Abstract
The major histocompatibility complex (MHC) on chromosome 6p is an established risk locus for ulcerative colitis (UC) and Crohn’s disease (CD). We aimed to better define MHC association signals in UC and CD by combining data from dense single nucleotide polymorphism (SNP) genotyping and from imputation of classical HLA types, their constituent SNPs and corresponding amino acids in 562 UC, 611 CD, and 1,428 control subjects. Univariate and multivariate association analyses were performed, controlling for ancestry. In univariate analyses, absence of the rs9269955 C allele was strongly associated with risk for UC (P = 2.67×10−13). rs9269955 is a SNP in the codon for amino acid position 11 of HLA-DRβ1, located in the P6 pocket of the HLA-DR antigen binding cleft. This amino acid position was also the most significantly UC-associated amino acid in omnibus tests (P = 2.68×10−13). Multivariate modeling identified rs9269955-C and 13 other variants in best predicting UC versus control status. In contrast, there was only suggestive association evidence between the MHC and CD. Taken together, these data demonstrate that variation at HLA-DRβ1, amino acid 11 in the P6 pocket of the HLA-DR complex antigen binding cleft is a major determinant of chromosome 6p association with ulcerative colitis.
Keywords: inflammatory bowel disease genetics, major histocompatibility complex, ulcerative colitis
Introduction
The major histocompatibility complex (MHC) on chromosome 6p contains the highly polymorphic human leukocyte antigen (HLA) genes and other immunoregulatory genes.1, 2 Genetic variants in the MHC have been associated with susceptibility for many infectious and immune-mediated diseases including the inflammatory bowel diseases (IBD), ulcerative colitis (UC) and Crohn’s disease (CD).3, 4 Features of the MHC such as dense gene clustering with broad linkage disequilibrium, extensive polymorphism, and heterogeneity among different populations have made localization of causal variants challenging.2
HLA polymorphisms were the focus of attention in several IBD candidate gene association studies of relatively small sample size and meta-analyses of these studies found HLA associations in UC that were mostly different from those found in CD.3–5 Subsequently, linkage between IBD and the chromosome 6p IBD3 locus was found in genome-wide linkage scans6–8. Recent genome-wide association studies (GWAS) have confirmed the MHC as one of 47 UC loci and 71 CD loci with significant evidence for association (P < 5×10−8).9, 10 The most significant association signal in a recent meta-analysis of six GWAS that included 6,687 UC cases and 19,718 controls of European ancestry was at a single nucleotide polymorphism (SNP) in the MHC class II region (rs9268853, P = 1.35×10−55).10 In contrast, the most significant MHC association signal in a meta-analysis of six CD GWAS that included a similar combined sample size (6,333 CD cases and 15,056 controls) was less significant than the UC signal and was located in the MHC class III region near the lymphotoxin A (LTA) locus (rs1799964, P = 3.98×10−11).9, 10
Here, we explore the MHC association signal in the discovery stage of a new UC and CD GWAS with excellent coverage (>10,000 SNPs) across the extended MHC. We used our MHC SNP data and an existing reference dataset to impute classical HLA allele types, their constituent SNPs, and corresponding amino acids in our UC, CD and control samples. This allowed us to evaluate if the observed SNP associations in the MHC can be explained by variation specifically in the classical HLA genes.
Results
Analysis of genotyped MHC SNPs in IBD
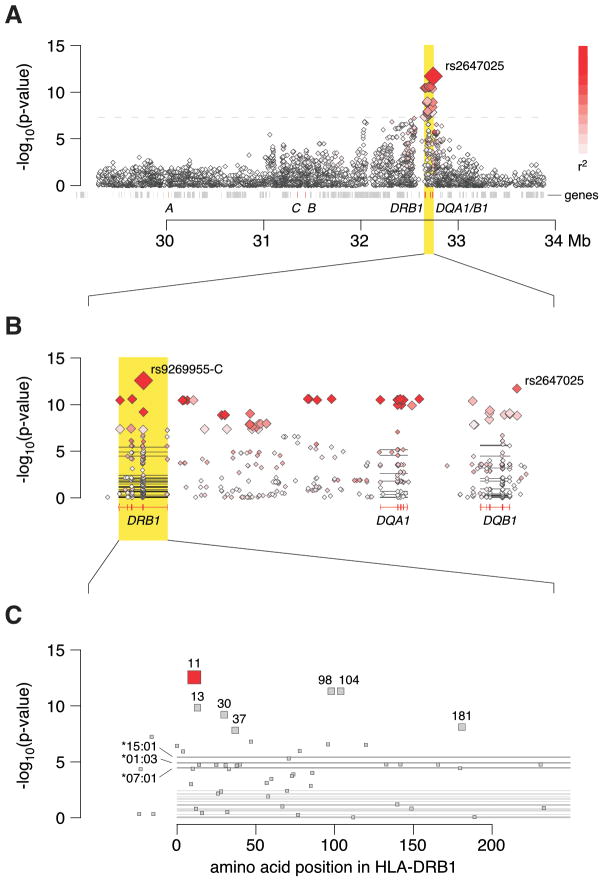
First, we tested 10,347 genotyped SNPs in the MHC region from 29,299 to 33,884 kb on chromosome 6 using NCBI36/hg18 coordinates for association with UC and CD with ileal involvement. Among 35 SNPs that reached genome-wide significance (P < 5 × 10−8) in the UC analysis, the most significant SNP was rs2647025 (OR=1.95 [1.62–2.35, 95% confidence interval (CI)] for the G allele; P = 1.94×10−12), located in the promoter region of HLA-DQB1 (Figure 1A). This SNP is correlated with rs9268853 (r2 = 0.63 in HapMap 3-CEU11), which was the MHC region SNP with the most significant association in a recent UC GWAS meta-analysis10, and it is also correlated with rs2395185 (r2 = 0.60 in our dataset), which was the MHC region SNP with the most significant association in the NIDDK IBD Genetics Consortium UC GWAS12, both at distances of > 200 kb.
Figure 1.
Major histocompatibility complex regional association plots for ulcerative colitis. (A) Association results for genotyped SNPs from the Illumina Omni1-quad BeadChip. The intensity of the red shading indicates the strength of the pairwise r2 correlation to the most associated SNP, rs2647025. (B) Association results for both genotyped (◇ symbols) and imputed (■ symbols) nucleotides focused in on the region of peak association in panel A. Horizontal lines represent the classical HLA alleles in this region. The intensity of the red shading indicates the strength of the pairwise r2 correlation to the most associated SNP marker, rs9269955-C. (C) Association results for imputed amino acids in HLA-DRβ1.
