Abstract
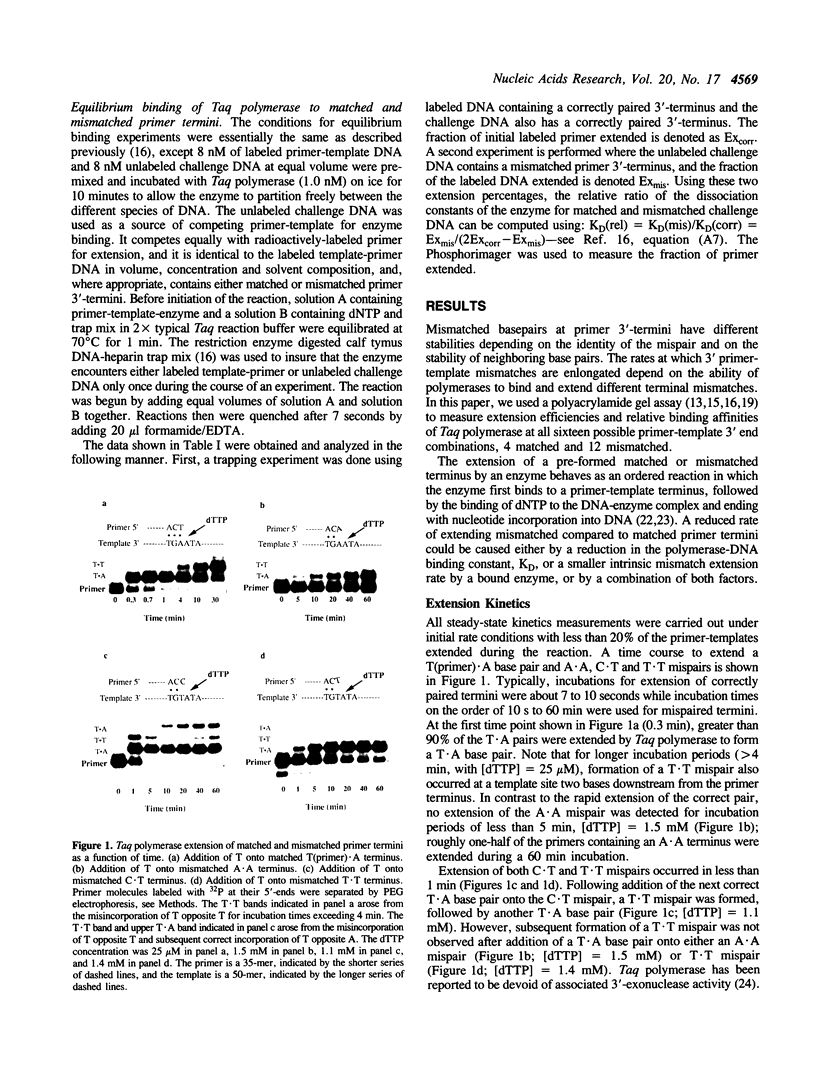
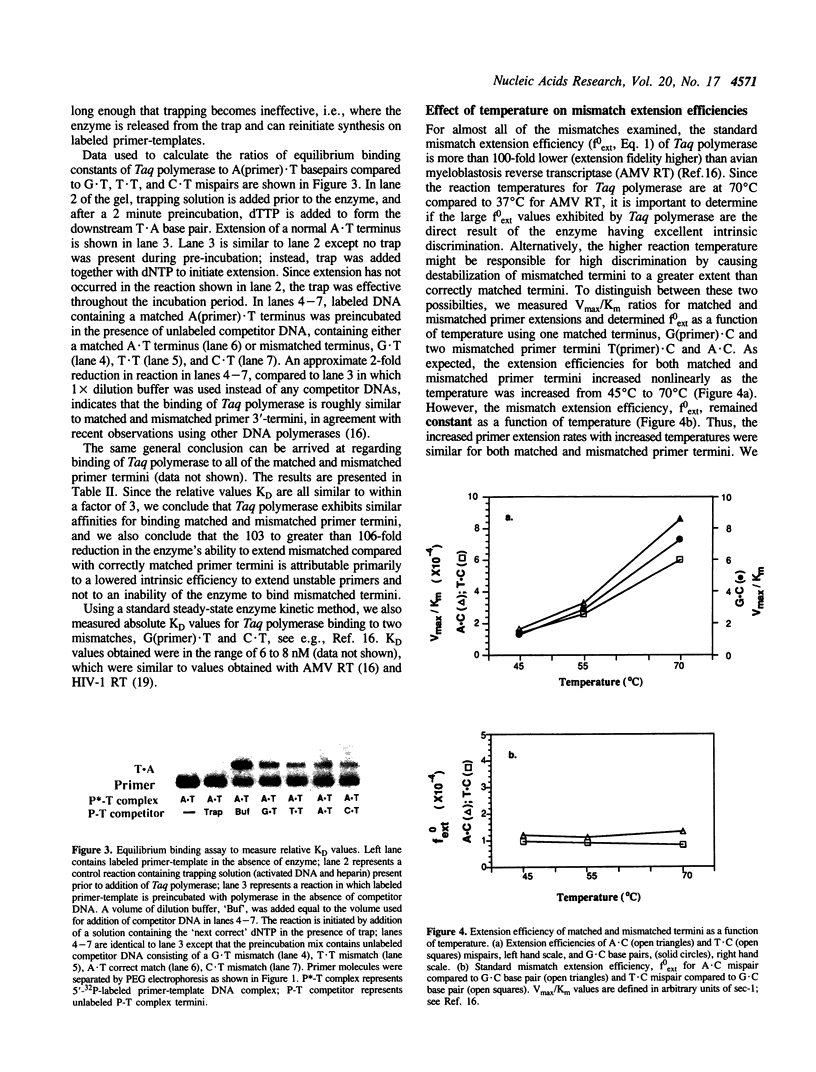
Thermus aquaticus (Taq) DNA polymerase was used to measure the extension efficiency for all configurations of matched and mismatched base pairs at template-primer 3'-termini. The transition mispairs, A(primer).C, C.A, G.T, and T.G were extended 10(-3) to 10(-4)-fold less efficiently than their correctly paired counterparts. Relative efficiencies for extending transversion mispairs were 10(-4) to 10(-5) for T.C and T.T, about 10(-6) for A.A, and less than 10(-6) for G.A, A.G, G.G and C.C. The transversion mispair C(primer).T was extended with high efficiency, about 10(-2) compared to a correct A.T basepair. The unexpected ease of extending the C.T mismatch was not likely to have been caused by primer-template misalignment. Taq polymerase was observed to bind with similar affinities to each of the correctly paired and mispaired primer-template 3'-ends. Thus, the failure of Taq polymerase to extend mismatches efficiently appears to be an intrinsic property of the enzyme and not due to an inability to bind to 3'-terminal mispairs. For almost all of the mispairs, C.T being the exception, Taq polymerase exhibits about 100 to 1000-fold greater discrimination against mismatch extension compared to avian myeloblastosis reverse transcriptase and HIV-1 reverse transcriptase which extend most mismatched basepairs permissively. Relative mismatch extension efficiencies for Taq polymerase were measured at 45 degrees C, 55 degrees C and 70 degrees C and found to be independent of temperature. The mispair extension data should be important in designing experiments using PCR to distinguish between sequences that vary by a single nucleotide.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Erlich H. Polymerase chain reaction strategy. Annu Rev Biochem. 1992;61:131–156. doi: 10.1146/annurev.bi.61.070192.001023. [DOI] [PubMed] [Google Scholar]
- Boulard Y., Cognet J. A., Gabarro-Arpa J., Le Bret M., Sowers L. C., Fazakerley G. V. The pH dependent configurations of the C.A mispair in DNA. Nucleic Acids Res. 1992 Apr 25;20(8):1933–1941. doi: 10.1093/nar/20.8.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton S., Huang M. M., Cai H., Arnheim N., Goodman M. F. Base mispair extension kinetics. Binding of avian myeloblastosis reverse transcriptase to matched and mismatched base pair termini. J Biol Chem. 1992 Feb 5;267(4):2633–2639. [PubMed] [Google Scholar]
