFR family, and the PI3K-AKT cell survival pathway. In another study, Bean et al33 showed MET amplification in 21% of patients with acquired resistance to gefitinib or erlotinib and only in 3% of untreated patients, confirming that MET could be a relevant therapeutic target for some individuals with acquired resistance to EGFR-TKIs.
The conflicting data on the prognostic value of EGFR together with the relevance of MET as a potential target against NSCLC and the absence of data on MET gene copy number effect on survival led us to conduct a study aiming to evaluate the prognostic effect of EGFR/MET in NSCLC patients.
PATIENTS AND METHODS
Patient Selection
This retrospective study was conducted in a cohort of 447 NSCLC patients that received a radical resection of a primary NSCLC at Istituto Clinico Humanitas IRCCS, Rozzano, Italy, during 2000 to 2004. The only criteria used for patient selection included availability of tumor tissue from primary lung cancer and survival data. Paraffin-embedded tumor specimens were used to construct a tissue microarray with 600-μm diameter cores. Each patient was represented by three tissue cores. An adhesive-coated tape system (Instrumedics, Hackensack, NJ) was used for sectioning the tumor array blocks at 4 μm. The study was approved by the local ethics committee and was conducted in accordance with ethical principles stated in the most recent version of the Declaration of Helsinki or the applicable guidelines on good clinical practice, whichever represented the greater protection of the individuals.
FISH Analyses
Unstained 4-μm sections from each of the three tissue microarray were submitted to dual-color FISH assays using the EGFR/CEP7 probe set from Abbott Molecular (Abbott Park, IL) and a MET/CEP7 probe cocktail prepared with the in house developed MET DNA (RP 11-95I20 BAC clone) labeled with SpectrumRed and the SpectrumGreen CEP7 (Abbott Molecular). The FISH assays were performed according to protocol previously described,5 including pretreatment with 2× sodium chloride-sodium citrate buffer at 75°C and digestion with proteinase K for 7 to 15 minutes each, codenaturation at 85°C for 15 minutes, hybridization for approximately 36 hours, and rapid posthybridization washes with 2× sodium chloride-sodium citrate buffer/0.4 nonyl-phenoxyl-polyethoxylethanol. Signals were enumerated in at least 50 tumor nuclei per core, using epifluorescence microscope with single interference filters sets for green (FITC), red (Texas red) and blue (DAPI) as well as dual (red/green) and triple (blue, red, green) band pass filters. For each core, the mean and standard deviation of copy number per cell of each tested DNA sequence, the percentage of cells with ≤ 2, 3, and ≥ 4 copies of the EGFR and MET genes, and the ratio EGFR/CEP7 and MET/CEP7 were calculated. When heterogeneous results were detected among the three tested cores, the core with the highest mean copy number was used to represent the patient in the statistic analyses. For documentation, images were captured using a CCD camera and merged using dedicated software (CytoVision; Genetix USA, Boston, MA).
Mutation Analysis
Genomic DNA was extracted from tumors and normal lung tissues according to standard procedures. Genetic analysis of exons 19 and 21of the EGFR gene was performed by PCR–single-strand conformation polymorphism and direct sequencing as previously described,37 with the following modifications. Sample were amplified in triplicate and run side by side in order to exclude artifactual sequence changes which are common events in formalin-fixed, paraffin-embedded samples.38–39 Samples harboring mutations were reamplified in duplicates, using the same experimental conditions, purified and subjected to bidirectional dye-terminator sequencing with the same primers employed for amplification. Sequencing fragments were detected by capillary electrophoresis using the ABI Prism 3100 DNA analyzer (Applied Biosystems, Foster City, CA). Sequence chromatograms were analyzed by Mutation Surveyor 2.61 (SoftGenetics, State College, PA), followed by manual review.
Statistical Analyses
The primary end point was to assess whether EGFR and MET gene copy number affected survival of surgically resected NSCLC. Overall survival (OS), calculated from the time of diagnosis to patient death or last contact, and the 95% CIs were evaluated by survival analysis using Kaplan-Meier method.40 Associations with clinical characteristics were compared by Fisher's exact test or χ2 test. OS for the groups with negative and positive biomarkers were compared using the log-rank test. Statistical significance was set at lower than .05 for each analysis. Multivariable analysis was performed using Cox proportional hazards regression analysis with a step-down procedure method. The criterion for variable removal was the likelihood ratio statistic based on the maximum partial likelihood estimates (default P value of .10 for removal from the model). Observations were independent and the proportional hazard assumption was respected. All statistical analyses were performed using SPSS version 11.5.1 (SPSS Italia srl, Bologna, Italy).
RESULTS
Patient Characteristics
A total of 447 surgically resected NSCLC patients were included in this analysis. As presented in Table 1, the majority of patients were male (83.4%), former (51.9%) or current (35.1%) smokers, with moderately or poorly differentiated tumors (grade 2 and 3). The median age was 66 years. All patients received radical surgery, with evidence of pathologic stage III in 34.7% and stage IV in 6% of cases. Patients with metastatic disease received surgery following or at the same time of single brain or lung lesion removal.
Table 1.
