Abstract
Subsequent to the discovery that RNA can have site specific cleavage activity, there has been a great deal of interest in the design and testing of trans-acting catalytic RNAs as both surrogate genetic tools and as therapeutic agents. We have been developing catalytic RNAs or ribozymes with target specificity for HIV-1 RNA and have been exploring chemical synthesis as one method for their production. To this end, we have chemically synthesized and experimentally analyzed chimeric catalysts consisting of DNA in the non-enzymatic portions, and RNA in the enzymatic core of hammerhead type ribozymes. Substitutions of DNA for RNA in the various stems of a hammerhead ribozyme have been analyzed in vitro for kinetic efficiency. One of the chimeric ribozymes used in this study, which harbors 24 bases of DNA capable of base-pairing interactions with an HIV-1 gag target, but maintains RNA in the catalytic center and in stem-loop II, has a sixfold greater kcat value than the all RNA counterpart. This increased activity appears to be the direct result of enhanced product dissociation. Interestingly, a chimeric ribozyme in which stem-loop II (which divides the catalytic core) is comprised of DNA, exhibited a marked reduction in cleavage activity, suggesting that DNA in this region of the ribozyme can impart a negative effect on the catalytic function of the ribozyme. DNA-RNA chimeric ribozymes transfected by cationic liposomes into human T-lymphocytes are more stable than their all-RNA counterparts. Enhanced catalytic turnover and stability in the absence of a significant effect on Km make chimeric ribozymes favorable candidates for therapeutic agents.
Full text
PDF


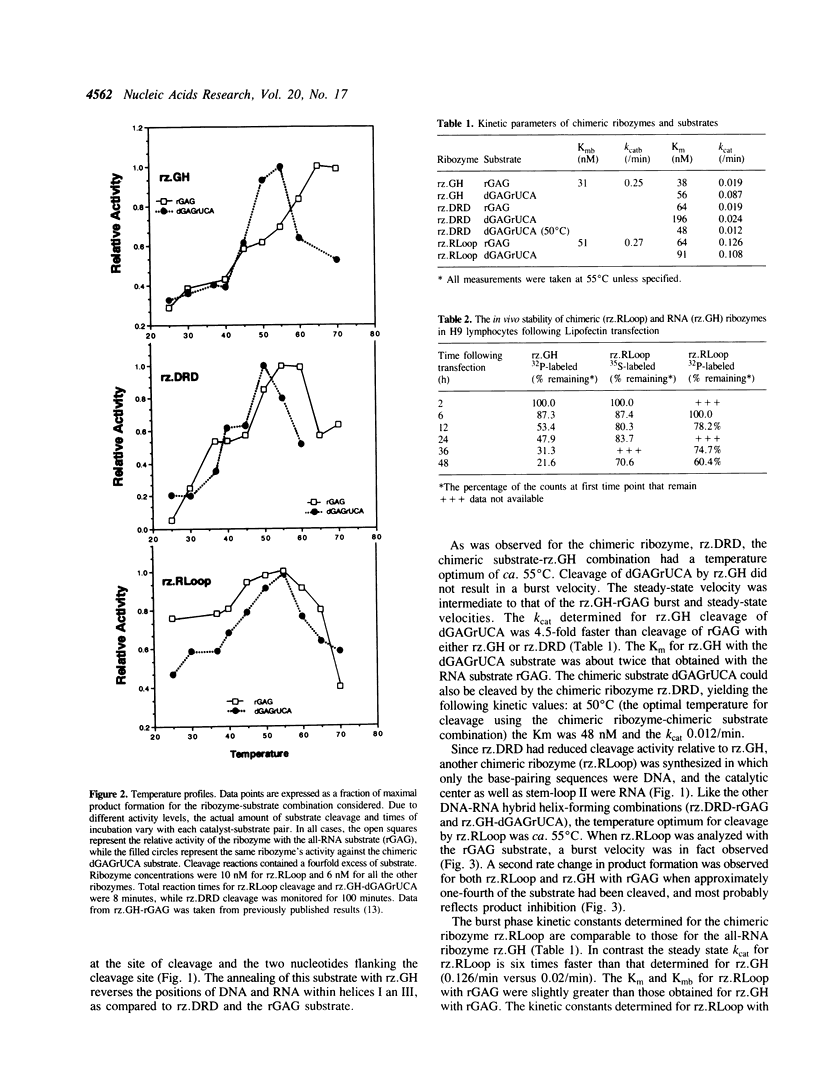
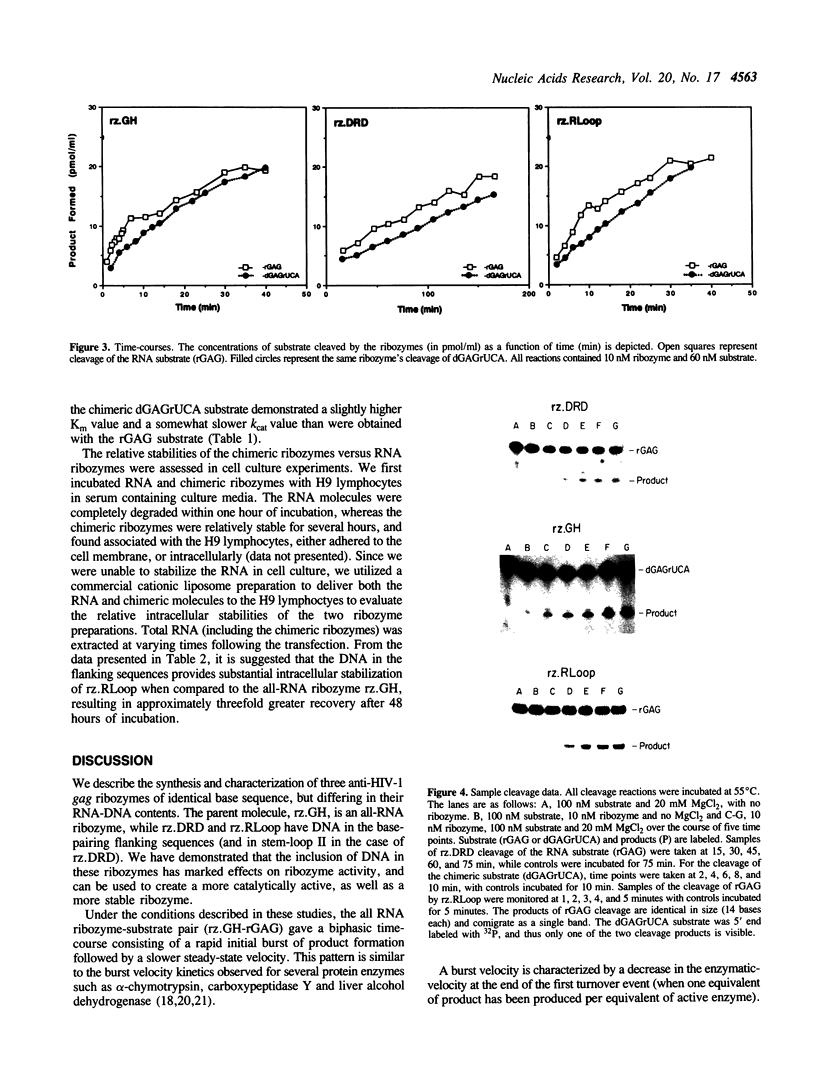


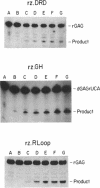
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender M. L., Kézdy F. J., Wedler F. C. Alpha-chymotrypsin: enzyme concentration and kinetics. J Chem Educ. 1967 Feb;44(2):84–88. doi: 10.1021/ed044p84. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Ribozymes and their medical implications. JAMA. 1988 Nov 25;260(20):3030–3034. [PubMed] [Google Scholar]
- Chou S. H., Flynn P., Reid B. High-resolution NMR study of a synthetic DNA-RNA hybrid dodecamer containing the consensus pribnow promoter sequence: d(CGTTATAATGCG).r(CGCAUUAUAACG). Biochemistry. 1989 Mar 21;28(6):2435–2443. doi: 10.1021/bi00432a014. [DOI] [PubMed] [Google Scholar]
- Cotten M. The in vivo application of ribozymes. Trends Biotechnol. 1990 Jul;8(7):174–178. doi: 10.1016/0167-7799(90)90168-w. [DOI] [PubMed] [Google Scholar]
