Abstract
Pathogen recognition is the first step of immune reactions. In animals and plants, direct or indirect pathogen recognition is often mediated by a wealth of fast-evolving receptors, many of which contain ligand-binding and signal transduction domains, such as leucine-rich or tetratricopeptide repeat (LRR/TPR) and NB-ARC domains, respectively. In order to identify candidates potentially involved in algal defense, we mined the genome of the brown alga Ectocarpus siliculosus for homologues of these genes and assessed the evolutionary pressures acting upon them. We thus annotated all Ectocarpus LRR-containing genes, in particular an original group of LRR-containing GTPases of the ROCO family, and 24 NB-ARC–TPR proteins. They exhibit high birth and death rates, while a diversifying selection is acting on their LRR (respectively TPR) domain, probably affecting the ligand-binding specificities. Remarkably, each repeat is encoded by an exon, and the intense exon shuffling underpins the variability of LRR and TPR domains. We conclude that the Ectocarpus ROCO and NB-ARC–TPR families are excellent candidates for being involved in recognition/transduction events linked to immunity. We further hypothesize that brown algae may generate their immune repertoire via controlled somatic recombination, so far only known from the vertebrate adaptive immune systems.
Keywords: brown alga, Ectocarpus, exon shuffling, resistance gene analogue, innate immunity, adaptive immunity
Introduction
Brown algae are predominant primary producers in cold and temperate coastal ecosystems. In particular, kelps form remarkable underwater canopies and are exploited commercially. Like any other living organism, the brown algae are plagued by diseases caused by fungi, oomycetes, bacteria, or viruses. However, little is known about the molecular mechanisms underpinning their immunity (Potin et al. 2002). Most molecular studies have been conducted on animals and plants, which diverged from the brown algae early in the eukaryotic evolution (Keeling et al. 2005), and it is therefore unclear to which extent the defense mechanisms might or might not be conserved between these lineages. Recently, however, we described the existence of differential susceptibility of clonal Ectocarpus sp. strains to the oomycete pathogen Eurychasma dicksonii, which points to the probable existence of genetically determined disease-resistance mechanisms in this alga (Gachon et al. 2009).
In all organisms studied, the onset of the immune reactions relies on successful pathogen recognition, followed by signal transduction and the induction of defense effectors (Ronald and Beutler 2010). The pathogen and the host thus engage into a molecular hide-and-seek game, which translates into a coevolutionary arms race between the pathogen's effectors and the host's receptors. Therefore, many pathogen receptors belong to rapidly evolving multigene families.
In plants, pathogen recognition is mediated by resistance genes and pattern recognition receptors, some of which directly recognize microbe-derived elicitors (e.g., the flagellin receptor FLS2, Boller and Felix 2009). Others monitor the integrity of endogenous proteins targeted by pathogens. These genes belong to families of up to several hundred members, which contain a Leucine-Rich Repeat (LRR) domain, coupled to various domains thought to be involved in signal transduction and/or interaction with other ligands (Maekawa et al. 2011). LRR domains are also prominent in animal proteins related to immunity, such as the well-characterized Toll-like receptors (Kumar et al. 2009) and the CATERPILLER family (Ting and Williams 2005). A direct role for LRR proteins in antigen recognition has also been recently uncovered or hypothesized in jawless fishes and mosquito, respectively (Han et al. 2008; Povelones et al. 2009).
In the above-mentioned families, the LRR domain is most often a key determinant of the pathogen recognition specificity. LRRs are 20–29 amino acid residue sequences defined by the core consensus LxxLxLxxNxL, which is important to sustain their typical β-sheet structure (Padmanabhan et al. 2009 and references therein). LRR domains are formed by the juxtaposition of a few to more than 40 individual repeats, with some of the variable amino acids between the structural leucine residues engaging in specific interactions with other ligands. Tetratricopeptide repeat (TPR) domains are less well known in the context of immunity but specialize in ligand binding too (Blatch and Lassle 1999). They are made of 34-amino acid–long repeats, characterized by eight loosely conserved residues that dictate the folding of two antiparallel α-helices. TPRs further assemble into a superhelix delimiting an amphipathic groove, where variable solvent-exposed residues bind specific ligands.
The LRR domains of plant resistance genes typically evolve new ligand-binding specificities under diversifying selection, via a combination of mechanisms ranging from point mutations of variable residues, variations in repeat numbers, gene duplication, and other rearrangements (Ellis et al. 2000; Friedman and Baker 2007). In jawless fishes, the somatic recombination of LRRs within antigen receptors generates a diverse immune repertoire, a process long thought to be restricted to the immunoglobulin-mediated adaptive immunity of vertebrates (Pancer et al. 2004).
Because of their involvement in pathogen recognition in both animal and plant systems, we set out to annotate the LRR-containing genes in the genome of Ectocarpus siliculosus, the first fully sequenced multicellular stramenopile. We also searched the genome for other homologues of plant and animal genes involved in immunity, which led us to identify a family of NB-ARC domain containing proteins. We further focused our attention on the genes that might exhibit signs of rapid evolution, in order to pinpoint candidates likely to be involved in immune defenses. One such prominent group of genes that emerged from our analysis turned out to belong to the ROCO family. This family is defined by a conserved core composed of a GTPase domain (Roc) coupled with a unique COR domain serving as a dimerization module (Bosgraaf and Van Haastert 2003; Gotthardt et al. 2008; Marin et al. 2008). The core may be decorated by a variety of additional domains, including LRR, Ankyrin or WD40 repeats, or kinase domains. The most thoroughly studied representative is the human LRRK2 protein, owing to defects in this protein leading to the onset of Parkinson's disease (Daniëls et al. 2011), but its exact cellular function remains unclear. Additional ROCO proteins that have been studied include the metazoan DAPK1 implicated in apoptosis, the Arabidopsis TRN1 involved in patterning processes during early leaf development, GbpC from Dictyostelium discoideum involved in chemotaxis, or Pats1 from the same species probably participating in cytokinesis (Marin et al. 2008).
We found that the LRR and TPR domains of ROCO and NB-ARC–TPR genes, respectively, exhibit signs of diversifying selection, and evolve by means of a highly unusual, if not unique, exon-shuffling mechanism. Although mechanistically distinct, the latter is functionally reminiscent of the cassette mechanisms underpinning vertebrate (mammal and cyclostome) adaptive immune systems. Therefore, we argue that brown algae might generate their immune repertoire via specific targeted somatic recombination, and in any case, that they do possess a suitable genomic mechanism that could easily be recruited to fulfill this function. Our findings therefore suggest that somatic variation of pathogen receptors might not be restricted to vertebrates, as is widely believed.