Patient Characteristics
| Characteristic | Total | % |
|---|---|---|
| Total | 447 | |
| Median age, years | 66 | |
| Range | 33-86 | |
| Sex | ||
| Male | 373 | 83.4 |
| Female | 74 | 16.6 |
| Smoking history | ||
| Never | 40 | 8.9 |
| Former | 232 | 51.9 |
| Current | 157 | 35.1 |
| Unknown | 18 | 4.1 |
| Histology | ||
| Adenocarcinoma + bronchioloalveolar | 241 | 53.9 |
| Squamous cell carcinoma | 139 | 31.1 |
| Other* | 67 | 15.0 |
| Pathologic stage | ||
| I | 166 | 37.1 |
| II | 99 | 22.2 |
| III | 155 | 34.7 |
| IV | 27 | 6.0 |
| Grade | ||
| I | 32 | 7.2 |
| II | 255 | 57.3 |
| III | 158 | 35.2 |
| Not defined | 2 | 0.3 |
Other histology included seven patients with large cell carcinoma, 28 cases with neuroendocrine differentiation, and 32 undifferentiated non–small-cell lung cancer.
Because adjuvant chemotherapy become a standard approach only after 2004, no postoperative therapy was delivered to patients in stage I-II, and only a minority of patients with stage III disease (18 cases) received adjuvant platinum-based chemotherapy. Patients with stage III disease and pathological evidence of N2 disease (n = 101) received postoperative mediastinal radiotherapy.
With a median follow-up of 40 months, in the whole population OS was 43.9 months. As expected, median survival was longer in stage I-II than in stage III-IV (not reached v 22.3 months; P < .0001). No significant difference in survival was observed according to sex, smoking history, histology, or grading.
EGFR FISH Results
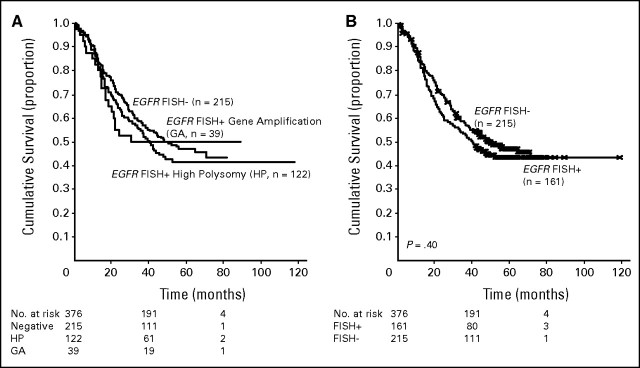
EGFR copy number was evaluated by FISH in 376 cases, and a total of 161 cases (42.8%) were considered as FISH positive according to criteria adopted for selection of patients candidate for EGFR-TKI therapy and described elsewhere.5 Positive patients included 39 cases with true gene amplification (10.4%) and 122 cases with high polysomy (32.4%). As illustrated in Figure 1A, median survival was longer in the EGFR FISH-negative (48.3 months) than in patients with high polysomy (40.7 months) or EGFR gene amplification (30.7 months). Patients with true EGFR gene amplification had shorter survival when compared with the other two groups combined (30.7 v 44.6 months), even if the difference was not statistically significant (P = .85). Moreover, as illustrated in Figure 1B, the difference in survival between EGFR FISH-positive and FISH-negative groups was not statistically significant (40.7 v 48.3 months; P = .4).
Fig 1.
(A) A total of 215 patients were epidermal growth factor receptor (EGFR) fluorescent in situ hybridization (FISH) negative (57.2%), and 161 were EGFR FISH positive (42.8%), including 122 with EGFR high polysomy (32.4%) and 39 with EGFR gene amplification (10.4%). Median survival was 48.3 months for the low polysomic group, 40.7 months for high polysomic, and 30.7 months for patients with gene amplification. (B) Patients with high polysomy or gene amplification (EGFR FISH+) had a median survival of 40.7 months, while patients with low polysomy (EGFR FISH−) had a median survival of 48.3 months. The difference was not statistically significant (P = .4).
We further analyzed the survival outcome of EGFR FISH-positive and FISH-negative patients according to clinical characteristics, and no significant difference was observed, as presented in Table 2.
Table 2.
Survival in EGFR FISH Positive and Negative Patients According to Clinical Characteristics
| Characteristic | EGFR FISH+ (No./month) | EGFR FISH− (No./month) | P |
|---|---|---|---|
| Sex | |||
| Female | 26/37.3 | 32/31.3 | .9 |
| Male | 135/42.0 | 183/51.8 | .4 |
| Smoking status | |||
| Never smokers | 13/40.7 | 17/NR | .3 |
| Smokers (former + current) | 141/42.0 | 189/47.6 | .7 |
| Histology | |||
| Adenocarcinoma + bronchioloalveolar | 87/40.7 | 111/44.6 | .6 |
| Other | 74/39.6 | 104/50.2 | .6 |
| Grade | |||
| I-II | 101/41.3 | 132/55.9 | .11 |
| III | 60/37.3 | 81/36.6 | .7 |
| Stage | |||
| I-II | 95/NR | 129/NR | .7 |
| III-IV | 66/20.5 | 86/25.6 | .4 |
Abbreviations: EGFR, epidermal growth factor receptor; FISH, fluorescent in situ hybridization; NR, not reached.