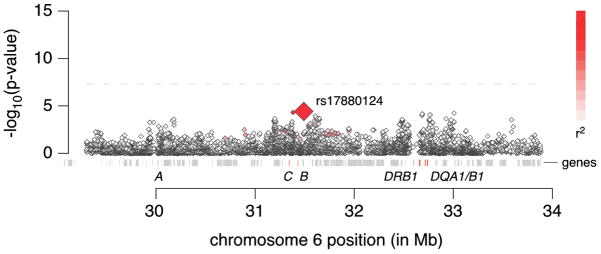
In contrast, there was only suggestive evidence for association between MHC region SNPs and CD with ileal involvement (Figure 2). The most significant association signal was found at rs17880124 (OR=2.23 [1.52–3.27, 95%CI] for the G allele; P = 3.82×10−5) which is located in an exon of the MHC class I polypeptide-related sequence A (MICA) gene. Of note, the association observed in UC was many orders of magnitude stronger than that in CD with ileal involvement despite a similar number of cases. Therefore, we focused on the UC signal through imputation of classical HLA alleles and their corresponding nucleotide and amino acid sequences.
Figure 2.
Major histocompatibility complex regional association plot for Crohn’s disease with ileal involvement. Association results are for genotyped SNPs from the Illumina Omni1-quad BeadChip. The intensity of the red shading indicates the strength of the pairwise r2 correlation to the most associated SNP, rs17880124.
Analysis of imputed classical HLA alleles in UC
The following imputed genetic markers were included in our UC vs. control analyses: 156 classical HLA alleles at four-digit resolution, 95 classical HLA allele groups at two-digit resolution, 1,765 binary SNP features at 1,573 nucleotide positions, and 561 binary HLA amino acid features at 357 amino acid positions. The most significant association signal in UC mapped to rs9269955 (Figure 1B), which is a tri-allelic SNP within the coding region of HLA-DRB1 (position 32,660,116 using NCBI36/hg18 coordinates). In combination with the nucleotide position directly adjacent to it (rs17878703 at position 32,660,115), rs9269955 determines the codon for amino acid position 11 of the HLA-DRβ1 protein, where six different amino acid alleles are observed in the population at large (Table 1). Chromosome 6 position 32,660,114 is the third position in this codon, and it is not known to be polymorphic. Rs9269955-C (to indicate the presence of the C allele) is associated with protection against UC (OR = 0.51 [0.43–0.61, 95% CI], P = 2.67×10−13). In combination with the adjacent rs17878703 alleles, rs9269955-C encodes three of the six observed amino acids (aspartic acid, valine, or glycine) at HLA-DRβ1 amino acid 11 (Table 1). This SNP is correlated with rs2395185 (r2 = 0.88 in our dataset), which was the MHC region SNP with the most significant association in the NIDDK IBD Genetics Consortium UC GWAS.12
Table 1.
Univariate results for DNA sequence (shown in order of positions 32,660,114 to 32,660,116) and codon determinants (shown in order of corresponding positions 32,660,116 to 32,660,114) for HLA-DRβ1, amino acid 11.
| Position | Allele | DNA sequence (Positions 32,660,114 – 32,660,116) | Codon (Positions 32,660,116 – 32,660,114) | Frequency (UC) | Frequency (Controls) | OR (95% CI) | P value |
|---|---|---|---|---|---|---|---|
| rs9269955 (position 32,660,116) | C | --C | 0.188 | 0.300 | 0.51 (0.43–0.61) | 2.67 × 10−13 | |
| A | --A | 0.451 | 0.431 | 1.11 (0.96–1.28) | 1.51 × 10−1 | ||
| G | --G | 0.362 | 0.268 | 1.52 (1.31–1.77) | 5.56 × 10−8 | ||
| rs17878703 (position 32,660,115) | T | -T- | 0.003 | 0.011 | 0.25 (0.07–0.81) | 2.14 × 10−2 | |
| C | -C- | 0.092 | 0.139 | 0.61 (0.48–0.77) | 3.38 × 10−5 | ||
| A | -A- | 0.238 | 0.266 | 0.86 (0.72–1.01) | 6.97 × 10−2 | ||
| G | -G- | 0.667 | 0.584 | 1.46 (1.26–1.70) | 9.25 × 10−7 | ||
| HLA-DRβ1, amino acid 11 | Asp | ATC | GAU | 0.003 | 0.011 | 0.25 (0.07–0.81) | 2.14 × 10−2 |
| Val | AAC | GUU | 0.093 | 0.151 | 0.55 (0.44–0.70) | 1.11 × 10−6 | |
| Gly | ACC | GGU | 0.092 | 0.139 | 0.61 (0.48–0.77) | 3.38 × 10−5 | |
| Ser | AGA | UCU | 0.451 | 0.431 | 1.11 (0.96–1.28) | 1.52 × 10−1 | |
| Leu | AAG | CUU | 0.145 | 0.115 | 1.32 (1.07–1.63) | 8.98 × 10−3 | |
| Pro | AGG | CCU | 0.216 | 0.153 | 1.48 (1.24–1.77) | 1.61 × 10−5 |
DNA, deoxyribonucleic acid; UC, ulcerative colitis; OR, odds ratio; CI, confidence interval; A, adenine; C, cytosine; G, guanine; T, thymine; U, uracil; Asp, aspartic acid; Val, valine; Gly, glycine; Ser, serine, Leu, leucine; Pro, proline.
To analyze the role of specific amino acid positions in the HLA genes in UC, we conducted omnibus tests for association with degrees-of-freedom equal to the number of distinct residues for that amino acid position minus one (Table 2). The most significant finding was for HLA-DRβ1 amino acid 11 (P = 2.68×10−13), consistent with the results noted above (Figure 1C). Several other amino acid associations were highly significant including other amino acid positions in HLA-DRβ1, HLA-DQα1 or HLA-DQβ1 (Table 2).
Table 2.
Omnibus amino acid tests for ulcerative colitis versus control. Amino acid positions with omnibus P < 5 × 10−8 are shown.
| HLA amino acid position | Codon middle nucleotide position (chromosome 6, NCBI36/hg18) | Degrees of freedom | Omnibus P value |
|---|---|---|---|
| HLA-DRβ1, amino acid 181 | 32,657,335 | 1 | 7.48 × 10−9 |
| HLA-DRβ1, amino acid 104 | 32,657,566 | 1 | 4.70 × 10−12 |
| HLA-DRβ1, amino acid 98 | 32,657,584 | 1 | 4.68 × 10−12 |
| HLA-DRβ1, amino acid 37 | 32,660,037 | 4 | 1.46 × 10−8 |
| HLA-DRβ1, amino acid 30 | 32,660,058 | 5 | 6.01 × 10−10 |
| HLA-DRβ1, amino acid 13 | 32,660,109 | 5 | 1.39 × 10−10 |
| HLA-DRβ1, amino acid 11 | 32,660,115 | 5 | 2.68 × 10−13 |
| HLA-DQα1, amino acid 47 | 32,717,191 | 3 | 2.73 × 10−10 |
| HLA-DQα1, amino acid 50 | 32,717,200 | 2 | 2.95 × 10−11 |
| HLA-DQα1, amino acid 53 | 32,717,209 | 2 | 2.12 × 10−11 |
| HLA-DQα1, amino acid 175 | 32,717,988 | 2 | 2.28 × 10−10 |
| HLA-DQα1, amino acid 215 | 32,718,464 | 1 | 5.95 × 10−12 |
| HLA-DQβ1, amino acid 185 | 32,737,733 | 1 | 8.62 × 10−11 |
Because these results highlighted HLA-DRβ1 amino acid 11, we further analyzed the six amino acids at this position and the corresponding classical HLA-DRB1 allele groups at two-digit resolution (Table 3). The three amino acids (aspartic acid, valine, and glycine) encoded by the rs9269955-C allele in combination with the adjacent rs17878703 alleles, are all associated with protection against development of UC.