- Detera S. D., Becerra S. P., Swack J. A., Wilson S. H. Studies on the mechanism of DNA polymerase alpha. Nascent chain elongation, steady state kinetics, and the initiation phase of DNA synthesis. J Biol Chem. 1981 Jul 10;256(13):6933–6943. [PubMed] [Google Scholar]
- Ehlen T., Dubeau L. Detection of ras point mutations by polymerase chain reaction using mutation-specific, inosine-containing oligonucleotide primers. Biochem Biophys Res Commun. 1989 Apr 28;160(2):441–447. doi: 10.1016/0006-291x(89)92452-2. [DOI] [PubMed] [Google Scholar]
- Erlich H. A., Gelfand D., Sninsky J. J. Recent advances in the polymerase chain reaction. Science. 1991 Jun 21;252(5013):1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- Fisher P. A., Korn D. Ordered sequential mechanism of substrate recognition and binding by KB cell DNA polymerase alpha. Biochemistry. 1981 Aug 4;20(16):4560–4569. doi: 10.1021/bi00519a008. [DOI] [PubMed] [Google Scholar]
- Gibbs R. A., Nguyen P. N., Caskey C. T. Detection of single DNA base differences by competitive oligonucleotide priming. Nucleic Acids Res. 1989 Apr 11;17(7):2437–2448. doi: 10.1093/nar/17.7.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta R. D., Benkovic P., Benkovic S. J. Kinetic mechanism whereby DNA polymerase I (Klenow) replicates DNA with high fidelity. Biochemistry. 1988 Sep 6;27(18):6716–6725. doi: 10.1021/bi00418a012. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Alexander P. S. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986 Jan 5;261(1):160–166. [PubMed] [Google Scholar]
- Kwok S., Kellogg D. E., McKinney N., Spasic D., Goda L., Levenson C., Sninsky J. J. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990 Feb 25;18(4):999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Cui X., Arnheim N. Direct electrophoretic detection of the allelic state of single DNA molecules in human sperm by using the polymerase chain reaction. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4580–4584. doi: 10.1073/pnas.87.12.