MET FISH Results
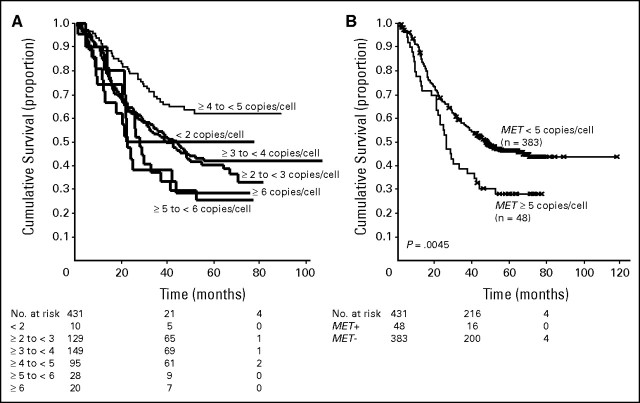
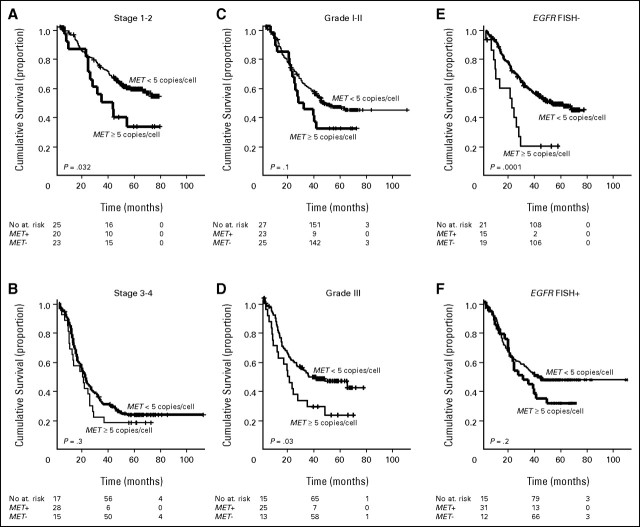
MET was evaluated in 435 cases, and the median mean MET gene copy number was 3.27 (mean range, 1.5 to 21.9 copies per cell). Patients were classified in six groups according to the ascending MET gene copy number, as illustrated in Figure 2A. Because the survival outcome of patients with a mean MET gene copy number per cell higher than 5 and higher than 6 was similar and worse than the other four groups with a mean copy number lower than 5, we considered as MET FISH-positive all cases with mean ≥ 5 copies per cell and negative all cases with mean fewer than 5 copies per cell. Using this cutoff, a total of 48 patients (11.1%) resulted MET FISH positive, including 18 cases (4.1%) with true gene amplification. Among patients with gene amplification, the median mean gene copy number was 10.28 (range, 4.87 to 27.50). MET FISH status was not associated with sex, smoking status, or histology. In contrast, MET FISH-positive status was significantly associated with grade 3 (P = .016) and with advanced stage (P = .01). Among patients with MET gene amplification, three had stage I disease, three had a stage II, nine had stage III, and three had stage IV. As shown in Figure 2B, in the whole population MET FISH-positive patients had a significant shorter survival than MET FISH-negative (25.8 v 47.5 months; P = .005). As illustrated in Figures 3A to 3D (Appendix and Appendix Figs A1 to A5, online only), MET FISH-positive patients had shorter survival than MET-negative irrespective of stage and grade, even if the difference was statistically significant only in patients with stage I-II (P = .032) and in individuals with grade 3 disease (P = .03).
Fig 2.
(A) Survival outcome of patients with a mean of ≥ 5 copies of MET/cell was similar to the outcome of patients with a mean of ≥ 6 copies of MET/cell, and shorter than survival of patients with less than 5 copies of MET/cell. Based on these results, mean of ≥ 5 copies of MET/cell qualified the result as MET+. Median survival was 22.3 months for the 10 patients with mean MET copy number less than 2; 44.4 months in 129 individuals with mean MET copy number ≥ 2 and less than 3; 38.6 months for 149 patients with mean MET copy number ≥ 3 and less than 4; not reached for 95 patients with mean MET copy number ≥ 4 and less than 5; 28.1 months for 28 individuals with MET copy number ≥ 5 and less than 6; 21.4 months for 20 patients with mean copy number ≥ 6. (B) Patients MET fluorescent in situ hybridization (FISH)+ (n = 48) had a median survival of 25.8 months versus 47.5 months in MET FISH− (n = 383). This difference was statistically significant (P = .0045).
Fig 3.
Survival of MET fluorescent in situ hybridization (FISH)+ and MET FISH− according to stage, grade, and epidermal growth factor receptor (EGFR) FISH. (A) In stage I-II disease, 20 patients were MET FISH+ and 233 were MET FISH−. Median survival was not reached in MET FISH− and was 33.0 months in MET FISH+. This difference was statistically significant (P = .032). (B) In stage III-IV disease, MET FISH+ patients (n = 28) had shorter survival (20.8 v 22.8 months) than MET FISH− (n = 150). This difference was not statistically significant (P = .3). (C) In grade 1-2 disease, MET FISH+ (n = 23) had shorter survival (28.1 v 48.8 months) than MET FISH− (n = 250). The difference did not reach the statistical significance (P = .1). (D) In grade 3 disease, MET FISH+ (n = 25) had significantly shorter survival (20.8 v 38.1 months) than MET FISH− (n = 131). The difference was statistically significant (P = .03). (E) EGFR FISH negative/MET FISH− (n = 197) had significantly longer survival (55.9 v 22.6 months; P = .0001) than EGFR FISH−/MET FISH+ (n = 15). (F) Among patients EGFR FISH+, survival was longer in MET FISH− (n = 128) than in the 31 patients MET FISH+ (median survival, 42.6 v 28.9 months) even if the difference was not statistically significant (P = .2).
