Abstract
Background:
The role of mitochondrial DNA (mtDNA) mutations in the development of breast cancer is largely unknown. In this study, we investigated the frequency and pattern of mutations in the D310 region, the most commonly mutated region in mtDNA, in a series of breast lesions.
Methods:
Using capillary electrophoresis, we genotyped the D310 sequence of neoplastic epithelial cells from 23 patients with synchronous ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC), 26 patients with IDC only and 29 patients with DCIS only.
Results:
A majority of DCIS (68.4%) and IDC (71.4%) lesions harbour different D310 sequences compared with their matched normal control. Specific D310 sequences were more frequently identified in tumour samples (77.1% of DCIS and 75.5% of IDC) compared with normal tissues (35.3% of normal; P<0.0001). No difference was identified between DCIS lesions with synchronous IDC and those from pure DCIS cases. In five cases, histologically normal tissue adjacent to tumour was found to share D310 sequences with the tumour, while normal tissue taken further away did not.
Conclusion:
Although D310 alterations do not seem to be related to DCIS progression, they were found in histologically normal cells adjacent to tumour. This suggests a field of genetically altered cells, thus D310 mutations could represent a potential marker for the clonal expansion of premalignant breast cancer cells.
Keywords: DCIS, mitochondrial DNA, D310 mutation, biomarker, clonal growth
Breast cancer is one of the leading causes of cancer death amongst women in the United States and Canada. The multistep development model of breast cancer involves the progression from premalignant hyperplastic lesions, to pre-invasive carcinoma (ductal carcinoma in situ (DCIS)), to invasive ductal carcinoma (IDC). In all, 14–50% of DCIS cases will progress to IDC after resection (Erbas et al, 2006). At present, the mechanism of DCIS progressing to IDC remains unknown, and there is a great need for markers that could reliably predict which cases of DCIS will relapse as an invasive carcinoma. It has been proposed that DCIS harbouring 17q22.24 gains are associated with a higher risk of progression to invasive disease (Iakovlev et al, 2008). Gains in this region were also associated with higher histological grade, large IDC size, lymphatic/vascular invasion, and lymph node metastasis. An important link has been made between p53 mutations and DCIS of high histological grade. P53 missense mutations occur at highest frequency in high-grade DCIS, but not in areas of hyperplasia or normal breast epithelium, suggesting that p53 mutations occur before the development of IDC (Done et al, 2001). Recently, mitochondrial DNA (mtDNA) depletion- and mutation-induced oxidative stress have been shown to be linked to increased tumourigenicity and an invasive phenotype, suggesting that mtDNA mutations may serve as markers for cancer progression (Amuthan et al, 2001).
Human mtDNA is a double-stranded, circular DNA molecule of 16.5 k. In contrast to nuclear DNA, mtDNA has hundreds to thousands of copies per cell. To date, mtDNA mutations have been detected in fine-needle aspirates (Parrella et al, 2001) and bodily fluids of cancer patients (Fliss et al, 2000). Many mtDNA mutations have been described in various forms of cancer, most of them in the regulatory region or D-loop, the replication origin for the heavy strand of mtDNA. Within the D-loop, the majority of mutations occur within a poly-C repeat stretch termed D310 (Fliss et al, 2000; Ha et al, 2002; Lievre et al, 2006). D310 mutations are highly prevalent in human tumours, including gastric, colorectal, bladder, lung, gallbladder, and breast cancers. It has been proposed as a marker for early tumour progression in head and neck cancer (Ha et al, 2002). In breast cancer, mutations in the D-loop have been associated with clinicopathological data, such as poor disease-free survival, later onset age, and estrogen and progesterone receptor-negative tumours (Tseng et al, 2006). However, questions still remain as to at which stage of breast cancer these D310 mutations occur, and whether they can be used as a marker for breast cancer progression.