Table 3.
Ulcerative colitis versus control associations for HLA-DRβ1 amino acid 11 residues and corresponding classical HLA-DRB1 alleles. The multivariate best model for HLA-DRβ1 amino acid 11 residues alone was identified with stepwise regression. UC, ulcerative colitis; OR, odds ratio; CI, confidence interval; Asp, aspartic acid; Gly, glycine; Leu, leucine; Pro, proline; Ser, serine; Val, valine.
| Amino Acid at HLA-DRβ1 position 11 | Corresponding HLA-DRB1 group | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Amino Acid | Frequency (UC) | Frequency (Controls) | Univariate | Multivariate | HLA-DRB1 group | Frequency (UC) | Frequency (Controls) | OR (95% CI) | P value | ||
| OR (95% CI) | P value | OR (95% CI) | P value | ||||||||
| Asp | 0.003 | 0.011 | 0.25 (0.07–0.81) | 2.14 × 10−2 | 0.21 (0.06–0.69) | 1.03 × 10−2 | HLA-DRB1*09 | 0.003 | 0.011 | 0.24 (0.07–0.81) | 2.11 × 10−2 |
| Gly | 0.092 | 0.139 | 0.61 (0.48–0.77) | 3.38 × 10−5 | 0.55 (0.43–0.69) | 7.53 × 10−7 | HLA-DRB1*07 | 0.092 | 0.139 | 0.61 (0.48–0.77) | 3.38 × 10−5 |
| Leu | 0.145 | 0.115 | 1.32 (1.07–1.63) | 8.98 × 10−3 | HLA-DRB1*01 | 0.145 | 0.115 | 1.32 (1.07–1.63) | 8.98 × 10−3 | ||
| Pro | 0.216 | 0.153 | 1.48 (1.24–1.77) | 1.61 × 10−5 | HLA-DRB1*15 | 0.193 | 0.122 | 1.64 (1.36–1.98) | 2.87 × 10−7 | ||
| HLA-DRB1*16 | 0.023 | 0.031 | 0.75 (0.47–1.20) | 2.34 × 10−1 | |||||||
| Ser | 0.451 | 0.431 | 1.11 (0.96–1.28) | 1.52 × 10−1 | HLA-DRB1*03 | 0.102 | 0.107 | 0.93 (0.74–1.16) | 5.05 × 10−1 | ||
| HLA-DRB1*08 | 0.027 | 0.029 | 0.93 (0.61–1.43) | 7.49 × 10−1 | |||||||
| HLA-DRB1*11 # | 0.155 | 0.130 | 1.30 (1.06–1.60) | 1.11 × 10−2 | |||||||
| HLA-DRB1*12 | 0.026 | 0.018 | 1.59 (0.97–2.61) | 6.57 × 10−2 | |||||||
| HLA-DRB1*13 | 0.110 | 0.118 | 0.94 (0.75–1.18) | 6.09 × 10−1 | |||||||
| HLA-DRB1*14 | 0.031 | 0.030 | 1.02 (0.68–1.54) | 9.11 × 10−1 | |||||||
| Val | 0.093 | 0.151 | 0.55 (0.44–0.70) | 1.11 × 10−6 | 0.50 (0.40–0.64) | 2.14 × 10−8 | HLA-DRB1*04 | 0.092 | 0.138 | 0.62 (0.48–0.78) | 6.93 × 10−5 |
| HLA-DRB1*10 | 0.001 | 0.014 | 0.06 (0.01–0.46) | 6.99 × 10−3 | |||||||
All HLA-DRB1*11 alleles are associated with serine at HLA-DRβ1 amino acid position 11, except HLA-DRB1*11:22 and HLA-DRB1*11:30, which are associated with valine and leucine, respectively.
Among 28 imputed classical HLA-DRB1 alleles tested at four-digit resolution, three were significantly associated with UC (DRB1*15:01, OR = 1.59 [1.31–1.93, 95% CI], P = 3.68×10−6; DRB1*01:03, OR = 38.39 [7.50–196.60, 95% CI], P = 1.20×10−5; DRB1*07:01, OR = 0.61 [0.48–0.77, 95% CI] P = 3.38×10−5).
Because the above findings highlighted HLA-DRB1 association in UC, we then evaluated the quality of our classical HLA-DRB1 allele imputation at two-digit resolution by performing HLA-DRB1 genotyping via SSO probes and also next-generation sequencing using genomic DNA from 384 of our study subjects. This analysis demonstrated that the imputation procedure we applied was 98.8% accurate (see Supplementary Materials).
We next determined the most parsimonious model to explain the association of HLA-DRβ1 amino acid 11 with UC using forward stepwise model selection for the six observed amino acids. The best model included only three of the six amino acids: valine, glycine and aspartic acid. The overall P value for this best model was 3.60×10−13 as compared to a P value of 2.68×10−13 for the full model that included all six amino acid alleles, suggesting that most of the association signal for UC at this position can be accounted for by only these three amino acids. Of note, valine, glycine and aspartic acid are the same three amino acids encoded by the most significant SNP allele, rs9269955-C, when it is combined with the adjacent rs17878703 SNP alleles. This provides good internal validation between these different analytic approaches and highlights that variation at HLA-DRβ1 amino acid 11 explains much of the HLA association with UC.
UC versus control best multivariate model
When we performed analyses conditioned on including either rs9269955-C or the HLA-DRβ1 amino acid 11 variants, there were residual UC versus control association signals due to effects of other variants in the HLA region. This finding is consistent with prior observations in UC that multiple independent association signals exist in the MHC. We used a forward stepwise model selection procedure to select the best set of markers to predict UC (Table 4). This best model has an overall P value of 4.28×10−40 and includes rs9269955-C and 13 other markers that span the chromosome 6 region from 29.45 to 33.81 Mb.
Table 4.