MET Gene Copy Number and EGFR
To further explore the prognostic impact of MET gene copy number in a context of EGFR gene gain or mutation, we analyzed MET according to EGFR status. Patients with adenocarcinoma with or without bronchioloalveolar features and with stage I-II disease (n = 144) were screened for EGFR mutations, and 14 (9.7%) harbored a deletion in exon 19 (seven patients) or the L858R mutation in exon 21 (seven patients). No difference in survival was observed between patients with or without mutation (67.1 v not reached; P = .99). Interestingly, no patient with EGFR mutation was MET FISH positive, and among EGFR wild type, MET FISH-positive patients had significantly shorter survival than MET FISH-negative patients (P = .005).
MET FISH-positive patients were more frequently EGFR FISH positive, and that association was statistically significant (P < .0001). EGFR FISH negative/MET FISH negative had significantly longer survival (55.9 v 22.6 months; P = .0001) than EGFR FISH negative/MET FISH positive (Fig 3E). Among patients EGFR FISH positive, survival was longer in MET FISH-negative than in MET FISH positive (42.6 v 28.9 months) even if the difference was not statistically significant (P = .2; Fig 3F).
Univariable and Multivariable Analyses
Table 3 presents the results of the univariate and multivariate survival analyses that included sex, stage, smoking history, grade, histology, EGFR FISH, and MET FISH. In the univariate analysis, only stage I-II (hazard ratio [HR], 0.40), and MET FISH-negative status (HR, 0.59) were significantly associated with a reduced risk of death. The multivariate model confirmed that patients with stage I or II and patients MET FISH-negative had a lower risk of death than patients with stage III or IV (HR, 0.42; P < .0001) or MET FISH+ (HR, 0.66; P = .04). In order to explore the possible impact of EGFR mutations, we further analyzed the risk of death according to the above mentioned categories in the 144 sequenced patients. In this model, that excluded patients with stage III-IV disease with nonadenocarcinoma histology, MET FISH-negative status was the only variable significantly associated with a reduced risk of death (HR, 0.32; 95%CI, 0.14 to 0.71; P = .006).
Table 3.
Univariable and Multivariable Analyses on Survival
| Variable | Category | Univariable |
Multivariable |
||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| Sex | Male/female | 0.91 | 0.66 to 1.26 | .6 | 0.87 | 0.57 to 1.33 | .5 |
| Smoking | Smoker/never smoker | 1.28 | 0.81 to 2.04 | .3 | 1.24 | 0.69 to 2.25 | .5* |
| Histology | Adenocarcinoma†/other | 0.97 | 0.75 to 1.25 | .8 | 1.02 | 0.76 to 1.38 | .9 |
| Grade | I + II/III | 0.78 | 0.60 to 1.01 | .06 | 0.89 | 0.66 to 1.20 | .4 |
| Stage | I + II/III + IV | 0.40 | 0.31 to 0.51 | < .0001‡ | 0.42 | 0.31 to 0.56 | < .0001‡ |
| MET FISH | < 5/≥ 5 copies | 0.59 | 0.41 to 0.85 | .005‡ | 0.66 | 0.43 to 0.99 | .04‡ |
| EGFR FISH | Negative/positive | 0.89 | 0.67 to 1.17 | .34 | 1.02 | 0.74 to 1.39 | .9 |
Abbreviations: HR, hazard ratio; FISH, fluorescent in situ hybridization; EGFR, epidermal growth factor receptor.
Sex, smoking status, histology, grade, and EGFR FISH variables were removed from the model at the last step of multivariable analysis.
Adenocarcinoma category also included bronchioloalveolar histology.
Statistically significant.
DISCUSSION
In this study we evaluated the prognostic role of two of the most relevant biomarkers in NSCLC, and to our knowledge, report for the first time that MET increased gene copy number represents a negative prognostic factor in radically resected NSCLC. MET FISH-positive patients had a significantly shorter survival than MET FISH-negative patients, and this negative prognostic effect was independent of any other clinical or biologic characteristic, including those factors classically associated with sensitivity to EGFR-TKIs.4–6
MET has gained considerable interest through its apparent deregulation by overexpression or mutation in various cancers, including NSCLC.41 Several anti-MET drugs are under evaluation in clinical trials, and the interest around these compounds has consistently increased since an interaction between EGFR and MET has been observed.42 In addition, two recent studies showed that MET amplification is responsible for EGFR-TKI acquired resistance in approximately 20% of patients.33,36 Although the great interest on MET, only few small studies evaluated whether this target could influence patient survival.25,27,43–47 In the study conducted by Cheng et al43 in 45 NSCLC patients, overexpression of circulating c-MET was significantly associated with early recurrence. In another study conducted in 106 NSCLC, Beau-Faller et al44 showed that adenocarcinoma patients with MET amplification had a trend for poor prognosis. Our study, conducted in a large series of non–small-cell lung cancer, provides clear evidence that MET is a negative prognostic factor, further supporting anti-MET strategies in this disease. Nevertheless, we cannot rule out the hypothesis that the poor prognosis detected by the MET FISH assay is not due to a broad level of genomic instability, since the assay only investigated changes in chromosome 7. A relevant aspect of our study was the significant association with EGFR gene copy number, similarly to what has been recently described by our group in colorectal cancer.48 We found that approximately 20% of EGFR FISH-positive patients were also MET positive, raising the possibility that some EGFR FISH-positive individuals could be resistant to EGFR-TKI due to MET over-representation. Nevertheless, recent findings suggested that only high levels of MET amplification could impair EGFR-TKI sensitivity.34 In this study, among EGFR FISH-positive patients, only three patients had MET amplification at equal or higher than levels previously reported in EGFR-TKI resistant models.33,36 Moreover, the lack of association between MET FISH-positive and EGFR mutations and the rarity of coamplification of MET and EGFR genes (only 1.1% in our cohort) suggest that chromosome 7 polysomy was responsible for the significant association of MET FISH and EGFR FISH status. The low incidence of MET gene amplification observed in our study is consistent with previous FISH studies showing that MET amplification is a rare event in NSCLC, occurring in 3% to 7% of patients.33–35
Another interesting finding was the association of MET gene copy number with tumor stage and grade. This aspect raises the possibility that testing MET genomic status in tumor biopsies collected at the time of primary diagnosis could not be reliable to predict MET gene status in a tumor progressing after an initial treatment.