Materials and Methods
Identification and Manual Curation of the Ectocarpus LRR and NB-ARC–Containing Genes
LRR and NB-ARC domains were identified in predicted Ectocarpus proteome using Interproscan (Cock et al. 2010) and reciprocal Blast searches. The corresponding gene models were refined manually according to all relevant available data (expressed sequence tags [ESTs], TILING array, and alignment with orthologues and paralogues) and can be accessed via the Ectocarpus genome database (Cock et al. 2010). The genome assembly was aligned to the individual sequence reads deposited in the NCBI Trace Archive database to confirm the predicted gene structures. Particular attention was paid to the intron–exon structure within the LRR (respectively TPR) domain. Additional LRRs/TPRs were identified in the genomic sequence by a combination of sequence signature searches (in particular, but not exclusively, for the LRR core consensus LxxLxLxxN and plant-specific motif GxIPxxL and for fragments of the TPR-associated sequences: GKYxEA[E/D]PL[Y/F]xxxxxxxxGx[D/E]xxx[V/I]AxxLxNxAxLLxxQ. Loci displaying frame shifts, stop codons, and/or deletions (missing conserved exons) within the predicted coding sequence were classified as pseudogenes. We refined the models of these pseudogenes in order to match the exon/introns arrangement of related intact genes as closely as possible, but these suggested models may contain an artificial first exon (to provide an initiation codon) and/or artificial introns or intron borders (to skip the disruptive sites), as indicated in the Ectocarpus genome database. Additional LRR (respectively TPR)-encoding exons interrupted by a stop codon or lacking a correct splicing site were annotated as “inactivated exons” (supplementary table 1, Supplementary Material online).
Protein Sequence Analyses, Alignments, and Phylogeny of the Ectocarpus ROCO and NB-ARC Family
Conserved regions of the Ectocarpus ROCO proteins were approximately delimited by BlastP searches (Altschul et al. 1997) and multiple alignments with ClustalX (Thompson et al. 1997). The boundaries of the Roc (GTPase) and COR domains characteristic of ROCO proteins were identified using several rounds of PSI-Blast searches (Altschul et al. 1997) with representative ROCO sequences from Ectocarpus and several other species (human LRRK2, Chlorobium tepidum ROCO—NP_662411) against a custom local database. The PSI-Blast results were confronted with the solved protein structure of the C. tepidum ROCO and the multiple alignment reported by Gotthardt et al. (2008). Even using PSI-Blast, we were unable to demonstrate convincingly that the region downstream of the strand β11 of the COR domain (as defined for the C. tepidum protein) is truly homologous throughout the whole ROCO family, so for subsequent phylogenetic analysis, we trimmed the sequences at the highly conserved motif between the helix α8 and the strand β11. Likewise, we trimmed the N-terminal regions up to a conserved motif directly upstream of the helix α0 preceding the Roc domain.
A multiple alignment was built using PROMALS3D (Pei et al. 2008) aided with the crystal structure of the C. tepidum Roc–COR tandem (PDB accession number 3DPU). The alignment was further polished manually using GeneDoc (Nicholas KB and Nicholas HB 1997). The alignment (supplementary fig. 1, Supplementary Material online) includes most Ectocarpus ROCO proteins, except the fragmentary Esi0027_0052 locus, and the highly divergent Esi0307_0011. Representatives from other taxa were added to provide an outgroup.
For a phylogenetic analysis, poorly conserved regions were removed, resulting in 261 positions suitable for tree inference. Except for the very fragmentary Esi0102_0087, pseudogenes were kept in the alignment, even though they were often rather incomplete (up to 102 missing characters). A tree was inferred with the maximum likelihood (ML) method using RAxML 7.0.4 (Stamatakis et al. 2008) at the CIPRES portal (http://www.phylo.org/sub_sections/portal/), using the rapid bootstrap heuristics (100 bootstrap replicates) and final thorough tree search under the WAG+F+Γ4+I substitution model. An additional ML bootstrap analysis (100 replicates) was run at the ATCG server (http://www.atgc-montpellier.fr/phyml/) employing PhyML 3.0 (Guindon and Gascuel 2003) and the LG+Γ4+I substitution model. Branch support values were calculated from the resulting bootstrap trees using the “consense” program in the Phylip 3.62 package (Felsenstein 2005).
A similar procedure was followed for the NB-ARC domain of NBR-ARC–TPR loci, except that the alignment (supplementary fig. 2A, Supplementary Material online) was not aided with a 3D structure. Additionally, poorly conserved positions were removed from the alignments with Gblocks (Castresana 2000) (supplementary fig. 2A, Supplementary Material online) before running the PhyML 3.0 analysis with the same parameters as above.
Evolutionary Analysis of LRR and TPR Exons
Evolutionary analyses were essentially performed following the procedure described in detail by Lynn, Higgs, et al. (2004) and Lynn, Lloyd, et al. (2004). Briefly, amino acid sequences were aligned using MUSCLE (Edgar 2004). Neighbor joining (NJ) trees were inferred with MEGA4, with distances calculated using the Poisson correction as the basic substitution model and Γ distribution modeling among-site rate variation. We estimated the shape parameter of the latter (α) with the REV model, implemented in Baseml (PAML version 4.2; Yang 2007), performed 1,000 bootstrap replicates, and collapsed the branches of the tree with a bootstrap value below 50%. Nucleotide sequences were aligned by feeding the corresponding protein alignments into a copygaps Perl script that maintains the gaps and removes any columns in the nucleotide alignments that has more than three gaps. We checked for substitution saturation by calculating synonymous substitution rates (dS) between the aligned nucleotide sequences using the modified Nei-Gojobori method of Yang and Nielsen (2000).
In order to test for evidence of positive selection, NJ trees and nucleotide alignments were used as input for the CODEML and CODEMLSITES programs (Yang 2007). These tests compare the rates of synonymous (dS) and nonsynonymous (dN) mutations among the nucleotide sequences. Theoretically, selectively neutral nonsynonymous mutations are fixed at the same rate as synonymous mutations, resulting in an ω ratio (dN/dS) = 1. Most genes exhibit ω values <1 (purifying selection), whereas ω values >1 are more unusual and indicative of positive selection, that is, nonsynonymous mutations being retained at a higher rate than the expected under neutral selection. CODEML specifically tests for variable selective pressures among lineages in the phylogeny by looking for significant differences in ω ratios, while CODEMLSITES uses site-specific codon substitution models, allowing for the detection of variable selective pressures among amino acid positions.
CODEML calculates log-likelihood values for two evolutionary scenarios: one presumes an equal ω ratio for all branches in the phylogeny and the second (free-ratios model) permits an independent ω ratio along different branches. The log-likelihood values for each model are then compared by a likelihood ratio test (LRT) and assigned a P value. Finally, posterior Bayesian probabilities are estimated for codon substitutions in each branch of the phylogenetic tree. CODEMLSITES relies on site-specific models of codon substitution to assess whether any of the six progressively more complex models is significantly better at explaining the data observed. As for CODEML, log-likelihood values are estimated for each model and compared by LRTs. The empirical Bayes method further allows to pinpoint codons subjected to positive selection under the relevant models.
Detection of Selective Constraints by a Sliding-Window Maximum Parsimony Analysis
We also applied the SWAPSC method (Fares 2004) to test for positive selection. The latter is an improved Kimura-based algorithm (Li 1993), which infers an optimum codon-window size and slides it along the alignment to detect positive selection at individual amino acid sites along each branch of the input tree.
3D Modeling of Ectocarpus LRR Domain—Location of Positively Selected Sites
We hypothesized that the LRR domain folds separately from the GTPase domain and can therefore be modeled independently from the Roc–COR module. Homology-based 3D modeling was performed using the Swiss-Model server and the associated software PDB Viewer (Kiefer et al. 2009), according to the developer's instructions. The predicted structure was displayed using Rasmol version 2.7.5.