Ulcerative colitis versus control association for MHC marker terms in best model identified with stepwise regression.
| Marker | Chromosome 6 position (NCBI36/hg18) | Gene | A1 | A2 | Frequency (UC) | Frequency (controls) | Univariate | Multivariate | ||
|---|---|---|---|---|---|---|---|---|---|---|
| P value | OR (95% CI) | P value | OR (95% CI) | |||||||
| rs9269955-C | 32,660,116 | HLA-DRB1 | Absent | Present | 0.812 | 0.700 | 2.67 × 10−13 | 1.95 (1.63–2.33) | 9.07 × 10−4 | 5.97 (2.08–17.17) |
| rs1049414 | 33,056,585 | BRD2 | A | G | 0.730 | 0.678 | 3.51 × 10−5 | 1.43 (1.21–1.69) | 1.84 × 10−5 | 1.53 (1.26–1.85) |
| rs440454 | 32,035,321 | SKIV2L | A | G | 0.339 | 0.247 | 1.57 × 10−7 | 1.51 (1.29–1.76) | 2.38 × 10−8 | 2.35 (1.74–3.17) |
| rs9273363 | 32,734,250 | HLA-DQA1/HLA-DQB1 | C | A | 0.835 | 0.752 | 1.44 × 10−8 | 1.71 (1.42–2.06) | 1.55 × 10−8 | 2.15 (1.65–2.81) |
| rs2844677 | 31,063,338 | MUC21 | G | A | 0.965 | 0.930 | 3.60 × 10−6 | 2.36 (1.64–3.39) | 2.01 × 10−3 | 1.83 (1.25–2.69) |
| rs1136759-T | 32,660,109 | HLA-DRB1 | Present | Absent | 0.184 | 0.276 | 6.35 × 10−10 | 0.57 (0.47–0.68) | 6.52 × 10−3 | 4.39 (1.51–12.76) |
| rs915654 | 31,646,476 | NFKBIL1/LTA | A | T | 0.382 | 0.330 | 4.70 × 10−4 | 1.31 (1.13–1.52) | 6.69 × 10−6 | 1.49 (1.25–1.77) |
| rs28435656 | 31,988,616 | C2 | G | A | 0.787 | 0.843 | 1.27 × 10−4 | 0.71 (0.59–0.84) | 6.99 × 10−6 | 2.27 (1.59–3.24) |
| rs7772982 | 29,448,986 | OR5V1/OR12D3 | C | T | 0.214 | 0.179 | 3.45 × 10−3 | 1.31 (1.09–1.57) | 4.65 × 10−4 | 1.41 (1.16–1.72) |
| rs3135391 | 32,518,965 | HLA-DRA | A | G | 0.181 | 0.115 | 1.18 × 10−6 | 1.61 (1.33–1.96) | 8.12 × 10−6 | 1.95 (1.45–2.61) |
| rs1130380-C | 32,740,672 | HLA-DQB1 | Absent | Present | 0.495 | 0.562 | 4.04 × 10−4 | 0.78 (0.67–0.89) | 2.85 × 10−4 | 1.47 (1.19–1.80) |
| rs6933763 | 32,830,830 | HLA-DQA2/HLA-DQB2 | G | A | 0.133 | 0.085 | 4.32 × 10−7 | 1.81 (1.44–2.29) | 3.41 × 10−4 | 1.61 (1.24–2.08) |
| rs9266196-C | 31,432,808 | HLA-B | Present | Absent | 0.374 | 0.329 | 4.72 × 10−3 | 1.23 (1.07–1.43) | 1.93 × 10−3 | 1.35 (1.12–1.64) |
| rs6457740 | 33,805,103 | IP6K3 | G | A | 0.755 | 0.710 | 5.86 × 10−3 | 1.25 (1.07–1.47) | 4.82 × 10−3 | 1.28 (1.08–1.53) |
Markers are listed according to the order in which they came into the model. The frequencies and odds ratios are given for the A1 allele. For markers with more than two alleles, presence or absence of the specified allele was compared. The reference sequence gene is listed for intragenic markers and the two flanking reference sequence genes are listed for intergenic markers. A1, allele 1; A2, allele 2; OR, odds ratio; CI, confidence interval; A, adenine; C, cytosine; G, guanine; T, thymine.
UC versus CD with ileal involvement best multivariate model
In order to compare HLA associations between UC and CD with ileal involvement, we performed an analysis using UC subjects as cases and CD with ileal involvement subjects as controls. Initial association analyses for all markers in our study were performed and then we applied stepwise model selection to determine the best model for a UC versus CD with ileal involvement comparison (Table 5A). The model that was selected included 11 markers and had an overall model P value of 4.48×10−33. Not unexpectedly, there was no overlap between these markers and those that were chosen in the UC versus control best model described above (Table 4).
Table 5.
Best model association results for ulcerative colitis versus Crohn’s disease with ileal involvement (Table 5A), ulcerative colitis versus control using markers from 5A (Table 5B) and Crohn’s disease with ileal involvement versus control using markers from 5A (Table 5C) for MHC marker terms identified with stepwise regression.
|
Table 5A. Ulcerative colitis versus Crohn’s disease with ileal involvement.
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Chromosome 6 position (NCBI36/hg18) | Gene(s) | A1 | A2 | Frequency (UC) | Frequency (Ileal CD) | Univariate | Multivariate | ||
| P value | OR (95% CI) | P value | OR (95% CI) | |||||||
| rs2647025 | 32743927 | HLA-DQB1/HLA-DQA2 | G | A | 0.836 | 0.682 | 2.00 × 10−16 | 2.35 (1.92–2.88) | 2.84 × 10−13 | 2.24 (1.80–2.78) |
| rs16899682 | 31551678 | HCG26/MICB | C | G | 0.031 | 0.014 | 5.09 × 10−3 | 2.34 (1.29–4.23) | 5.27 × 10−4 | 3.14 (1.64–6.00) |
| rs2257269 | 31431332 | HLA-B | G | A | 0.634 | 0.541 | 6.66 × 10−6 | 1.48 (1.25–1.75) | 1.88 × 10−4 | 1.43 (1.19–1.73) |
| rs41544112 | 32737898 | HLA-DQB1 | C | T | 0.966 | 0.939 | 3.93 × 10−3 | 1.81 (1.21–2.72) | 7.52 × 10−4 | 2.12 (1.37–3.27) |
| rs3130609 | 33097499 | HLA-DOA/HLA-DPA1 | C | T | 0.984 | 0.949 | 1.86 × 10−5 | 3.25 (1.89–5.56) | 2.05 × 10−3 | 2.45 (1.39–4.34) |
| rs16899168 | 31366666 | HLA-C/HLA-B | G | A | 0.977 | 0.956 | 7.83 × 10−3 | 1.93 (1.19–3.12) | 8.02 × 10−3 | 2.02 (1.20–3.39) |
| rs210134 | 33648187 | ZBTB9/BAK1 | G | A | 0.731 | 0.678 | 3.39 × 10−3 | 1.31 (1.09–1.57) | 9.68 × 10−4 | 1.39 (1.14–1.68) |
| HLA-B, amino acid 99-Y | 31432174 | HLA-B | Present | Absent | 0.994 | 0.977 | 2.12 × 10−3 | 4.01 (1.65–9.74) | 5.29 × 10−3 | 3.77 (1.48–9.57) |
| rs3130559 | 31205280 | PSORS1C1 | C | T | 0.819 | 0.750 | 4.45 × 10−5 | 1.53 (1.25–1.87) | 4.43 × 10−3 | 1.38 (1.11–1.72) |
| rs2256974 | 31663371 | LST1 | A | C | 0.201 | 0.151 | 1.52 × 10−3 | 1.42 (1.14–1.76) | 1.23 × 10−3 | 1.48 (1.17–1.89) |
| rs3135365 | 32497233 | BTNL2/HLA-DRA | C | A | 0.237 | 0.186 | 4.87 × 10−3 | 1.32 (1.09–1.61) | 2.90 × 10−3 | 1.40 (1.12–1.74) |
| Table 5B. Ulcerative colitis versus control.