The second biomarker evaluated in this study was EGFR. Recently, a large meta-analysis showed that EGFR expression was not a prognostic factor in NSCLC.13 Three studies evaluated the prognostic impact of EGFR gene copy number determined by FISH, and all showed no association with patient survival.22–24 Results from large phase III trials comparing an EGFR-TKI versus placebo, the only trials able to discriminate between prognostic versus predictive value of a biomarker, clearly showed that EGFR gene copy number represents a predictive factor.8–9 Nevertheless, large phase III trials comparing EGFR-TKI plus chemotherapy versus chemotherapy alone, also suggested that EGFR increased gene copy number could contribute to a better prognosis.11–12 Because of the confusion among clinicians on the role of EGFR gene gain, and because in previous study we evaluated the prognostic impact of EGFR FISH using a different scoring system,22 in this study we analyzed EGFR gene copy number using the same criteria described for selection of patients candidate for EGFR-TKI therapy.5 Our results, obtained in a large patient population, demonstrated that EGFR gene copy number has a limited effect on patient survival. Although EGFR amplified patients had a trend for shorter survival, the difference with nonEGFR-amplified individuals was not significant and the whole group of EGFR FISH-positive patients had similar survival than EGFR FISH-negative patients, confirming that EGFR gene gain is not a favorable prognostic factor.
In conclusion, this study shows that MET increased gene copy number is an independent prognostic factor in surgically resected NSCLC, supporting anti-MET therapeutic strategies in selected patients. EGFR gene gain has no prognostic role in NSCLC, further supporting its role as predictive factor for improved survival in NSCLC patients exposed to EGFR-TKIs.
Acknowledgment
We are indebted to the Cytogenetics Core of the University of Colorado Cancer Center (Aurora, CO) and the Chieti University (Italy) for technical assistance.
Appendix
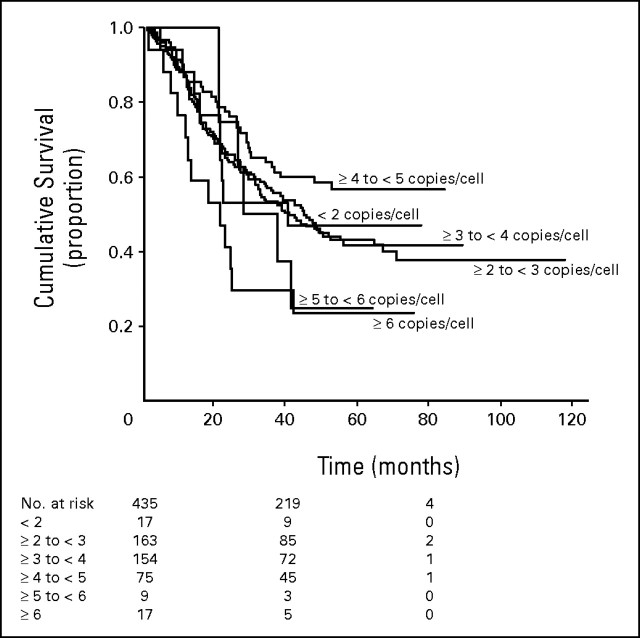
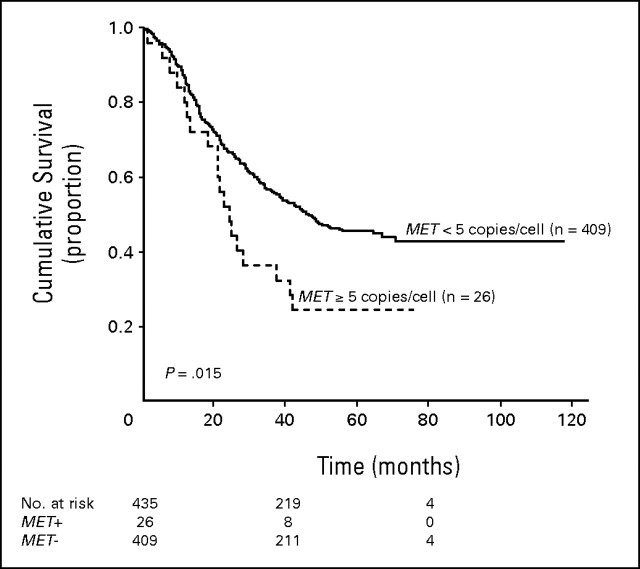
Heterogeneity among cores, defined as more than 1 copy number difference in the mean, was observed in 58 patients (13.3%), but most of the differences did not impacted the final pattern classification. In 18 patients (4.1%) presenting tumor cores classified individually as positive and negative for MET genomic gain, the highest mean was used to represent the individual. In order to assess the impact of tumor heterogeneity, we further evaluated the survival outcome of the whole patient population based on the average MET gene copy number in the three tissue microarraycores. This analysis confirmed that MET fluorescent in situ hybridization (FISH)-positive patients had shorter survival than MET FISH-negative. Figures A1 and A2 show the outcome of resected patients with non–small-cell lung