Mutations in the D310 region have been used to track clonal growth of cancer cells. In normal tissues from the general population, there are between seven and nine cytosine residues in the D310 region (Anderson et al, 1981). A recent study examining D310 mutations in head and neck carcinoma indicated that normal tissues adjacent to tumour had a propensity towards expansion/deletion of this region similar to cancerous tissues. These mutations can be used as markers of clonal growth of precancerous cells (Ha et al, 2002). It remains to be seen whether D310 mutations may be used as a marker for clonal growth in breast cancer.
The aims of our study were therefore to evaluate the frequency of mtDNA D310 mutation in tumour adjacent normal tissues and tumour tissues of breast cancer patients, and to assess the relationship between the D310 mutations and the progression of pre-invasive to invasive breast carcinoma.
Material and methods
Patient selection
Breast tumour and normal tissues were collected from 78 breast cancer patients selected from the files of the Department of Pathology at the University Health Network (Toronto, Canada) between 2004 and 2008. The patients were diagnosed either with DCIS only (n=29), IDC only (n=26) or synchronous DCIS and IDC (n=23). The study was approved by the UHN Research Ethics Board.
Tissue preparation and DNA extraction
About 15–20 5-μm-thick sections from formalin-fixed, paraffin-embedded tissue blocks were used for DNA extraction using the standard protocols. Briefly, sections were deparafinised, stained in hematoxylin for 30 s, and microdissected using a stereomicroscope. Areas of DCIS, IDC, and normal epithelium were selected by a breast pathologist (SJD). Microdissected cells were incubated in lysis buffer at 56 °C, and DNA was extracted using the QIAmp DNA Mini Kit (Qiagen, Mississauga, Canada) according to the manufacturer's recommendations.
Genotype data from 48 samples were excluded due to poor DNA quality, which contained higher than threshold background signal. The final sample size is 131, as indicated in Supplementary Table 1.
D310 sequence genotyping
For amplification, 50–100 ng of DNA was used in a final 20 μl master mix containing 1 × final concentration of 10 × PCR buffer, 0.8 mM of each dNTP, 0.3 μM of each primer, and 0.8 units of HotStarTaq DNA polymerase (Qiagen). The forward primer was 5′- (WellRED D4 dye)-GCCACTTTCCACACAG-3′ (259–275 nt; Proligo Primers and Probes, Boulder, CO, USA), and the reverse primer was 5′-TGGTTAGGCTGGTGTTAGGG-3′ (368–387 nt; Operon Biotechnologies, Huntsville, AL, USA). Total DNA was subject to PCR amplification as follows on a Gene Amp PCR system 9700 automated sequencer (Applied Biosystems, Foster City, CA, USA): 95 °C for 15 min, 94 °C for 30 s, 58.9 °C for 30 s, 72 °C for 30 s, repeat all steps for 34 more times, and 72 °C for 3 min. Finally, PCR products were purified by adding ExoSAP-IT reagents (USB, Cleveland, OH, USA). Amplification was confirmed by 2% agarose gel electrophoresis. For genotyping, 2 μl underwent capillary electrophoresis on a CEQ 8000 Genetic Analysis System (Beckman Coulter, Fullerton, CA, USA) with a dilution of 1/45 as described previously (Legras et al, 2008). The results of D310 genotyping were analysed using the Fragment Analysis v.4.0 (Beckman Coulter).