Results
Identification of Resistance Gene Candidates in Ectocarpus: The LRR-Containing Gene Family and NB-ARC–TPR Proteins
In line with their usual abundance and diversity in other eukaryotic genomes, 251 proteins are predicted to contain LRRs in Ectocarpus (supplementary table 1, Supplementary Material online). They are structurally extremely diverse, with a predicted size from less than 50 to more than 2,000 amino acids. A small fraction is well conserved with clear orthologues in other eukaryotes (e.g., dynein light chain, U2 small nuclear ribonucleoprotein A, nischarin, regulator of protein phosphatase 1), or with uncharacterized proteins of related organisms such as oomycetes (Esi0145_0047, Esi0383_0010). Some genes present a clearly modular structure, with additional conserved domains mostly related to signal transduction and protein–protein interactions (e.g., EF hand, calmodulin-binding motif, kinase, and GTPase), others only exhibit an LRR motif, with no apparent sequence conservation to any known orthologue or additional functional domain. No Ectocarpus LRR protein contains any Toll/Interleukin-1 receptor, Nucleotide-Binding Site, or Coiled-Coil motif that would make it directly comparable to plant resistance genes or animal Toll-like receptors. Instead, we found 37 LRR-GTPases of the ROCO family (associated to 20 related pseudogenes, further referred to as ROCOs) and 15 LRR-kinases, the structure and the genome organization of which seemed compatible with a putative defense function, as explained in detail in the following sections and summarized in table 1.
Table 1.
Overview of the Genome Organization, Probable Posttranscriptional Regulation, and Evolutionary Pressures Acting on ROCO, LRR-Kinases, and NB-ARC–TPR Genes.
| Gene Clustering | Rapid Gene Birth and Death | Repetitive Exon Structure | Exon Shuffling | Probable miRNA Regulationa | Positive Selection on Repetitive Exons | Evolution of Original Ligand-Binding Specificities | |
| LRR-ROCO | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| LRR-kinase (type I) | — | — | ✓ | — | — | ✓ | ✓ |
| LRR-kinase (type II) | — | — | ✓ | — | — | ✓ | ✓ |
| NB-ARC–TPR | ✓ | ✓ | ✓ (partial) | ✓ | ✓ | ✓ | n.d. |
Note.—✓: present; —: absent; n.d.: not determined in this study.
Some repetitive LRR (respective TPR) exons predicted as miRNA targets by Cock et al. (2010).
Additionally, we identified a family of 24 proteins containing an NB-ARC domain (IPR002182), which was originally defined as a conserved motive between the plant resistance genes and animal apoptosis-related proteins Rpm1, Rpp5, Ced4, and Apaf1 (Van der Biezen and Jones 1998). In Ectocarpus, all NB-ARC domain containing proteins are fused to a C-terminal TPR domain, which is capable of establishing ligand-specific interactions, and might therefore be functionally equivalent to the LRR domains found in the plant resistance genes Rpm1 and Rpp5.
We thus asked the question whether these ROCO GTPases, LRR-kinases, and NB-ARC–TPR genes might represent good candidate defense genes, and further assessed their phylogenetic history, as well as the nature of the evolutionary pressures acting upon them.
Organization of the Ectocarpus ROCO Proteins
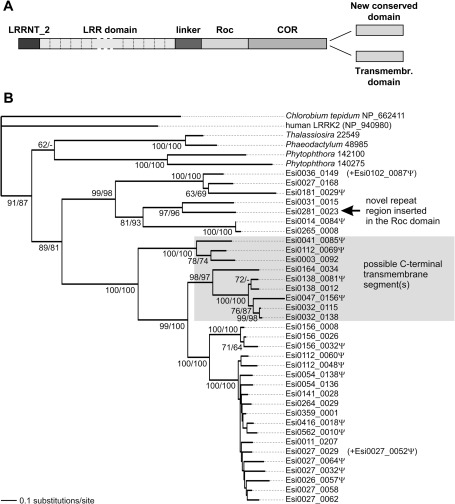
The overall structure of the Ectocarpus ROCO proteins is typical for the family and is very well conserved (fig. 1A). The N-terminal extremity (≈45 residues) is highly similar to the N-termini of other (non-ROCO) LRR proteins from Ectocarpus (e.g., Esi0191_0017) and gives a weak match to the Pfam profile LRRNT_2 (PF08263), a conserved region often found at the N-terminus of LRRs. Indeed, PSI-BlastT searches support the homology of the N-terminal region of Ectocarpus ROCO proteins to regions in LRR proteins in a wide variety of organisms (data not shown). The region downstream comprises up to 18 tandem LRRs arranged in a single continuous block, connected to the Roc GTPase domain by a linker. In most Ectocarpus ROCO proteins, the C-terminus after the COR domain is homologous to a corresponding region in ROCO proteins from some other species and perhaps represents a separate new domain (supplementary fig. 1B, Supplementary Material online). The other group is characterized by a shorter C-terminal region with one or two predicted transmembrane segments. It is therefore possible that these proteins are anchored by their C-termini to membranes, with the bulk of the protein (including the LRR region and the Roc and COR domains) most likely protruding to the cytoplasm.
FIG. 1.
The phylogenetic history of Ectocarpus ROCO proteins. (A) The general domain architecture of Ectocarpus ROCO proteins. (B) The ML tree presented was inferred from the conserved position of the central Roc–COR unit using RAxML (see Materials and Methods and supplementary fig. 1, Supplementary Material online). Bootstrap support values indicated for branches correspond to values obtained with RAxML rapid bootstrapping/thorough ML bootstrapping performed with PhyML. The tree was arbitrarily rooted with the ROCO protein from the bacterium Chlorobium tepidum. The letter Ψ attached to some Ectocarpus gene IDs indicates putative pseudogenes. The approximate position of two fragmentary pseudogenes (Esi0027_0052 and Esi0102_0087) is indicated with arrows near the genes that appear most similar to them.
Apart from incomplete sequences due to pseudogenization or interruptions in the genome assembly, there are two notable exceptions to this general ROCO structure. First, Esi0281_0023 is unusual in that it contains an insertion of a region (≈850 residues) representing an array of around 40 tandem repeats of a novel repeated motif (supplementary fig. 1C, Supplementary Material online). This repeated region is inserted into the GTPase domain between the conserved strand 5 and helix 4 (downstream of the highly conserved NKxD GTPase-specificity motif). Second, even though Esi0307_0011 contains an N-terminal LRR array followed by Roc and COR domains, it is highly dissimilar to the remaining family members in Ectocarpus and may be instead closer to the diatom ROCO proteins. In fact, it is the only Ectocarpus ROCO gene in which the LRR domain is not encoded by repetitive exons (see below). The distinctiveness of this protein makes it very difficult to obtain a reliable gene model in the absence of direct evidence from cDNA, and therefore was omitted from the alignment and the phylogenetic analysis presented below.
The Phylogenetic History of the Ectocarpus ROCO Family
To gain insight into the origins of the stunning expansion of the ROCO family genes in Ectocarpus, we conducted a phylogenetic analysis based on an alignment of the conserved Roc–COR unit (fig. 1B). The resulting tree indicates that the Ectocarpus ROCO family forms a monophyletic group to the exclusion of ROCOs from other species, including those from other stramenopiles (diatoms and Phytophthora). The backbone topology within the Ectocarpus family is very well resolved and reflects a series of successive gene duplications up to the most recent bursts of duplications in some terminal branches. Notably, loci located on the same genome scaffold (e.g., on scaffolds 27, 32, 112, 138, or 156) generally show close phylogenetic relationship, suggesting that the family mostly expands via local gene duplications. Interestingly, the proteins exhibiting the putative C-terminal transmembrane segment(s) fall into two distinct clades nested among the proteins with the conserved C-terminal domain shared with some other ROCO proteins outside Ectocarpus (see above). This may suggest that the C-terminal transmembrane anchor was recruited twice independently as a replacement of the original C-terminal domain. The protein Esi0281_0023 is unusual due to the novel repeat region inserted into the Roc domain. This locus is closely related to Esi0031_0015, which is apparently devoid of it, so the insertion/expansion of the repeat region seems to be a relatively recent event, further underscoring the dynamic evolution of the Ectocarpus ROCO family.