cancer according to the average MET gene copy number in three tissue microarray cores. In Figure A1, survival outcome of patients with a mean of ≥ 5 copies of MET/cell was similar to the outcome of patients with a mean of ≥ 6 copies of MET/cell, and shorter than survival of patients with fewer than 5 copies of MET/cell. Based on these results, mean of ≥ 5 copies of MET/cell qualified the result as MET+. Median survival was 40.4 months for the 17 patients with mean MET copy number lower than 2; 44.9 months in 163 individuals with mean MET copy number ≥ 2 and lower than 3; 40.7 months for 154 patients with mean MET copy number ≥ 3 and lower than 4; not reached for 75 patients with mean MET copy number ≥ 4 and lower than 5; 28.1 months for nine individuals with MET copy number ≥ 5 and lower than 6; 21.4 months for 17 patients with mean copy number ≥ 6. In Figure A2, MET FISH-positive patients (n = 26) had a median survival of 24.1 months versus 46.5 months in MET FISH-negative patients (n = 409). This difference was statistically significant (P = .015).
Fig A1.
Survival outcome based on the average MET gene copy number in the three tissue microarray cores. Survival of patients with a mean of ≥ 5 copies of MET/cell was similar to the outcome of patients with a mean of ≥ 6 copies of MET/cell, and shorter than survival of patients with < 5 copies of MET/cell. Based on these results, mean of ≥ 5 copies of MET/cell qualified the result as MET fluorescent in situ hybridization positive. Median survival was 40.4 months for the 17 patients with mean MET copy number < 2; 44.9 months in 163 individuals with mean MET copy number ≥ 2 and < 3; 40.7 months for 153 patients with mean MET copy number ≥ 3 and < 4; not reached for 75 patients with mean MET copy number ≥ 4 and < 5; 28.1 months for nine individuals with MET copy number ≥ 5 and < 6; and 21.4 months for 17 patients with mean copy number ≥ 6.
Fig A2.
Survival outcome based on the average MET gene copy number in the three tissue microarray cores. Patients who were MET fluorescent in situ hybridization (FISH) positive (n = 26) had a median survival of 24.1 v 46.5 months in those who were MET FISH negative (n = 409). This difference was statistically significant (P = .015).
Fig A3.
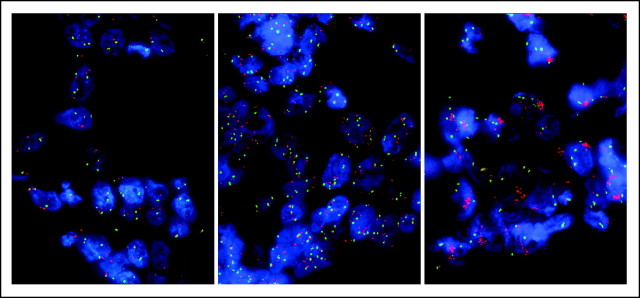
MET fluorescent in situ hybridization negative (low copy number, left) and positive high copy number by polysomy (center) and gene amplification (right).
Fig A4.
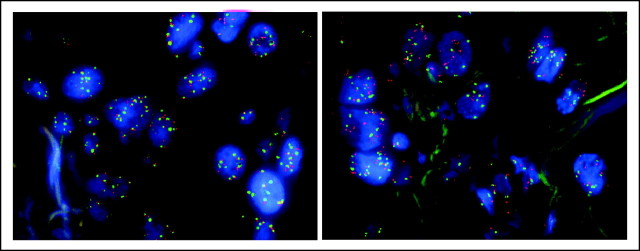
Section from a patient with non–small-cell lung cancer showing epidermal growth factor receptor fluorescent in situ hybridization (FISH) high polysomy (left) and MET FISH high polysomy (right).
Fig A5.
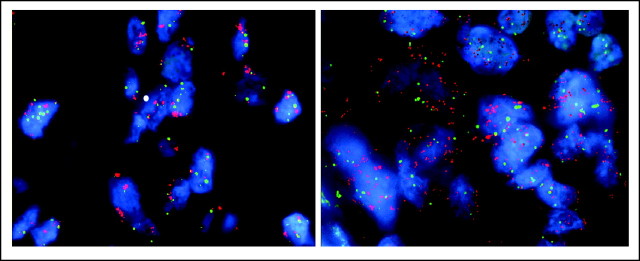
Section from a patient with non–small-cell lung cancer showing epidermal growth factor receptor fluorescent in situ hybridization (FISH) gene amplification (left) and MET FISH gene amplification (right).