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley M. J., Bennett S. E., Mosbaugh D. W. Characterization of the 5' to 3' exonuclease associated with Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990 Dec 25;18(24):7317–7322. doi: 10.1093/nar/18.24.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelman L. V., Boosalis M. S., Petruska J., Goodman M. F. Nearest neighbor influences on DNA polymerase insertion fidelity. J Biol Chem. 1989 Aug 25;264(24):14415–14423. [PubMed] [Google Scholar]
- Mendelman L. V., Petruska J., Goodman M. F. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J Biol Chem. 1990 Feb 5;265(4):2338–2346. [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Newton C. R., Graham A., Heptinstall L. E., Powell S. J., Summers C., Kalsheker N., Smith J. C., Markham A. F. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989 Apr 11;17(7):2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrino F. W., Loeb L. A. Differential extension of 3' mispairs is a major contribution to the high fidelity of calf thymus DNA polymerase-alpha. J Biol Chem. 1989 Feb 15;264(5):2898–2905. [PubMed] [Google Scholar]
- Petruska J., Goodman M. F., Boosalis M. S., Sowers L. C., Cheong C., Tinoco I., Jr Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sowers L. C., Fazakerley G. V., Kim H., Dalton L., Goodman M. F. Variation of nonexchangeable proton resonance chemical shifts as a probe of aberrant base pair formation in DNA. Biochemistry. 1986 Jul 15;25(14):3983–3988. doi: 10.1021/bi00362a002. [DOI] [PubMed] [Google Scholar]
- Tindall K. R., Kunkel T. A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988 Aug 9;27(16):6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- Wong I., Patel S. S., Johnson K. A. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991 Jan 15;30(2):526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- Wu D. Y., Ugozzoli L., Pal B. K., Wallace R. B. Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle cell anemia. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2757–2760. doi: 10.1073/pnas.86.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Goodman M. F. Comparison of HIV-1 and avian myeloblastosis virus reverse transcriptase fidelity on RNA and DNA templates. J Biol Chem. 1992 May 25;267(15):10888–10896. [PubMed] [Google Scholar]