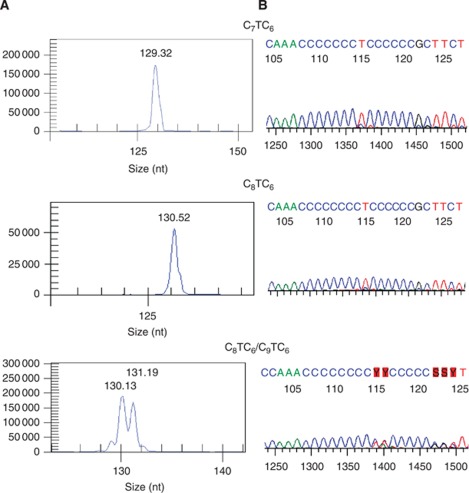
Products of 128, 129, 130, 131, 132, and 133 bp were observed when the D310 sequence contained, respectively, 6, 7, 8, 9, 10, or 11 cytosines. Sequences were thus denoted as C6, C7, C8, C9, C10, and C11 for the purpose of this study. Any mtDNA sequences that differed between tumour and matched normal tissue mtDNA were scored as a somatic mutation. Mutations affecting all copies of the mtDNA were defined as homoplasmic, whereas those which were present only in a proportion of copies of mtDNA were defined as heteroplasmic. Thus, a coexistence of two or more peaks on the electropherogram indicated heteroplasmy (Figure 1). More than 85% of samples were genotyped in duplicate to validate the results.
Figure 1.
Concordance between data from genotyping (A) and sequencing (B). Fragment 129 bp represents C7TC6, fragment of 130 bp represents C8TC6 and a mixture of fragments (heteroplasmy) 130 and 131 bp represent C8TC6 and C9TC6.
Direct sequencing of the D310 region
A 414-bp fragment of the D-loop including the D310 sequence was amplified and sequenced as described previously (Legras et al, 2008). Briefly, 50–100 ng of DNA were used in a final 25 μl master mix containing 1 × PCR buffer, 250 μM of dNTP, 0.5 μM of each primer and three units of AmpliTaq Gold 360 DNA polymerase (Applied Biosystems). The forward primer was 5′-ACCTACGTTCAATATTACAGGCG-3′ and the reverse primer was 5′-CTGTGGGGGGTGTCTTTGGGG-3′ (Operon Biotechnologies). Total DNA was subject to a step-down PCR protocol as described previously (Legras et al, 2008). PCR products were purified using a GenElute PCR cleanup kit (Sigma-Aldrich, St Louis, MO, USA).
Purified PCR products were sequenced and analysed using a Big Dye Terminator cycle sequencing kit (Applied Biosystems) on an ABI PRISM 3130XL Genetic Analyser (Applied Biosystems). Sequence data were analysed by Sequence Scanner v.1.0 (Applied Biosystems).
Statistical analysis
A χ2 test was used to compare the distributions of D310 polymorphism in histologically normal and tumour samples. Also, it was used to compare the prevalence of mutant alleles and heteroplasmy between normal and tumour samples. Fisher's exact test was used to compare the frequency and type of mutation between DCIS and IDC samples that have matched normal samples, as well as between DCIS samples from various cases. The software SAS Analytics (SAS, Cary, NC, USA) was used to perform all statistical tests.
Results
High frequency of somatic D310 mutations in Ductal carcinoma in situ and IDC samples
In order to ensure the consistency and validity of our D310 genotyping methodology, >85% samples were genotyped in duplicate using another set of independently microdissected samples. Results obtained with the duplicate samples were identical to the original sample in all cases. In addition, our methodology was also validated by performing direct sequencing of the D310 region on a subset of samples (n=15). We found that our genotyping method correlated with direct sequencing in 100% of the cases (Figure 1).
Using this D310 genotyping methodology, we investigated the presence of somatic mutations in DCIS and IDC samples by comparing the genotype of the tumours with that of corresponding adjacent normal tissue. In cases where the adjacent normal tissue shared at least one allele with the tumour, another normal tissue control was selected at least 2 cm away from tumour. A total of 19 DCIS and 14 IDC samples were genotyped along with their matching normal. As seen in Table 1, somatic mutations (defined as different mtDNA sequences between tumour and normal tissue) occurred in 68.4% of DCIS samples (13 out of 19) and 71.4% of IDC samples (10 out of 14). Supplementary Table 2 lists the samples scored as somatic mutation. The frequencies of mutation were not significantly different between DCIS and IDC samples. Insertion or deletion of cytosine(s) occurred at approximately similar frequency, and was not significantly different between tumour samples (P> 0.05).