Footnotes
Supported in part by Italian Association for Cancer Research (F.C.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Federico Cappuzzo, Marileila Varella-Garcia
Financial support: Federico Cappuzzo, Armando Santoro, Marileila Varella-Garcia
Administrative support: Federico Cappuzzo
Provision of study materials or patients: Federico Cappuzzo, Antonio Marchetti, Annarita Destro, Luigi Terracciano, Massimo Roncalli, Marco Alloisio
Collection and assembly of data: Federico Cappuzzo, Margaret Skokan, Sujatha Gajapathy, Lara Felicioni, Maela Del Grammastro, Maria Grazia Sciarrotta, Fiamma Buttitta, Matteo Incarbone, Luca Toschi, Giovanna Finocchiaro
Data analysis and interpretation: Federico Cappuzzo, Elisa Rossi, Marileila Varella-Garcia
Manuscript writing: Federico Cappuzzo, Marileila Varella-Garcia
Final approval of manuscript: Federico Cappuzzo, Antonio Marchetti, Margaret Skokan, Elisa Rossi, Sujatha Gajapathy, Maela Del Grammastro, Maria Grazia Sciarrotta, Fiamma Buttitta, Matteo Incarbone, Luca Toschi, Giovanna Finocchiaro, Annarita Destro, Luigi Terracciano, Massimo Roncalli, Marco Alloisio, Armando Santoro, Marileila Varella-Garcia
REFERENCES
- 1.Cohen S. Isolation of mouse submaxillary gland protein accelerating incisor eruption and eyelid opening in the new-born animal. J Biol Chem. 1962;237:1555–1562. [PubMed] [Google Scholar]
- 2.Pirker R, Szczesna A, von Pawel J, et al. FLEX: A randomized, multicenter, phase III study of cetuximab in combination with cisplatin/vinorelbine (CV) versus CV alone in the first-line treatment of patients with advanced non-small cell lung cancer (NSCLC). J Clin Oncol. 2008;26(suppl):6s. abstr 3. [Google Scholar]
- 3.Shepherd FA, Rodrigues Pereira J, et al. Erlotinib in previously treated non-small cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 6.Miller VA, Kris MG, Shah N, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 7.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 9.Zhu CQ, da Cunha Santos G, Ding K, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada clinical trial group study BR21. J Clin Oncol. 2008;26:4268–4275. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch FR, Herbst RS, Olsen C, et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol. 2008;26:3351–3357. doi: 10.1200/JCO.2007.14.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch FR, Varella-Garcia M, Bunn PA, et al. Fluorescence in situ hybridization (FISH) subgroup analysis of TRIBUTE: A phase III trial of erlotinib plus carboplatin and paclitaxel in NSCLC. Clin Cancer Res. 2008;14:6317–6373. doi: 10.1158/1078-0432.CCR-08-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell DW, Lynch TJ, Haserlat SM, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: Molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–8092. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura H, Kawasaki N, Taguchi M, et al. Survival impact of epidermal growth factor receptor overexpression in patients with non-small cell lung cancer: A meta-analysis. Thorax. 2006;61:98–99. doi: 10.1136/thx.2005.042275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veale D, Kerr N, Gibson GJ, et al. The relationship of quantitative epidermal growth factor receptor expression in non-small cell lung cancer to long term survival. Br J Cancer. 1993;68:162–165. doi: 10.1038/bjc.1993.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastorino U, Andreola S, Tagliabue E, et al. Immunocytochemical markers in stage I lung cancer: Relevance to prognosis. J Clin Oncol. 1997;15:2858–2865. doi: 10.1200/JCO.1997.15.8.2858. [DOI] [PubMed] [Google Scholar]
- 16.Rusch V, Klimstra D, Venkatraman E, et al. Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non-small cell lung cancer but does not predict tumor progression. Clin Cancer Res. 1997;3:515–522. [PubMed] [Google Scholar]
- 17.Fontanini G, Laurentiis M, Vignati S, et al. Evaluation of epidermal growth factor-related growth factors and receptors and of neoangiogenesis in completely resected stage I-IIIA non-small-cell lung cancer: Amphiregulin and microvessel count are independent prognostic indicators of survival. Clin Cancer Res. 1998;4:241–249. [PubMed] [Google Scholar]
- 18.Pfeiffer P, Nexo E, Bentzen SM, et al. Enzyme-linked immunosorbent assay of epidermal growth factor receptor in lung cancer: Comparisons with immunohistochemistry, clinicopathological features and prognosis. Br J Cancer. 1998;78:96–99. doi: 10.1038/bjc.1998.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu XL, Zhu XZ, Shi DR, et al. Study of prognostic predictors for non-small cell lung cancer. Lung Cancer. 1999;23:143–152. doi: 10.1016/s0169-5002(99)00009-4. [DOI] [PubMed] [Google Scholar]
- 20.Selvaggi G, Novello S, Torri V, et al. Epidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non-small-cell lung cancer. Ann Oncol. 2004;15:28–32. doi: 10.1093/annonc/mdh011. [DOI] [PubMed] [Google Scholar]