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Chromosome 6 position (NCBI36/hg18) | Gene(s) | A1 | A2 | Frequency (UC) | Frequency (controls) | Univariate | Multivariate | ||
| P value | OR (95% CI) | P value | OR (95% CI) | |||||||
| rs2647025 | 32743927 | HLA-DQB1/HLA-DQA2 | G | A | 0.836 | 0.730 | 1.94 × 10−12 | 1.95 (1.62–2.35) | 1.95 × 10−10 | 1.88 (1.55–2.29) |
| rs16899682 | 31551678 | HCG26/MICB | C | G | 0.031 | 0.018 | 2.63 × 10−2 | 1.67 (1.06–2.62) | 1.29 × 10−3 | 2.20 (1.36–3.56) |
| rs2257269 | 31431332 | HLA-B | G | A | 0.634 | 0.575 | 8.59 × 10−4 | 1.29 (1.11–1.49) | 1.59 × 10−2 | 1.22 (1.04–1.43) |
| rs41544112 | 32737898 | HLA-DQB1 | C | T | 0.966 | 0.958 | 2.66 × 10−1 | 1.24 (0.85–1.82) | 1.58 × 10−1 | 1.33 (0.90–1.97) |
| rs3130609 | 33097499 | HLA-DOA/HLA-DPA1 | C | T | 0.984 | 0.963 | 4.89 × 10−4 | 2.50 (1.49–4.18) | 8.72 × 10−3 | 2.04 (1.20–3.47) |
| rs16899168 | 31366666 | HLA-C/HLA-B | G | A | 0.977 | 0.966 | 8.06 × 10−2 | 1.50 (0.95–2.36) | 5.99 × 10−2 | 1.56 (0.98–2.49) |
| rs210134 | 33648187 | ZBTB9/BAK1 | G | A | 0.731 | 0.713 | 2.09 × 10−1 | 1.11 (0.94–1.30) | 8.98 × 10−2 | 1.15 (0.98–1.36) |
| HLA-B, amino acid 99-Y | 31432174 | HLA-B | Present | Absent | 0.994 | 0.983 | 5.29 × 10−3 | 3.38 (1.44–7.97) | 5.31 × 10−3 | 3.47 (1.45–8.32) |
| rs3130559 | 31205280 | PSORS1C1 | C | T | 0.819 | 0.800 | 1.60 × 10−1 | 1.14 (0.95–1.37) | 4.35 × 10−1 | 1.08 (0.89–1.31) |
| rs2256974 | 31663371 | LST1 | A | C | 0.201 | 0.164 | 3.26 × 10−3 | 1.31 (1.09–1.57) | 9.22 × 10−3 | 1.29 (1.06–1.56) |
| rs3135365 | 32497233 | BTNL2/HLA-DRA | C | A | 0.237 | 0.183 | 1.89 × 10−3 | 1.31 (1.10–1.55) | 2.55 × 10−4 | 1.40 (1.17–1.67) |
| Table 5C. Crohn’s disease with ileal involvement versus control.
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Chromosome 6 position (NCBI36/hg18) | Gene(s) | A1 | A2 | Frequency (Ileal CD) | Frequency (controls) | Univariate | Multivariate | ||
| P value | OR (95% CI) | P value | OR (95% CI) | |||||||
| rs2647025 | 32743927 | HLA-DQB1/HLA-DQA2 | G | A | 0.682 | 0.730 | 2.54 × 10−3 | 0.79 (0.68–0.92) | 7.86 × 10−3 | 0.81 (0.69–0.94) |
| rs16899682 | 31551678 | HCG26/MICB | C | G | 0.014 | 0.018 | 2.53 × 10−1 | 0.72 (0.41–1.27) | 1.57 × 10−1 | 0.66 (0.37–1.17) |
| rs2257269 | 31431332 | HLA-B | G | A | 0.541 | 0.575 | 4.53 × 10−2 | 0.87 (0.75–1.00) | 9.01 × 10−2 | 0.88 (0.76–1.02) |
| rs41544112 | 32737898 | HLA-DQB1 | C | T | 0.939 | 0.958 | 7.67 × 10−3 | 0.66 (0.49–0.90) | 6.42 × 10−3 | 0.65 (0.47–0.88) |
| rs3130609 | 33097499 | HLA-DOA/HLA-DPA1 | C | T | 0.949 | 0.963 | 7.74 × 10−2 | 0.74 (0.53–1.03) | 2.76 × 10−1 | 0.83 (0.58–1.17) |
| rs16899168 | 31366666 | HLA-C/HLA-B | G | A | 0.956 | 0.966 | 1.03 × 10−1 | 0.75 (0.52–1.06) | 4.19 × 10−2 | 0.69 (0.48–0.99) |
| rs210134 | 33648187 | ZBTB9/BAK1 | G | A | 0.678 | 0.713 | 4.79 × 10−2 | 0.86 (0.74–1.00) | 3.85 × 10−2 | 0.85 (0.73–0.99) |
| HLA-B, amino acid 99-Y | 31432174 | HLA-B | Present | Absent | 0.977 | 0.983 | 2.84 × 10−1 | 0.77 (0.47–1.25) | 2.24 × 10−1 | 0.73 (0.45–1.21) |
| rs3130559 | 31205280 | PSORS1C1 | C | T | 0.750 | 0.800 | 4.98 × 10−4 | 0.75 (0.64–0.88) | 4.24 × 10−3 | 0.78 (0.66–0.93) |
| rs2256974 | 31663371 | LST1 | A | C | 0.151 | 0.164 | 4.53 × 10−1 | 0.93 (0.77–1.12) | 4.32 × 10−1 | 0.92 (0.76–1.13) |
| rs3135365 | 32497233 | BTNL2/HLA-DRA | C | A | 0.186 | 0.183 | 7.93 × 10−1 | 0.98 (0.82–1.17) | 6.01 × 10−1 | 0.95 (0.79–1.15) |
Markers are listed according to the order in which they came into the model. The frequencies and odds ratios are given for the A1 allele. For markers with more than two alleles, presence or absence of the specified allele was compared. The reference sequence gene is listed for intragenic markers and the two flanking reference sequence genes are listed for intergenic markers. A1, allele 1; A2, allele 2; UC, ulcerative colitis; Ileal CD, Crohn’s disease with ileal involvement; OR, odds ratio; CI, confidence interval; A, adenine; C, cytosine; G, guanine; T, thymine; Y, tyrosine.