Table 1. Frequency and type of somatic mutations in the D310 region in ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC) samples.
|
Type of mutation
|
||||
|---|---|---|---|---|
| Samples | Total # of samples | # Of samples with mutation (%) a | Deletions (%) b | Insertions |
| DCIS | 19 | 13 (68.4) | 8 (61.5) | 5 |
| IDC | 14 | 10 (71.4) | 4 (40.0) | 6 |
| Total | 33 | 23 (69.7) | 12 (52.2) | 11 |
A mutation was scored when the D310 allele in tumour differed from the allele found in its matched normal sample.
Fisher's exact test for differences in mutation rate: P>0.05.
Fisher's exact test for differences in the prevalence of deletion mutation: P>0.05.
D310 sequences C6, C7, C10, and C11 are more common in tumour samples
We subsequently analysed the genotype of the D310 sequence in mtDNA extracted from 48 cases of pre-invasive breast cancer (29 cases from patients with DCIS only and 19 cases from patients with synchronous DCIS and IDC) and 49 cases of invasive breast cancer (26 cases from patients with IDC only and 23 cases from patients with DCIS and IDC). We compared the distributions of D310 sequences extracted from tumour tissues to sequences extracted from normal tissue controls. Polymorphisms in the D310 sequence in a normal human population consist of variations in the number of cytosine repeats, which range most commonly between seven and nine cytosine repeats (Ha et al, 2002). For the purpose of this study, D310 sequences C6TC6 to C11TC6 were therefore termed as C6 to C11, according to the number of cytosine residues.
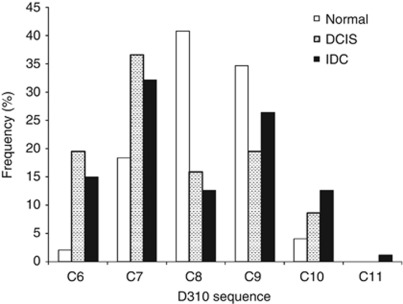
In our patient series, we have found that C8 and C9 were the more frequent polymorphisms detected in normal control tissues (Figure 2). Sequences C6, C7, C10, and C11 were, on the other hand, found more frequently in tumours (P=0.0002): 64.6% (53 out of 82) of all sequences in DCIS samples, and 60.9% (53 out of 87) of all sequences in IDC samples, respectively. In comparison, these sequences are only detected in 24.5% (12 out of 49) of all sequences in normal samples.
Figure 2.
Distribution of D310 sequences in normal (n=49), DCIS (n=82), and IDC (n=87) samples. Frequency of alleles C6, C7, C10, and C11 are significantly higher in DCIS and IDC samples than normal (χ2 test, P=0.0002). Multiple sequences may be found in one tissue sample (heteroplasmy).
Because of the high abundance of C8 and C9 sequences in the normal samples and the general population, it is reasonable to consider C8 and C9 as normal alleles. We hypothesise that any variation from these sequences is likely due to either somatic mutations or germ-line polymorphisms. The high abundance of sequences C6, C7, C10, and C11 in tumour samples further supports their derivation from mutations. Table 2 shows that 77.1% of DCIS and 75.5% of IDC samples contain at least one mutant allele compared with only 35.3% of histologically normal samples (P<0.0001), confirming that mutant alleles are present in higher frequency in tumours. Tumour samples were also found to be more heteroplasmic than normal samples (Table 2). The D310 sequence was heteroplasmic in 38.2% (13 out of 34) of normal samples, 58.3% of DCIS (28 out of 48), and 65.3% of IDC (32 out of 49; P=0.045).
Table 2. Prevalence of D310 mutant alleles (C6, C7, C10, or C11) and frequency of heteroplasmy in the three groups of samples.
| Samples | # Of samples with at least one mutant allele (%) a | # Of samples with heteroplasmy (%) b | Total |
|---|---|---|---|
| Normal | 12 (35.3) | 13 (38.2) | 34 |
| DCIS | 37 (77.1) | 28 (58.3) | 48 |
| IDC | 37 (75.5) | 32 (65.3) | 49 |
| Total | 86 (65.7) | 73 (55.7) | 131 |
χ2 test, P<0.0001.