- 21.Swinson DE, Cox G, O'Byrne KJ. Coexpression of epidermal growth factor receptor with related factors is associated with a poor prognosis in non-small-cell lung cancer. Br J Cancer. 2004;91:1301–1307. doi: 10.1038/sj.bjc.6602149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 23.Jeon YK, Sung SW, Chung JH, et al. Clinicopathologic features and prognostic implications of epidermal growth factor receptor (EGFR) gene copy number and protein expression in non-small cell lung cancer. Lung Cancer. 2006;54:387–398. doi: 10.1016/j.lungcan.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki S, Dobashi Y, Sakurai H, et al. Protein overexpression and gene amplification of epidermal growth factor receptor in non small cell lung carcinomas: An immunohistochemical and fluorescence in situ hybridization study. Cancer. 2005;103:1265–1273. doi: 10.1002/cncr.20909. [DOI] [PubMed] [Google Scholar]
- 25.Ichimura E, Maeshima A, Nakajima T, et al. Expression of c-met/HGF receptor in human non-small cell lung carcinomas in vitro and in vivo and its prognostic significance. Jpn J Cancer Res. 1996;87:1063–1069. doi: 10.1111/j.1349-7006.1996.tb03111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsao MS, Liu N, Chen JR, et al. Differential expression of Met/hepatocyte growth factor receptor in subtypes of non-small cell lung cancers. Lung Cancer. 1998;20:1–16. doi: 10.1016/s0169-5002(98)00007-5. [DOI] [PubMed] [Google Scholar]
- 27.Masuya D, Huang C, Liu D, et al. The tumour–stromal interaction between intratumoral c-Met and stromal hepatocyte growth factor associated with tumour growth and prognosis in non-small-cell lung cancer patients. Br J Cancer. 2004;90:1555–1562. doi: 10.1038/sj.bjc.6601718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara T, Ooi A, Kobayashi M, et al. Amplification of c-myc, K-sam, and c-met in gastric cancers: Detection by fluorescence in situ hybridization. Lab Invest. 1998;78:1143–1153. [PubMed] [Google Scholar]
- 29.Miller CT, Lin L, Casper AM, et al. Genomic amplification of MET with boundaries within fragile site FRA7G and upregulation of MET pathways in esophageal adenocarcinoma. Oncogene. 2006;25:409–418. doi: 10.1038/sj.onc.1209057. [DOI] [PubMed] [Google Scholar]
- 30.Samuelson E, Levan K, Adamovic T, et al. Recurrent gene amplifications in human type I endometrial adenocarcinoma detected by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2008;181:25–30. doi: 10.1016/j.cancergencyto.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Birchmeier C, Birchmeier W, Gherardi E, et al. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 32.Lutterbach B, Zeng Q, Davis LJ, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67:2081–2088. doi: 10.1158/0008-5472.CAN-06-3495. [DOI] [PubMed] [Google Scholar]
- 33.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cappuzzo F, Skokan M, Gajapathy S, et al. MET increased gene copy number and primary resistance to gefitinib therapy in never smokers or EGFR FISH positive non-small cell lung cancer (NSCLC) patients. Ann Oncol. doi: 10.1093/annonc/mdn635. epub ahead of print on October 3, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 37.Marchetti A, Martella C, Felicioni L, et al. EGFR mutations in non–small cell lung cancer: Analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 38.Marchetti A, Felicioni L, Buttitta F. Assessing EGFR mutations. N Engl J Med. 2006;354:526–528. doi: 10.1056/NEJMc052564. [DOI] [PubMed] [Google Scholar]
- 39.Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non–Small-Cell Lung Cancer Working Group: Standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 40.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1985;53:457–481. [Google Scholar]
- 41.Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 42.Bonine-Summers AR, Aakre ME, Brown KA, et al. Epidermal growth factor receptor plays a significant role in hepatocyte growth factor mediated biological responses in mammary epithelial cells. Cancer Biol Ther. 2007;4:561–570. doi: 10.4161/cbt.6.4.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng TL, Chang MY, Huang SY, et al. Overexpression of circulating c-met messenger RNA is significantly correlated with nodal stage and early recurrence in non-small cell lung cancer. Chest. 2005;128:1453–1460. doi: 10.1378/chest.128.3.1453. [DOI] [PubMed] [Google Scholar]
- 44.Beau-Faller M, Ruppert AM, Voegeli AC, et al. MET gene copy number in non-small cell lung cancer: Molecular analysis in a targeted tyrosine kinase inhibitor naïve cohort. J Thorac Oncol. 2008;4:331–339. doi: 10.1097/JTO.0b013e318168d9d4. [DOI] [PubMed] [Google Scholar]
- 45.Tokunou M, Niki T, Eguchi K, et al. C-MET expression in myofibroblasts: Role in autocrine activation and prognostic significance in lung adenocarcinoma. Am J Pathol. 2001;158:1451–1463. doi: 10.1016/S0002-9440(10)64096-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siegfried JM, Weissfeld LA, Singh-Kaw P, et al. Association of immunoreactive hepatocyte growth factor with poor survival in resectable non-small cell lung cancer. Cancer Res. 1997;57:433–439. [PubMed] [Google Scholar]
- 47.Lo Muzio L, Farina A, Rubini C, et al. Effect of c-Met expression on survival in head and neck squamous cell carcinoma. Tumour Biol. 2006;27:115–121. doi: 10.1159/000092716. [DOI] [PubMed] [Google Scholar]
- 48.Cappuzzo F, Varella-Garcia M, Finocchiaro G, et al. Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients. Br J Cancer. 2008;99:83–89. doi: 10.1038/sj.bjc.6604439. [DOI] [PMC free article] [PubMed] [Google Scholar]