Rapid Gene Birth and Death in the ROCO and NB-ARC–TPR Families
Twenty-one of the 37 ROCO-containing (56%) loci and 12 of the 24 NB-ARC–TPR (50%) loci are organized in clusters of physically related genes and pseudogenes. These proportions are in sharp contrast with the overall scarcity of tandem and short-range duplications (5%) in the Ectocarpus genome. The latter only contains 823 tandem duplications (defined as two homologous genes within a distance of 20–30 genes) among the 16,377 predicted protein-coding loci (Cock et al. 2010). Additionally, at least a half of the ROCO loci (supplementary table 1, Supplementary Material online) are to a varying extent disrupted by frame shifts, stop codons, and/or deletions, while 5 of the 24 NB-ARC loci probably encode pseudogenes. This suggests that the ROCO and NB-ARC families undergo relatively high gene birth and death rates. In contrast, only 2 of the 15 (13%) LRR-kinases are tandemly duplicated, and we did not identify any obvious pseudogene among them.
Highly Dynamic LRR Exon Shuffling Underpins Variability of ROCO but Not LRR-Kinase Genes
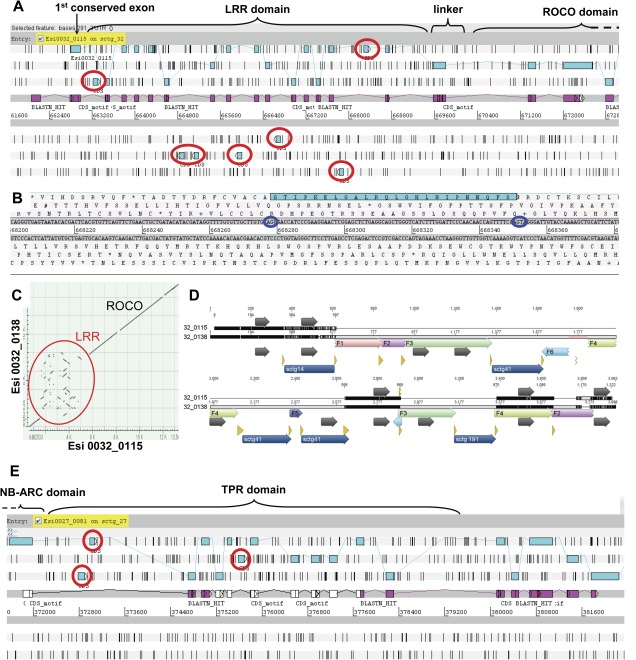
The LRR domain of all but one ROCO proteins exhibits a striking repetitive intron–exon structure, whereby each LRR is encoded by a single 72-nucleotide (24-amino acid) long exon (“type I LRR exon”: GxxPxxLxxxxxLxxLxLxxNxLx, fig. 2A). Additionally, intervening introns are sometimes interspersed with closely related, noncoding LRRs, as judged by either the interruption of EST support or their reverse orientation. Remarkably, the acceptor and donor splicing sites of intronic LRRs located on the noncoding strand are frequently conserved, suggesting a very recent origin (fig. 2B). We identified similar intact and/or inactivated (as judged by nonsense mutations or the loss of suitable splicing sites) LRRs on the noncoding strand of 19 ROCO loci (supplementary table 1, Supplementary Material online) pointing to the generality of the LRR exon shuffling across the ROCO family. Searches for similar noncoding LRRs over the entire corresponding scaffolds revealed that they are physically restricted to introns within the LRR domain.
FIG. 2.
The genomic organization of Ectocarpus ROCO and NB-ARC–TPR genes reveals intense shuffling of LRR (respectively TPR)-encoding exons. (A) Genomic organization of the Esi0032_0115 ROCO locus. The predicted protein sequence (joined blue rectangles) is supported throughout by ESTs (mapped in pink on the coding strand). Additional noncoding LRR exons in the intronic regions appear as blue rectangles circled in red. (B) Zoomed view of an intact noncoding type I LRR exon, with its conserved splicing sites circled in dark blue. (C) Nucleotide identity dot matrix between the two recently duplicated ROCO loci Esi0032_0115 and Esi0032_0138. Both gene sequences are almost identical, except for their LRR domain that appears highly recombined. (D) Alignment of the genomic sequences of Esi0032_0115 and Esi0032_0138, along the first 3 kb of their LRR domain. Highly conserved regions between the two genes are in black, unconserved regions are in light gray. LRR exons (on the coding strand only) are depicted with dark gray arrows. The pastel boxes represent homologous, locally rearranged sequence fragments (numbered F1–F6, originating from intragenic recombination), whereas dark blue boxes at the bottom represent sequences inserted from other genome scaffolds (sctg 14, sctg 41, and sctg 191, originating from intergenic recombinations). Recombination sites are highlighted with the yellow arrows. The pale red area around position 1777 in the Esi0032_0138 sequence corresponds to a short interruption in the genome assembly. (E) Genomic organization of the TPR domain of the Esi0027_0081 NB-ARC locus. The predicted protein sequence is depicted in blue, partially supported by ESTs (mapped in pink on the coding strand). Additional noncoding TPR exons (as judged by the absence of splicing sites, the presence of a frame shift, or of a stop codon) in the intronic regions appear as blue rectangles circled in red.
The interruption of EST support for LRR exons on the coding strand might be simply ascribed to alternative splicing. However, the presence of intact LRR exons on the noncoding strand suggests the occurrence of a highly dynamic rearrangement of LRR exons within the LRR domain, which results in an accordingly high variability both in LRR number and sequence. For example, the two recently duplicated genes Esi0032_0115 and Esi0032_0138 contain 13 and 17 LRR exons, respectively. Their LRR domain is indeed a recombination hot spot, as illustrated by the nucleotidic identity dot matrix in figure 2C. Multiple intra- and intergenic recombination events are traceable (fig. 2D). Intergenic recombinations involve other type I LRR loci, which may therefore act as a reservoir of diversity for ROCO proteins.
The situation is more complex for LRR-kinases, where two types of 24-residue repetitive exons were found. Five loci have “type I LRR exons” as described above for ROCO genes, whereas seven LRR-kinases contain type II LRR exons following the structure: xLxxNxLxxxxxGxxxxxxxLxxL (supplementary table 1, Supplementary Material online). Whereas each of these type II LRR exons encodes a 24-aa structural module, its boundaries are offset compared with the “type I” LRR exons. Hence, both types of exons have the potential to be reshuffled, but they cannot be mixed within a single domain. Despite extensive searches in the genomic sequence, we were unable to identify any reshuffled or inactivated type I or type II exons associated to any of the kinase loci.