- Dahm S. C., Uhlenbeck O. C. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991 Oct 1;30(39):9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- Douglas K. T., Nakagawa Y., Kaiser E. T. Mechanistic studies of carboxypeptidase Y. Kinetic detection of an acyl-enzyme intermediate in trimethylacetate esterase action. J Am Chem Soc. 1976 Dec 8;98(25):8231–8236. doi: 10.1021/ja00441a057. [DOI] [PubMed] [Google Scholar]
- Eisenthal R., Cornish-Bowden A. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem J. 1974 Jun;139(3):715–720. doi: 10.1042/bj1390715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Substrate sequence effects on "hammerhead" RNA catalytic efficiency. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Goodchild J., Kohli V. Ribozymes that cleave an RNA sequence from human immunodeficiency virus: the effect of flanking sequence on rate. Arch Biochem Biophys. 1991 Feb 1;284(2):386–391. doi: 10.1016/0003-9861(91)90313-8. [DOI] [PubMed] [Google Scholar]
- HOFSTEE B. H. Non-inverted versus inverted plots in enzyme kinetics. Nature. 1959 Oct 24;184:1296–1298. doi: 10.1038/1841296b0. [DOI] [PubMed] [Google Scholar]
- Hall K. B., McLaughlin L. W. Thermodynamic and structural properties of pentamer DNA.DNA, RNA.RNA, and DNA.RNA duplexes of identical sequence. Biochemistry. 1991 Nov 5;30(44):10606–10613. doi: 10.1021/bi00108a002. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Heidenreich O., Eckstein F. Hammerhead ribozyme-mediated cleavage of the long terminal repeat RNA of human immunodeficiency virus type 1. J Biol Chem. 1992 Jan 25;267(3):1904–1909. [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. DNA cleavage catalysed by the ribozyme from Tetrahymena. Nature. 1990 Mar 29;344(6265):405–409. doi: 10.1038/344405a0. [DOI] [PubMed] [Google Scholar]
- KEZDY F. J., BENDER M. L. The kinetics of the alpha-chymotrypsin-catalyzed hydrolysis of p-nitrophenyl acetate. Biochemistry. 1962 Nov;1:1097–1106. doi: 10.1021/bi00912a021. [DOI] [PubMed] [Google Scholar]
- Milman G., Langridge R., Chamberlin M. J. The structure of a DNA-RNA hybrid. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1804–1810. doi: 10.1073/pnas.57.6.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. W., Tinoco I., Jr Comparison of the kinetics of ribooligonucleotide, deoxyribooligonucleotide, and hybrid oligonucleotide double-strand formation by temperature-jump kinetics. Biochemistry. 1982 Oct 12;21(21):5289–5295. doi: 10.1021/bi00264a026. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Labuda D., Usman N., Yang J. H., Cedergren R. Relationship between 2'-hydroxyls and magnesium binding in the hammerhead RNA domain: a model for ribozyme catalysis. Biochemistry. 1991 Apr 23;30(16):4020–4025. doi: 10.1021/bi00230a029. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Wu T. F., Cousineau B., Ogilvie K. K., Cedergren R. Mixed deoxyribo- and ribo-oligonucleotides with catalytic activity. Nature. 1990 Apr 5;344(6266):565–567. doi: 10.1038/344565a0. [DOI] [PubMed] [Google Scholar]
- Perriman R., Delves A., Gerlach W. L. Extended target-site specificity for a hammerhead ribozyme. Gene. 1992 Apr 15;113(2):157–163. doi: 10.1016/0378-1119(92)90391-2. [DOI] [PubMed] [Google Scholar]
- Rossi J. J., Elkins D., Taylor N., Zaia J., Sullivan S., Deshler J. O. Exploring the use of antisense, enzymatic RNA molecules (ribozymes) as therapeutic agents. Antisense Res Dev. 1991 Fall;1(3):285–288. [PubMed] [Google Scholar]
- Rossi J. J., Sarver N. RNA enzymes (ribozymes) as antiviral therapeutic agents. Trends Biotechnol. 1990 Jul;8(7):179–183. doi: 10.1016/0167-7799(90)90169-x. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Scaringe S. A., Francklyn C., Usman N. Chemical synthesis of biologically active oligoribonucleotides using beta-cyanoethyl protected ribonucleoside phosphoramidites. Nucleic Acids Res. 1990 Sep 25;18(18):5433–5441. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore J. D., Gutfreund H. Transients in the reactions of liver alcohol dehydrogenase. Biochemistry. 1970 Nov 24;9(24):4655–4659. doi: 10.1021/bi00826a006. [DOI] [PubMed] [Google Scholar]
- Stawinski J., Strömberg R., Thelin M., Westman E. Studies on the t-butyldimethylsilyl group as 2'-O-protection in oligoribonucleotide synthesis via the H-phosphonate approach. Nucleic Acids Res. 1988 Oct 11;16(19):9285–9298. doi: 10.1093/nar/16.19.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Walker G. T. Deoxycytidine methylation does not affect DNA.RNA hybrid formation or B-A transitions of (dG)n.(dC)n sequences. Nucleic Acids Res. 1988 Apr 11;16(7):3091–3099. doi: 10.1093/nar/16.7.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Fujii S., van Boom J. H., van der Marel G. A., van Boeckel S. A., Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA--RNA hybrid helix joined to double helical DNA. Nature. 1982 Oct 14;299(5884):601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Perreault J. P., Labuda D., Usman N., Cedergren R. Mixed DNA/RNA polymers are cleaved by the hammerhead ribozyme. Biochemistry. 1990 Dec 25;29(51):11156–11160. doi: 10.1021/bi00503a002. [DOI] [PubMed] [Google Scholar]