We then used the 11 markers from the UC versus CD with ileal involvement best model to perform two further analyses: UC versus control and CD with ileal involvement versus control (Tables 5B and 5C). The model P value for UC versus control was 1.59×10−19 which is less significant than the P value of 4.28×10−40 for the unrestricted UC best model (Table 4). The model P value for CD with ileal involvement versus control was 1.42×10−5. Divergent effects for each UC versus CD with ileal involvement best model marker in the UC versus control compared to the CD with ileal involvement versus control analyses are apparent when the odds ratios for each marker are compared.
Discussion
The MHC locus demonstrates the strongest evidence for association to UC among 47 well-established UC loci identified in a GWAS meta-analysis10, and is also one of 71 well-established CD loci identified by GWAS meta-analysis.9 In order to better understand MHC association signals in UC and CD, we used dense MHC SNP data from the discovery stage of an ongoing, new UC and CD GWAS to impute classical HLA types, their constituent SNPs and corresponding amino acids, and we performed detailed analyses of the genotyped and imputed data.
Our univariate tests of binary SNP and SNP allele markers, and our omnibus tests of polymorphic HLA amino acid positions both highlighted HLA-DRβ1, amino acid position 11 as the MHC feature most significantly associated with UC. The C allele of rs9269955 was the SNP allele most significantly associated with UC (presence of rs9269955-C is associated with protection and absence is associated with risk for UC). In combination with the immediately adjacent SNP, it encodes the valine, glycine or aspartic acid amino acid residues at HLA-DRβ1, amino acid 11, which were all associated with protection against UC. Furthermore, in multivariate analysis, the most parsimonious model to explain the association with UC at amino acid 11 consisted of valine, glycine and aspartic acid as the only terms.
HLA-DRB1 has extensive polymorphism as demonstrated by its 928 alleles and the 704 proteins for which it codes (International Immunogenetics Information System/HLA Database: http://www.ebi.ac.uk/imgt/hla)13. Valine at amino acid 11 corresponds to the common DRB1*04 (DR4) or lower frequency DRB1*10 (DR10) allele groups, glycine to DRB1*07 (DR7), and aspartic acid to DRB1*09 (DR9). The HLA-DR4, -DR7 and -DR9 allele groups were associated with protection against UC in a meta-analysis of prior studies.3 They almost always occur on haplotypes carrying the HLA-DRB4 gene which encodes the DR53 antigen, and HLA-DRB4*01:01 has been associated with protection against UC in Japan.14 In addition, the previously reported HLA-DR2 association with risk for UC3, 5 is consistent with our observation that proline at position 11 in HLA-DRβ1 is associated with risk for UC. Based on the complementary findings from our different analyses and their correlation with results from prior studies, we conclude that variation at amino acid position 11 of HLA-DRβ1 is a major determinant of chromosome 6p association with ulcerative colitis.
The potential biological significance of the UC association of amino acid position 11 relates to the peptide binding specificity of HLA class II molecules and their role in antigen presentation to T cells.15, 16 The three-dimensional structure of the class II molecule HLA-DR1 heterodimer (DRA/DRB1*0101) has been well characterized and its peptide binding groove has been shown to be determined by polymorphic molecules that form nine pockets with different chemical and size characteristics.15, 17 In one of these pockets (P6), amino acid position 11 appears to be the only variable residue and thus determines the binding specificity of that pocket.18 Of note, hydrophobic amino acid residues at DRβ1 amino acid 11 were found to be associated with protection against development of sarcoidosis.19 This finding suggests that such hydrophobic interactions could affect peptide binding in the P6 pocket.19 We therefore hypothesize that variation at the amino acid position 11 of HLA-DRβ1 could have an effect on peptide binding in the HLA-DR complex antigen binding cleft that alters risk for the development of UC.
It is important to note that the MHC association signal in UC is complex and not completely explained by amino acid position 11 in HLA-DRβ1. In fact, our forward stepwise model selection identified 13 other terms besides rs9269955-C. This model is highly significant with an overall P value of 4.28×10−40, but it will need to be validated in additional large cohorts.
Included in our model was another missense SNP allele in HLA-DRB1, the T allele of rs1136759. rs1136759 and two adjacent flanking SNPs encode variation at HLA-DRβ1, amino acid 13, which is located in the P4 pocket of the HLA-DR complex antigen binding cleft. The finding that two of the terms in the best model for prediction of UC risk relate to the HLA-DRβ1 complex antigen binding cleft emphasizes the probable importance of HLA-DRB1 in the pathogenesis of UC. Four other MHC class II loci variants, including SNPs in HLA-DQB1 (rs1130380-C) and HLA-DRA (rs3135391), between HLA-DQA1 and HLA-DQB1 (rs9273363), and between HLA-DQA2 and HLA-DQB2 (rs6933763), were associated with UC in our multivariate model. The HLA-DRB, -DQB and -DPB genes are all highly polymorphic and encode β-chains of the class II molecule αβ heterodimer while the α-chains are encoded by the HLA-DQA, -DPA genes and -DRA genes.4
Three polymorphisms in MHC class III loci (rs440454, rs28435656, and rs915654) were included as terms in our UC versus control model. The MHC class III region is one of the most gene dense regions in the human genome. Two of the SNPs in our model, rs440454 and rs28435656, are in linkage disequilibrium (r2 = 0.54 in HapMap 3-CEU11) and located in an MHC class III segment that contains four genes within 30 kb including superkiller viralicidic activity 2-like (SKIV2L) and RD RNA binding protein (RDBP).20 rs440454 is in perfect linkage disequilibrium (r2 = 1.0 in HapMap 3-CEU11) with SNP rs419788 that was associated with risk for lupus.21 rs28435656 is located in the complement component 2 (C2) gene which is located immediately adjacent to the region that includes SKIV2L and RDBP. Finally, rs915654 is located 5 prime to the lymphotoxin A (LTA) locus which has been associated with CD and diabetes.22 All these findings suggest a role for MHC class III genes in UC pathogenesis which warrants further investigation.