Across the genome, 46 of the 164 other LRR-containing loci (non-ROCO, nonkinase) contain type I LRR exons, of which 13 exhibit intronic LRRs on the noncoding strand (supplementary table 1, Supplementary Material online). A further 8 loci contain type II LRR exons, none of which exhibits any tangible sign of exon shuffling (inactivated exons and/or intact exons on the noncoding strand). Overall, this suggests that type I LRR exons are more widespread and more extensively reshuffled across the genome compared with the type II LRR exons, with ROCO loci being key players in this process.
NB-ARC–TPR Loci Also Exhibit Signs of Exon Shuffling, Albeit to a Lesser Extent than ROCO Genes
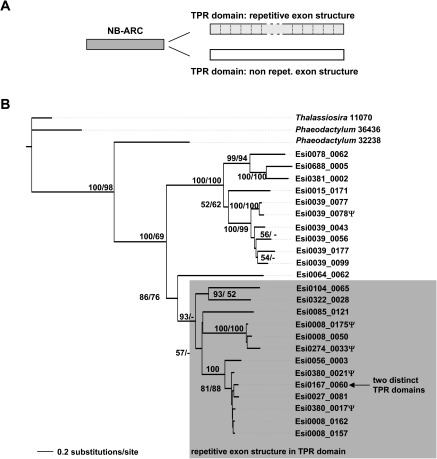
Similar to ROCO genes, 13 of the 24 NB-ARC predicted proteins also exhibit a repetitive structure, whereby the TPR domain is encoded by 42-aa long exons, each specifying a 34-residue TPR and a linker sequence (fig. 3A). An alignment of the NB-ARC domain of all 24 loci shows that the genes with a repetitive TPR exon structure group into a single clade (fig. 3B). As for the repetitive LRR exons of the ROCO loci, we identified inactivated TPR exons in the intervening introns (as judged by nonsense mutations, the loss of any suitable splicing site, or a shorter length leading to the truncation of the 34-aa TPR repeat; fig. 2E; supplementary table 1, Supplementary Material online). However, in contrast to the LRR exons of the ROCO loci, all but one inactivated TPR exons were restricted to the coding strand of the corresponding locus. We could not identify any TPR exon on the noncoding strand of NB-ARC loci.
FIG. 3.
Ectocarpus NB-ARC genes containing repetitive TPR exons fall into a single clade. (A) The general domain architecture of the Ectocarpus NB-ARC–TPR proteins. (B) ML tree obtained based on the alignment of the NB-ARC domains. Bootstrap support values indicated for branches correspond to values obtained with RAxML rapid bootstrapping/thorough ML bootstrapping performed with PhyML. Genes containing a TPR domain made of repetitive 42-amino acid–long exons are highlighted in gray. The letter Ψ following locus names indicates putative pseudogenes.
Exon Shuffling Is Restricted to TPR and Type I LRR-Containing Genes in the Ectocarpus Genome
We screened the whole genome in order to determine whether exon shuffling might underpin Ectocarpus proteome evolution or rather reflect a physiologically relevant specialization of the LRR- and TPR-containing genes. For this purpose, we relied on the fact that exon shuffling events (or exon indels) can only be retained in a gene if they do not introduce a frame shift in the encoded protein. Exon shuffling therefore only involves adjacent, so-called “phase 0” exons, the length of which is a multiple of three nucleotides. Hence, for each Ectocarpus locus, we divided the length of each exon by three, computed the rest of this Euclidean division, and deduced the number of successive Phase 0 exon pairs. As the vast majority of known shuffling cases involve exons of identical length (Patthy 1999), we further filtered successive Phase 0 exon pairs of identical length. NB-ARC–TPR and ROCO genes feature prominently in the resulting output, along with other repetitive TPR and type I LRR loci (supplementary table 2, Supplementary Material online). Only a very small number of non-TPR/non-LRR loci appeared as potential candidates for exon shuffling, nearly all of which we could ascribe to artefactual automated gene predictions. Altogether, this analysis strongly suggests that the exon shuffling in the Ectocarpus genome is overwhelmingly restricted to TPR- and type I LRR-containing loci.
Exon Shuffling Extensively Remodels the LRR and TPR Domains of ROCO and NB-ARC–TPR Genes and Obscures Their Paralogy Relationship
We aligned the individually extracted LRR (respectively TPR) exons of ROCO, NB-ARC, and LRR-kinase loci in an attempt to trace their origin (supplementary fig. 3, Supplementary Material online). As expected, the resulting trees reveal extremely little conservation of exon order or paralogy relationships even between recently duplicated ROCO and NB-ARC genes. Local duplications within the LRR/TPR domains of a single locus are identifiable on the alignments of the individual exons (supplementary fig. 3A and B, Supplementary Material online; LRR: Esi0041_0085, Esi0032_138, Esi0264_0029, Esi0112_0060, Esi0138_0012, and Esi0027_0062; TPR: Esi0008_0162, Esi0104_0065, Esi0322_0028, and Esi0380_0021). In contrast, paralogy relationships between type I and type II LRR exons of duplicated kinase genes are more easily traceable than for the LRR exons of ROCO genes (supplementary fig. 3C and D, Supplementary Material online), as illustrated for example by the exons 1–4 of Esi0107_0028, Esi0009_0077, and Esi0009_0083.
LRR and TPR Exons of Ectocarpus ROCO, NB-ARC–TPR, and LRR-Kinase Proteins Exhibit Signs of Diversifying Selection
As alluded above, aligning the entire LRR (and to a lesser extent, TPR) domains between paralogous ROCO and TPR–NB-ARC genes proved meaningless. Therefore, despite some limitations highlighted in the discussion, we searched for evidence of positive selection on individually extracted exons instead. We thus compared five independent exon data sets: 1) the 222 (72 nucleotide, 24-amino acid–long) type I LRR exons extracted from the ROCO loci; 2) the 124 (126 nucleotide, 42-amino acid–long) TPR exons extracted from the 13 NB-ARC loci with repetitive TPR exons; 3) the 242 (72 nucleotide, 24-amino acid–long) type I LRR exons extracted from both ROCO and LRR-kinase genes; 4) the 21 shorter (102 nucleotide, 34-amino acid–long) TPR exons extracted from the repetitive domains of NB-ARC loci (so-called TPR-like exons because the truncation leads to an interruption of the TPR structural motif, and therefore to a probable loss of function); 5) the 44 (72 nucleotide, 24-amino acid–long) type II LRR exons extracted from the LRR-kinase genes.
1) Estimated dS Rates Among LRR/TPR Exon Sequences
For each exon data set, we calculated the average codon-based evolutionary divergence over all sequence pairs using MEGA 4 (Tamura et al. 2007). In all instances, the average number of synonymous substitutions (dS) per synonymous site over all sequence pairs was well below two, which is the threshold above which the individual sequences would have to be excluded from further analysis to avoid the saturation effect of nucleotide substitution (supplementary table 3, Supplementary Material online; Yang and Nielsen 2000).
2) Evolutionary Analysis of the LRR and the TPR Exons Reveals Variable Selective Pressures Among Lineages
The main parameters estimated by CODEML for all exon data sets are detailed in the supplementary table 3, Supplementary Material online. The free-ratio model performed significantly better than the one-ratio model in all cases (P < 0.001), except for TPR-like exons (P < 0.1). In all five data sets, the free-ratio model predicted an ω > 1 in some lineages (supplementary fig. 3A–E, Supplementary Material online).