χ2 test, P=0.045.
D310 mutations do not appear to be associated with DCIS progression
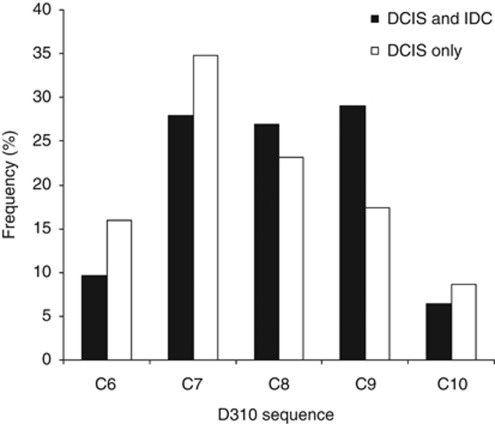
In the quest for genetic markers that may be able to identify DCIS cases with high risk of progression, we assessed the relationship between D310 mutations and the risk of IDC associated with DCIS. We compared the distribution of D310 genotypes between DCIS samples from synchronous DCIS and IDC cases, and DCIS samples from pure DCIS cases. The distribution of D310 sequences did not differ significantly between DCIS samples from synchronous DCIS and IDC cases, and DCIS only cases (P=0.59, Figure 3). In addition, the frequency of somatic mutations (any D310 sequences in tumour that differed from sequences of the matched normal tissue) was similar in DCIS samples from synchronous DCIS and IDC cases and pure DCIS cases (P>0.05, Supplementary Table 3).
Figure 3.
Distribution of D310 sequences in DCIS samples from synchronous DCIS and IDC cases and from pure DCIS cases. The frequency of mutant alleles was not significantly different between the two type of DCIS lesions (χ2 test, P=0.59).
Normal tissues adjacent to tumour have D310 mutations similar to tumour
DNA was extracted from 34 histologically normal tissues adjacent to tumour (same section) and from an additional 10 normal tissues taken at least 2 cm away from the tumour site. The additional normal tissues were selected based on the observation that in 10 out of 34 patients, adjacent normal tissue samples shared at least 1 allele with tumour samples. Genotype similarities found in both normal and tumour tissues mtDNA could be due to germ-line polymorphism, field cancerisation (genetic alterations in histologically normal cells due to the clonal expansion of cancer progenitor cells), or contamination from tumour cells during microdissection. For these cases, additional normal tissues further away from the tumour were selected and these were considered as the actual controls in the paired analyses.
To confirm the sequences from these samples, two independent microdissection and amplification experiments were performed. In 5 out of the 10 patients where normal adjacent tissue contained at least 1 allele that was also found in the tumour, normal tissue samples taken at least 2 cm away from the tumour site had different allele(s) than normal tissue adjacent to tumour sample (Table 3). In the other five cases, both normal tissue adjacent and further away from the tumour shared the same alleles. In the first five samples, our results were suggestive of a field cancerisation effect: adjacent normal tissue having an identical sequence to the tumour, but a different sequence compared with normal tissue further away from the tumour. The shortest distance between tumour and the adjacent normal tissue showing this phenomenon was measured under a microscope by a pathologist (SJD) on hematoxylin- and eosin-stained slides. The mean minimal distance between sampled adjacent normal and tumour tissues that shared alleles was 1.08±1.0 mm (n=5). This was significantly lower than the mean minimal distance in cases that did not display field cancerisation: 2.64±0.93 mm (n=5; P=0.03).