Another association of potential pathogenic interest identified in our UC versus control model is rs2844677, a synonymous SNP in the coding region of the mucin 21, cell surface associated (MUC21) gene. MUC21 is a recently identified gene that is expressed in normal colon among other tissues and produces a transmembrane mucin involved in cell adhesion.23, 24
In the last part of our analysis, we compared MHC region association signals between UC and CD with ileal involvement. The finding that the 11 studied markers each had odds ratios with effects in opposite directions for the two IBD phenotypes together with the results from our initial association analysis in which the most significant associations in UC were different than those for ileal CD, demonstrates that the association signals for UC and ileal CD are quite different. This conclusion correlates with results of prior studies which have shown that the only consistent associations with risk for both UC and CD have been for HLA-DRB1*01:03 and HLA-B52.3, 4 In contrast, alleles of the HLA-DR2 split antigen DR15 have been associated in opposite directions with HLA-DRB1*15:01 associated with protection against CD and HLA-DRB1*15:02 associated with increased risk for UC.3, 5
In summary, we have performed detailed analyses to better understand MHC association signals in UC and CD. Our most significant finding is that a specific variation at amino acid position 11 of HLA-DRβ1, the only variable amino acid in the P6 pocket of the HLA-DR complex antigen binding cleft, explains a substantial portion of the MHC association signal and corresponds with several previously established classical HLA class II associations in UC. The observed alteration at amino acid position 11 of HLA-DRβ1 may affect peptide binding and result in an altered immune activation underlying protection against UC. We have also developed a novel multivariate model that further defines the contribution of MHC variation to risk for UC and highlights other genes of potential importance in UC pathogenesis. Finally, our multivariate modeling suggests different effects of MHC polymorphisms in UC and CD.
Materials and Methods
Study subjects
Our study sample included 574 UC, 630 CD with at least ileal involvement, and 1,508 control subjects of European ancestry that were recruited for genetic studies at the Cleveland Clinic or the University of Pittsburgh under institutional review board-approved protocols. All subjects provided written informed consent. IBD diagnoses and assessment of disease location were confirmed by IBD physicians via review of primary medical records using standard endoscopic, radiographic and histologic criteria.
Genotyping and quality control
Study subjects were genotyped using the Illumina Omni1-quad BeadChip (Illumina, San Diego, CA) at the Feinstein Institute for Medical Research of the North Shore-Long Island Jewish Health System. Data from samples with preliminary genotype call rates > 0.98 using cluster positions provided by Illumina were reclustered using the Illumina GenomeStudio software, and the new cluster positions were applied to all samples. Initial quality control of the genotyping data included removal of one sample from each pair with estimated identity-by-descent proportion > 0.10, removal of samples with genotype missing rates > 0.05, or with discordant SNP-determined and reported gender or ambiguous SNP-determined gender, and removal of SNPs with genotype missing rates > 0.05, minor allele frequencies in controls < 0.005, or Hardy-Weinberg P values in controls < 1×10−6. These quality control steps were performed using the PLINK software.25 Subsequently, tag SNPs with genotype missing rates < 0.1% and physical separation of at least 0.4 megabases (Mb) were used in spectral analysis of ancestry that identified 929 controls with a relatively homogenous ‘European’ ancestral background. Additional SNPs with minor allele frequencies < 0.005 or Hardy-Weinberg P values < 0.001 in these 929 controls were removed from the dataset.
Ancestry matching
To control for potential confounding due to variation in genetic ancestry, study subjects were grouped into 11 approximately homogenous clusters, based on genetic distances derived from GemTools.26, 27 Ancestry was inferred based on SNPs with genotype missing rates < 0.1% and a physical separation of at least 0.2 Mb. In all of the association analyses, we controlled for ancestry by including cluster membership as a blocking variable. The inflation across the genome-wide SNP data was minimal (genomic control lambda28 = 1.02 for UC vs. control and 1.03 for CD with ileal involvement vs. control), confirming that the samples were well matched.
Imputation of classical HLA, SNP, and amino acid allele dosages
We followed a previously described procedure29 to impute classical HLA alleles and their corresponding amino acid sequences in our cases and controls, using the genotyped SNPs in our GWAS as input. This imputation procedure is conceptually similar to HLA*IMP32 in that haplotype information across the region is used to predict classical HLA alleles based on genotyped SNPs. A prior study demonstrated empirical evidence that the imputations have good accuracy29 reaching comparable levels of accuracy to the work on which HLA*IMP is based.32
As the reference panel, we used a data set of 263 HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 classical alleles at four-digit resolution, 3,852 SNPs, and 372 amino acid positions in 2,767 unrelated founder individuals of European descent collected by the MHC Working Group of the Type 1 Diabetes Genetics Consortium.30 All variants were encoded as biallelic markers, allowing us to use standard tools for imputation. For variants with greater than two alleles, each allele was coded as present or absent, and analyzed in a separate test. We used default parameters for BEAGLE (http://faculty.washington.edu/browning/beagle/beagle.html): ten iterations of phasing/imputation, testing four pairs of haplotype pairs for each individual at each iteration. For each variant, we used the posterior probabilities of carrying 0 (AA), 1 (AB) or 2 (BB) copies to calculate the effective dosage for allele B (=2xPr(BB) + Pr(AB)). To obtain allele dosages for MHC region Omni1-quad SNPs, we used BEAGLECALL.31 Three iterations of BEAGLECALL were run, with increasing stringency of genotype calling filters (callthreshold=0.9 and missingcohort=0.1 in iteration 1, callthreshold=0.98 and missingcohort=0.02 in iteration 2, and callthreshold=0.985 and missingcohort=0.015 in iteration 3). We combined dosage information for markers in the Type 1 Diabetes Genetics Consortium reference panel with dosage information for additional Omni1-quad SNPs that appeared in both genome builds NCBI36/hg18 and GRCh37/hg19 into a combined set of genetic features in the MHC region from 29,299 to 33,884 kilobases (kb) on chromosome 6 using NCBI36/hg18 coordinates.
HLA-DRB1 imputation quality at two-digit resolution was assessed by sequence-specific oligonucleotide (SSO) probes and next-generation sequencing of genomic DNA collected from 384 of our study subjects (see Supplementary Materials).
Association analyses
Association analyses were performed using allele dosage data from 562 UC, 611 CD with ileal involvement, and 1,428 control samples that passed quality control. We examined the association between binary markers in the HLA region and UC versus control and CD with ileal involvement versus control using logistic regression with a log-additive model. Forward stepwise model selection was used to determine a set of markers in the post imputation data that jointly predicted disease versus control status, without including multiple markers that were in tight linkage disequilibrium. Markers with an allele frequency < 0.001 were excluded. The Bayesian Information Criterion (BIC) was used to find a model that balanced model complexity with parsimony. The stepwise procedure started by taking the best marker (lowest P value) into the regression model and iteratively adding markers until the BIC ceased to improve. This procedure was performed in R (http://www.r-project.org) using the “glm” and “step” functions.
For each polymorphic amino acid position in the HLA region we also conducted an omnibus test for association using multivariate logistic regression with degrees-of-freedom equal to the number of distinct residues for that amino acid position minus one. For the position yielding the smallest P value we used stepwise regression, limited to that position, to select a parsimonious model for the site.