The most extensive signs of adaptive selection were found for the type I exons of both ROCO and LRR-kinases, as well as the type II exons of LRR-kinases and TPR-like exons, with 27 of the 481 (5.6%), 25 of the 441 (5.6%), 5 of the 85 (5.8%), and 2 of the 39 (5.2%) lineages, respectively, exhibiting ω > 1. For TPR-like exons, the highest ω values are very close to 1 (1.16 and 1.03) and might just reflect divergence under neutral evolution, in line with the probable loss of function arising from their truncation. Remarkable, but less widespread evidence of positive selection was also found in 5 of the 245 (2.0%) lineages of the TPR exon data set. Overall, these data suggest that positive selection is mostly acting on some lineages of the type I and type II LRR exons of ROCO and LRR-kinase genes and to a lesser extent, on the TPR (and perhaps the TPR-like) exons extracted from the NB-ARC–TPR genes.
In all data sets, positive selection is spread across the entire trees and detected mostly on the terminal branches (ROCO and kinase type I LRRs: 21 of 27; type II LRRs: 3 of 5; TPRs: 4 of 5). It seems to act within and across the loci alike, with no general rule emerging. For example, diversifying selection was evidenced between exons of Esi0011_0207, whereas it was not found amongst any of the most recently duplicated TPR loci (Esi0104_0065, Esi0322_0028). The latter observation, however, might be ascribed to a limited sensitivity of the algorithms used for the detection of positive selection using short and highly similar sequences.
3) Variable Selective Pressures Among Amino Acid Sites
To test for positive selection at individual amino acid sites, CODEMLSITES was used to compare model M0 (one ratio) and M3 (discrete), M1 (neutral), and M2 (selection), and M7 (beta) and M8 (beta and ω) for all exon alignments (supplementary table 4, Supplementary Material online). All three models (M2, M3, and M8) that allow for selection were significantly favored over the other models (P < 0.001), except for the probably inactivated TPR-like exons. For the four other data sets, the sites predicted to be under positive selection with the posterior probabilities greater than 0.99 (highlighted in boldface in supplementary table 4, Supplementary Material online, and in red on fig. 4C) were in agreement between the models M2, M3, and M8, except the 20th residue of type I LRR exons (highlighted in green on fig. 4C), which was only supported under the model M8 by a posterior probability P = 0.886 (ω = 1.436 ± 0.178).
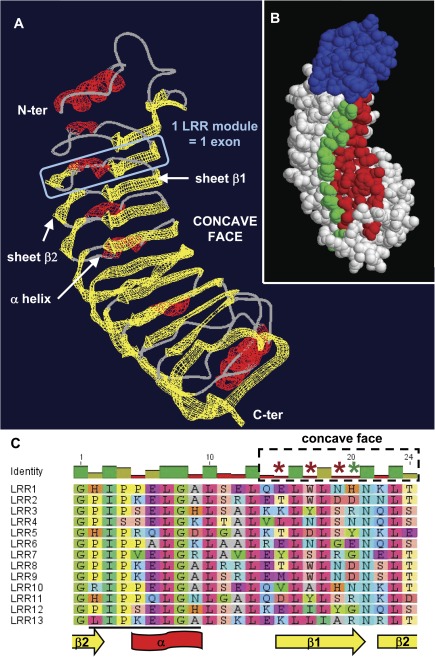
FIG. 4.
Positive selection acts on solvent-exposed and potentially ligand-binding residues of the LRR domains. (A) Homology-based 3D model of Esi0032_0115 N-terminus and LRR domain. The designation of β−β−α structures follows (Di Matteo et al. 2003). (B) Corresponding surface view of the polypeptide, with the potential location of positively evolving sites in type I LRR exons highlighted. Sites shown in red are those (15, 17, 19) where model M8 revealed posterior probabilities greater than 0.99, while site 20 (in green) was supported by a posterior probability of 0.88. All sites are predicted to be located on the concave, probably ligand-binding face of the domain. The N-terminal region is depicted of the protein in blue. (C) Alignment of the 13 exons composing the LRR domain, showing the conserved structural residues interspersed with variable amino acids. Asterisks highlight the four amino acid positions subject to positive selection, with the same color code as in (B). The region targeted by the esi_mir_cand_5 miRNA family (Cock et al. 2010) is underlined in black.
Many of these sites were also detected with SWAPSC (Fares 2004). Among the positively selected amino acid sites in each branch of the evolutionary trees, the average ω values were 5.895, 3.493, and 5.473 for LRRs from ROCO, TPR, and type II LRR exons, respectively.
In contrast, SWAPSC did not reveal any significant hint of positive selection in the TPR-like exons, and more surprisingly, for the entire (ROCO + kinase) type I LRR data set. The latter result might be ascribed to the addition of the rather divergent type I LRR exons of kinases (compared with the data set restricted to ROCOs), which leads to an increase in average Ks values (2.14). Additionally, in all five data sets, a small proportion of codons exhibited accelerated rates of nonsynonymous nucleotide substitutions but no definite conclusion could be reached regarding the existence or not of adaptive evolution (Fares 2004).
Variation in LRR Domain Length and Sequence Likely Supports Variation in Ligand-Binding Specificities of ROCO Proteins and LRR-Kinases
In order to assess the possible functional significance of our results, we attempted homology-based 3D modeling of a type I LRR domain, focusing on Esi0032_0115 because its sequence is well supported by the EST data and reflects exon shuffling. The protein of known structure most similar to the LRR domain of Esi0032_0115 is the Phaseolus polygalacturonase inhibitor (Di Matteo et al. 2003). Since Esi0032_0115 contains more LRRs than polygalacturonase-inhibiting protein (PGIP), we split the Ectocarpus LRR domain into two regions, modeled them independently using PGIP as a template, and then merged the two preliminary models together thanks to their overlap. No attempt was made to refine the model further by minimizing its free energy. Instead, repeated modeling attempts with alternative templates and slightly different conditions suggested that the LRR domain of Esi0032_0115 adopts a repetitive parallel beta-sheet structure, where each LRR exon encodes a β−β−α (fig. 4A) or a single β-strand structural module. Hence, a variation in the number of LRR exons would probably lead to the insertion/deletion of one structural unit, without affecting its overall organization.
Furthermore, the positively selected sites detected by the empirical Bayes analysis after the implementation of the most stringent model M8 are located on the concave face of the domain (fig. 4B and C), as observed in the plant PGIP protein and the variable lymphocyte receptors of jawless vertebrates. In the latter proteins, the concave face is the known ligand- or antigen-binding face of the LRR domain, and mutations of its hypervariable residues do affect the protein recognition specificities (Han et al. 2008; Casasoli et al. 2009). Hence, our observations point to the LRR domain of Ectocarpus ROCO proteins rapidly evolving new ligand-binding specificities under positive selection. This in turn suggests a functional specialization of these proteins in the recognition of highly variable ligands such as antigens, followed by the subsequent signal transduction via their GTPase activity.
Strikingly, sites 3 and 23 of type II LRR exons (subject to significant positive selection under the M8 model, supplementary table 4, Supplementary Material online) are structurally equivalent to the positively selected positions 19 and 15, respectively, of type I exons. This observation adds weight to the overall biological significance of our results and suggests that despite the apparent absence of any exon shuffling, the evolution of Ectocarpus type II LRR-kinases is also driven by the emergence of original ligand-binding specificities.
Discussion
Specific Limitations of the PAML and SWAPSC Analyses of Repetitive LRR and TPR Exons
The limitations of computational methods for detecting positive selection have already been extensively discussed (Fares 2004; Yang 2007). Additional difficulties arose from uncertainties about gene structures and widespread exon shuffling. Indeed, due to their high intron number and the presence of coding-like sequences within the intronic regions, extensive manual curation was required to reconstruct the likely structure of ROCO and NB-ARC–TPR proteins. On the other hand, the relatively low number of ESTs mapping on these genes, occasional interruptions in the genome assembly, combined to the potential existence of splicing variants as well as the shortness and high variability of LRR and TPR exons renders their structural prediction somewhat speculative.
Moreover, extensive exon shuffling precluded any meaningful alignment of the whole LRR and TPR domains of ROCO and NB-ARC proteins. We chose to bypass this issue by extracting individual exons from the gene sequences, in an attempt to improve the quality of the alignment underlying dN/dS calculations. A key limitation of this approach stems from the short length of the input sequences, which draws the applicability of the PAML and SWAPSC packages to their limit. In particular, our strategy may account for the unusually high number of infinite ω values observable on supplementary figure 3A–E, Supplementary Material online, and the insignificant SWAPSC results in the (ROCO + kinase) type I LRR exon data set. Additionally, the implementation of site-specific LRTs in CODEMLSITES effectively averages site-specific ω values across the branches of the tree. Hence, our exon-centered approach fails to capture the variability of evolutionary forces potentially at play across the different exons of individual LRR (respectively TPR) domains and thus has only little power to detect positively selected sites on a particular branch of the exon tree.
Despite these reservations, our parsimony-based SWAPSC results coincide with those obtained using the ML methods, with 3D modeling, and with observations made on the LRR domains of the other organisms. We take the global coherence of this data set as a strong indication of its biological significance.
Ectocarpus ROCOs and NB-ARC–TPRs Are Good Defense Gene Candidates
The Ectocarpus genome does not contain clear orthologues of plant resistance genes or animal pathogen receptors, but as alluded in the Introduction, Ectocarpus ROCOs LRR-kinase and NB-ARC–TPR genes encode domains widely known to participate in pathogen detection and subsequent signal transduction in other organisms. These genes exhibit the signs of evolution under diversifying selection (table 1). Additionally, Ectocarpus ROCO and NB-ARC–TPR genes are subject to high birth and death rates, an unusual evolutionary features not restricted to but shared by many animal and plant gene families involved in immunity (Nei and Rooney 2005).
In the jawless fishes, the LRR hyperdiversity underpinning the antigen recognition is generated via the somatic recombination of a cassette locus with a reservoir of variable related sequences. In Ectocarpus, exon shuffling likely provides a comparable and extremely efficient mechanism for the evolution of new ligand-binding specificities. The restriction of exon shuffling to LRR and TPR domains in the genome, and in particular, the fact that proteins (type I and type II exon LRR-kinases) with a modular LRR exon structure do not exhibit any sign of exon shuffling or physical clustering within the genome, strongly suggests that the latter reflect functional adaptations linked to the physiological function of the ROCO and NB-ARC proteins. Finally, 3D modeling of the Esi003_0115 LRR domain indicates that diversifying selection probably underpins the acquisition of original ligand-binding specificities, in line with a potential specialization in the recognition of highly variable ligands such as antigens.
Normally, many selective episodes date back to the birth of paralogous genes by duplication at an ancestral locus (Lynch and Conery 2000). Despite that, most positively selected LRR and TPR branches revealed by CODEML are terminal, pointing to an intense and recent positive selection acting upon these exons. It is striking that these recent episodes of positive selection of ROCO LRR and TPR exons often overlap similar more ancient events, as unveiled by the more sensitive SWAPSC analysis. In other words, those residues that originally conferred specificity to a hypothetical ligand were apparently altered repeatedly to provide novel binding functions later.
In conclusion, although the Ectocarpus ROCO and NB-ARC families are roughly ten times smaller than the ones encoding resistance gene analogues in plants, we think that the combination of such uncommon structural, regulatory, and evolutionary features makes them excellent candidates for being involved in recognition and/or transduction events linked to immunity (table 1). Our results will be instrumental for narrowing down a list of candidates for full functional characterization, greatly enhancing the prospect of identifying protein–protein interaction “hotspots” involved in Ectocarpus defense reactions.
Exon Shuffling in Ectocarpus ROCO and NB-ARC–TPR Genes Is Mediated by Specific, Targeted, Somatic Recombination
The molecular mechanism underpinning exon shuffling in the ROCO and the NB-ARC families was not investigated in depth here, but it exhibits striking properties. First, the apparent absence of exon shuffling in type II exons of the LRR-kinase family suggests that this process is not simply driven by the illegitimate recombination of homologous sequences (table 1). Second, whereas inactivated and reshuffled TPR exons are disproportionately restricted to the coding strand of the NB-ARC proteins, the abundance of reshuffled LRR exons on the noncoding strand of ROCO loci points to the existence of comparable, yet potentially distinct mechanisms acting on these two gene families. We have not found any trace of the RAG1 and RAG2 genes in the Ectocarpus genome and thus the transposition mechanism is probably different from the one described in the mammal lymphocytes. Exon shuffling in other organisms is often linked to the activation of nonautonomous transposable elements (TEs; e.g., Morgante et al. 2005; Hancks et al. 2009). However, for both gene families, our initial observations do not support the involvement of TEs in exon shuffling, so that we are currently investigating other possible hypotheses.
Intriguingly, reshuffled LRR (respectively TPR) exons of ROCO and NB-ARC proteins have been identified as the likely target of miRNA families isolated from Ectocarpus (fig. 2C; Cock et al. 2010). Therefore, exon shuffling may not only affect the ligand-binding specificities of any given protein but also the regulation of its expression. The functional significance of this observation remains to be investigated.
Are Brown Algae Capable of Adaptive Immunity?
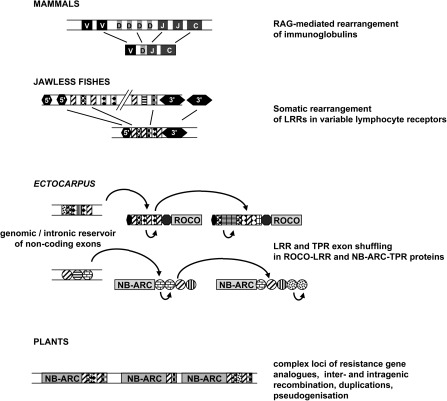
We do not yet have direct evidence of the involvement of ROCO and NB-ARC–TPR proteins in Ectocarpus immune responses. However, their remarkable structural (and possibly regulatory) features point them as the best candidates in its genome to fulfill a role in pathogen perception. We also hypothesize that excised, temporarily noncoding, LRR/TPR exons might constitute a reservoir evolving under relaxed selection before being recruited again into an LRR (respectively TPR) domain during the next recombination event (fig. 5). This combinatorial process would result in a tremendous, but controlled potential for somatic variation, leading to the emergence of a repertoire of new ligand-binding specificities upon the induction of recombination.
FIG. 5.