Finally, using stepwise regression we determined a model for differentiating UC and CD with ileal involvement. In this model, CD with ileal involvement subjects served as controls and UC subjects served as cases.
For each multivariate model, we provide the P value associated with the best model. This P value pertains to the null hypothesis that none of the terms in the model has any explanatory value, versus the alternative hypothesis that at least one term is associated with the phenotype. The degrees-of-freedom associated with this test equals the number of markers in the multivariate model.
Supplementary Material
Acknowledgments
Support: Supported by the National Institutes of Health grants DK068112 (J-PA), AG030653 (MIK), MH057881 (BD and KR), DK062420 (RHD) and DK076025 (RHD); a Crohn’s & Colitis Foundation of America Senior Research Award (RHD); Department of Defense grant W81XWH-07-1-0619 (MT); and funds generously provided by Kenneth and Jennifer Rainin, Gerald and Nancy Goldberg, and Victor and Ellen Cohn.
The authors would like to acknowledge Leonard Baidoo, MD and David Binion, MD for providing phenotypic information for some of the study subjects, the Feinstein Institute for Medical Research of the North Shore-Long Island Jewish Health System for Illumina Genotyping BeadChip processing, and the University of Pittsburgh Genomics and Proteomics Core Laboratories for HLA-DRB1 sequencing technical assistance.
Abbreviations
- BIC
Bayesian Information Criterion
- CI
confidence interval
- C2
complement component 2
- GWAS
genome-wide association study
- HLA
human leukocyte antigen
- kb
kilobases
- LTA
lymphotoxin A
- Mb
megabases
- MHC
major histocompatibility complex
- MICA
MHC class I polypeptide-related sequence A
- MUC21
mucin 21 cell surface associated
- NCBI
National Center for Biotechnology Information
- OR
odds ratio
- RDBP
RD RNA binding protein
- SKIV2L
superkiller viralicidic activity 2-like
- SNP
single nucleotide polymorphism
- SSO
sequence-specific oligonucleotide
Footnotes
Conflict of interests: The authors declare no conflict of interest.
References
- 1.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 2.Traherne JA. Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet. 2008;35:179–92. doi: 10.1111/j.1744-313X.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassinotti A, Birindelli S, Clerici M, Trabattoni D, Lazzaroni M, Ardizzone S, et al. HLA and autoimmune digestive disease: a clinically oriented review for gastroenterologists. Am J Gastroenterol. 2009;104:195–217. doi: 10.1038/ajg.2008.10. [DOI] [PubMed] [Google Scholar]
- 5.Stokkers PC, Reitsma PH, Tytgat GN, van Deventer SJ. HLA-DR and -DQ phenotypes in inflammatory bowel disease: a meta-analysis. Gut. 1999;45:395–401. doi: 10.1136/gut.45.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hampe J, Schreiber S, Shaw SH, Lau KF, Bridger S, Macpherson AJ, et al. A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am J Hum Genet. 1999;64:808–16. doi: 10.1086/302294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hampe J, Shaw SH, Saiz R, Leysens N, Lantermann A, Mascheretti S, et al. Linkage of inflammatory bowel disease to human chromosome 6p. Am J Hum Genet. 1999;65:1647–55. doi: 10.1086/302677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Heel DA, Fisher SA, Kirby A, Daly MJ, Rioux JD, Lewis CM, et al. Inflammatory bowel disease susceptibility loci defined by genome scan meta- analysis of 1952 affected relative pairs. Hum Mol Genet. 2004;13:763–70. doi: 10.1093/hmg/ddh090. [DOI] [PubMed] [Google Scholar]
- 9.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome- wide association study. Nature Genet. 2009;41:216–20. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–6. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshitake S, Kimura A, Okada M, Yao T, Sasazuki T. HLA class II alleles in Japanese patients with inflammatory bowel disease. Tissue Antigens. 1999;53:350–8. doi: 10.1034/j.1399-0039.1999.530405.x. [DOI] [PubMed] [Google Scholar]
- 15.Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–9. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 16.Janeway C, Travers P, Walport M, Shlomchik M. Immunobiology: The Immune System in Health and Disease. 6. Garland Science; New York: 2005. Antigen recognition by B-cell and T-cell receptors; pp. 103–134. [Google Scholar]
- 17.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–21. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 18.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–61. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 19.Foley PJ, McGrath DS, Puscinska E, Petrek M, Kolek V, Drabek J, et al. Human leukocyte antigen-DRB1 position 11 residues are a common protective marker for sarcoidosis. Am J Respir Cell Mol Biol. 2001;25:272–7. doi: 10.1165/ajrcmb.25.3.4261. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Shen L, Dangel AW, Wu LC, Yu CY. Four ubiquitously expressed genes, RD (D6S45)-SKI2W (SKIV2L)-DOM3Z-RP1 (D6S60E), are present between complement component genes factor B and C4 in the class III region of the HLA. Genomics. 1998;53:338–47. doi: 10.1006/geno.1998.5499. [DOI] [PubMed] [Google Scholar]
- 21.Fernando MM, Stevens CR, Sabeti PC, Walsh EC, McWhinnie AJ, Shah A, et al. Identification of two independent risk factors for lupus within the MHC in United Kingdom families. PLoS Genet. 2007;3:e192. doi: 10.1371/journal.pgen.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valdes AM, Thomson G, Barcellos LF. Genetic variation within the HLA class III influences T1D susceptibility conferred by high-risk HLA haplotypes. Genes Immun. 2010;11:209–18. doi: 10.1038/gene.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi Y, Kamata-Sakurai M, Denda-Nagai K, Itoh T, Okada K, Ishii-Schrade K, et al. Mucin 21/epiglycanin modulates cell adhesion. J Biol Chem. 2010;285:21233–40. doi: 10.1074/jbc.M109.082875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh Y, Kamata-Sakurai M, Denda-Nagai K, Nagai S, Tsuiji M, Ishii-Schrade K, et al. Identification and expression of human epiglycanin/MUC21: a novel transmembrane mucin. Glycobiology. 2008;18:74–83. doi: 10.1093/glycob/cwm118. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee AB, Luca D, Klei L, Devlin B, Roeder K. Discovering genetic ancestry using spectral graph theory. Genet Epidemiol. 2010;34:51–9. doi: 10.1002/gepi.20434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klei L, Kent B, Melhem N, Devlin B, Roeder K. GemTools: A fast and efficient approach to estimating genetic ancestry. 2011 http://arxiv.org/abs/1104.1162.
- 28.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 29.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–7. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown WM, Pierce J, Hilner JE, Perdue LH, Lohman K, Li L, et al. Overview of the MHC fine mapping data. Diabetes Obes Metab. 2009;11(Suppl 1):2–7. doi: 10.1111/j.1463-1326.2008.00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am J Hum Genet. 2009;85:847–61. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. American journal of human genetics. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.