Exon shuffling in the Ectocarpus ROCO and NB-ARC proteins could underpin the emergence of a pathogen recognition repertoire in a way comparable to the adaptive immune systems of vertebrates. The mammal antigen receptor repertoire is generated via the Variable-Diversity-Joining (VDJ) rearrangement of immunoglobulin segments, which is mediated by the RAG (recombination activating gene) transposon. In jawless fishes, somatic rearrangement of LRR genomic cassettes generates the variable lymphocyte receptors, which are structurally extremely similar to the LRR domain of the Ectocarpus ROCO proteins. In Ectocarpus, intronic and other LRRs scattered in the genome could similarly serve as a source of diversity for the LRR exon shuffling. The Ectocarpus ROCO and NB-ARC families also share some genomic features with plant resistance genes, in particular, physical clustering and high gene birth and death rates. Patterned rectangles and circles represent hypervariable ligand-binding LRR and TPR modules (respectively) subject to positive selection. Curved arrows represent exon shuffling events followed by divergence. Light gray rectangles highlight domains likely involved in signal transduction events triggered by recognition.
In conclusion, our findings unambiguously demonstrate that mechanistically, brown algae do possess the genomic toolbox necessary for sustaining a fully functional adaptive immune system, and that they may build up their immune repertoire by targeted somatic recombination, a mechanism so far believed to be restricted to vertebrates (Flajnik and Kasahara 2010; fig. 5). At first sight, somatic variation of antigen receptors would make little point in an organism devoid of any circulatory apparatus. However, its potential selective advantage should not be underestimated if combined to vegetative propagation. Indeed, all development stages of Ectocarpus have a propensity to parthenogenesis, via sporogenesis, gametogenesis, and/or thallus fragmentation. Thus somatic diversification might be combined to selection, leading to the rapid emergence of individuals capable of recognizing new pathotypes.
We have not yet investigated the timescale of exon shuffling in the ROCO and NB-ARC–TPR genes. Thus, it is possible that recombination of the LRR (respectively TPR) domains may not occur during the lifetime of a plantlet, so that these proteins might as well operate as part of an innate immune system. Under this hypothesis, Ectocarpus would give us a unique insight about how an adaptive immune system could emerge from an innate one, by recruiting a somatic recombination mechanism at a particular development stage or in a specific cell type.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
Anne-Flore Bonvalot (Ecole Polytechnique, France), J. Mark Cock (Centre National pour la Recherche Scientifique, Roscoff, France), Valérie Geffroy (Université Paris-Sud, France), Pierre Rouzé, Yves van de Peer (Plant Systems Biology, Ghent, Belgium), and three anonymous reviewers are gratefully acknowledged for stimulating discussions and constructive suggestions. This work was supported by the Natural Environment Research Council (Strategic Ocean Funding Initiative NE/F012705/1 and New Investigator grant NE/J00460X/1 to C.M.M.G.); an FP7 Marie Curie award (PERG03-GA-2008-230865 to C.M.M.G.); the Czech Science Foundation (grant number P305/10/0205 to M.E.); the FP7 “Capacities” Specific Programme ASSEMBLE (grant number 227799 to A.Z.), and a research bursary from the Scottish Association for Marine Science to A.Z.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Ann Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Bosgraaf L, Van Haastert PJM. Roc, a Ras/GTPase domain in complex proteins. Biochim Biophys Acta Mol Cell Res. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Casasoli M, Federici L, Spinelli F, Di Matteo A, Vella N, Scaloni F, Fernandez-Recio J, Cervone F, De Lorenzo G. Integration of evolutionary and desolvation energy analysis identifies functional sites in a plant immunity protein. Proc Natl Acad Sci U S A. 2009;106:7666–7671. doi: 10.1073/pnas.0812625106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouze P, et al. (77 co-authors) The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Daniëls V, Baekelandt V, Taymans JM. On the road to leucine-rich repeat kinase 2 signalling: evidence from cellular and in vivo studies. Neurosignals. 2011;19:1–15. doi: 10.1159/000324488. [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, De Lorenzo G, Tsernoglou D, Cervone F. The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defence. Proc Natl Acad Sci U S A. 2003;100:10124–10128. doi: 10.1073/pnas.1733690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Dodds P, Pryor T. Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol. 2000;3:278–284. doi: 10.1016/s1369-5266(00)00080-7. [DOI] [PubMed] [Google Scholar]
- Fares MA. SWAPSC: sliding window analysis to detect selective constraints. Bioinformatics. 2004;20:2867–2868. doi: 10.1093/bioinformatics/bth303. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. 2005. Distributed by the author. Seattle (WA): Department of Genome Sciences, University of Washington. Available from: http://evolution.genetics.washington.edu/phylip.html. [Google Scholar]
- Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AR, Baker BJ. The evolution of resistance genes in multi-protein plant resistance systems. Curr Opin Genet Dev. 2007;17:493–499. doi: 10.1016/j.gde.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Gachon CMM, Strittmatter M, Müller DG, Kleinteich J, Küpper FC. Detection of differential host susceptibility to the marine oomycete pathogen Eurychasma dicksonii by real-time PCR: not all algae are equal. Appl Environ Microbiol. 2009;75:322–328. doi: 10.1128/AEM.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt K, Weyand M, Kortholt A, Van Haastert PJ, Wittinghofer A. Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J. 2008;27:2239–2249. doi: 10.1038/emboj.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321:1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Ewing AD, Chen JE, Tokunaga K, Kazazian HH. Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009;19:1983–1991. doi: 10.1101/gr.093153.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Li W-H. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynn DJ, Higgs R, Gaines S, Tierney J, James T, Lloyd AT, Fares MA, Mulcahy G, O'Farrelly C. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics. 2004;56:170–177. doi: 10.1007/s00251-004-0675-0. [DOI] [PubMed] [Google Scholar]
- Lynn DJ, Lloyd AT, Fares MA, O'Farrelly C. Evidence of positively selected sites in mammalian alpha-defensins. Mol Biol Evol. 2004;21:819–827. doi: 10.1093/molbev/msh084. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol. 2011;12:818–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- Marin I, Van Egmond WN, Van Haastert PJM. The Roco protein family: a functional perspective. FASEB J. 2008;22:3103–3110. doi: 10.1096/fj.08-111310. [DOI] [PubMed] [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB. GeneDoc: a tool for editing and annotating multiple sequence alignments. 1997. Distributed by the author. Available from: http://www.nrbsc.org/gfx/genedoc/ [Google Scholar]
- Padmanabhan M, Cournoyer P, Dinesh-Kumar SP. The leucine-rich repeat domain in plant innate immunity: a wealth of possibilities. Cell Microbiol. 2009;11:191–198. doi: 10.1111/j.1462-5822.2008.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z, Amemiya CT, Ehrhardt GRA, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. [Google Scholar]
- Patthy L. Genome evolution and the evolution of exon-shuffling—a review. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potin P, Bouarab K, Salaün J-P, Pohnert G, Kloareg B. Biotic interactions of marine algae. Curr Opin Plant Biol. 2002;5:308–317. doi: 10.1016/s1369-5266(02)00273-x. [DOI] [PubMed] [Google Scholar]
- Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-rich repeat protein complex activates mosquito complement in defence against Plasmodium parasites. Science. 2009;324:258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald PC, Beutler B. Plant and animal sensors of conserved microbial signatures. Science. 2010;330:1061–1064. doi: 10.1126/science.1189468. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JPY, Williams KL. The CATERPILLER family: an ancient family of immune/apoptotic proteins. Clin Immunol. 2005;115:33–37. doi: 10.1016/j.clim.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JD. The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol. 1998;8:226–227. doi: 10.1016/s0960-9822(98)70145-9. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.