Table 3. D310 sequences in normal tissue sampled adjacent to and further away (>2 cm) from tumour in five cases showing ‘field cancerisation’.
| Patient | D310 seq. in DCIS | D310 seq. in IDC | D310 seq. in N | D310 seq. in NF |
|---|---|---|---|---|
| 62 | C9/C10 | C9/C10 | C9/C10 | C8 |
| 74 | C8/C9 | C9 | C9 | C8 |
| 82 | C9 | C9 | C9/C10 | C8 |
| 42 | C6/C7 | C6/C7 | C6/C7 | C7/C8 |
| 38 | C7/C8 | C7/C8 | C7 | C8 |
N=normal tissue adjacent to tumour; NF=normal tissue further away (>2 cm) from tumour; seq.=sequence.
Discussion
Mitochondrial DNA (mtDNA) mutations are highly prevalent in many cancers including breast cancer. In this study, we investigated the frequency and pattern of mutations in a regulatory region of the mtDNA referred to as the D-loop. Within the D-loop, a sequence of poly-C repeats (the D310 region) is a mutational hot spot. In breast cancer tissue, 72% of all D-loop mutations occur in the D310 region (Tseng et al, 2006), whereas in fine-needle aspirates from breast cancer patients, 58% of D-loop mutations occur in the D310 region (Parrella et al, 2001). In a previous study, D310 mutations have been found, along with three other mitochondrial mutations, in 0% (0/4) of pre-invasive (DCIS) and 34.1% (15/44) of invasive (IDC) breast cancers (Wang et al, 2006). In this study, the prevalence of D310 somatic mutations was investigated in a larger sample size of pre-invasive breast cancer, and our results demonstrated for the first time that these mutations occurred early in the development of breast cancer.
Analysis of D310 sequences in our patient series suggests a high D310 mutation frequency. In all, 68% (13/19) of DCIS and 71.4% (10/14) of IDC samples were mutated when compared with matched normal controls. Similarly, we found that 77.1% (37/48) of DCIS and 75.5% (37/49) of IDC samples contained at least one mutant allele (defined in our study as alleles found with the least frequency in normal tissues; i.e., C6, C7, C10, and C11 alleles; Table 2). These mutation frequencies were similar to those reported in other types of cancers: 61.5% (8/13) of squamous cell carcinoma in situ of the head and neck (Ha et al, 2002) and 70% (10/14) of lung cancers harboured a D-loop mutation (Fliss et al, 2000). In these studies, mutation was scored by comparing D310 sequences from tumour and corresponding normal tissue/ blood samples, similar to our work.
In this study, we did not find a correlation between the frequency of D310 mutations in breast cancer and the degree of dysplasia (DCIS and IDC cases have a similar frequency of mutations). In addition, no difference was observed between cases of DCIS associated with invasive carcinoma compared with those that were not (72.7% and 62.5%, respectively, Supplementary Table 3). Interestingly, identical D-loop sequences have been identified in pre-malignant and malignant lesions of the prostate and esophagus (Jeronimo et al, 2001; Miyazono et al, 2002). Taken together, these results show that D310 mutations likely occurred before DCIS transformation to IDC; perhaps during the transformation of atypical ductal hyperplasia to DCIS. Thus, the timing of events explains the lack of association between D310 mutations and DCIS progression.
As D310 mutations occur at high frequency in pre-malignant lesions, it is important to consider whether they have a causative role in breast carcinogenesis. Interestingly, associations have been made between D310 mutations and poor disease-free survival, a late onset age, and estrogen and progesterone-negative breast cancers (Tseng et al, 2006). Although the poly-C repeat is polymorphic in the normal population (mainly C8 and C9 alleles), certain alleles are found more frequently in tumour samples compared with normal tissues: C6, C7, C10, and C11 alleles. The D310 region is located within the mtDNA replication origin. D310 mutations most likely resulted from a combination of oxidative stress, low efficiency of the mtDNA mismatch repair mechanism, and poor proof reading by polymerase γ (Mambo et al, 2003). It is postulated to have a central role in the maintenance of mtDNA copy number. Increase/decrease in the number of cytosine residues in the poly-C region may affect the rate of DNA replication by impairing the binding of polymerase and other trans-acting factors (Fliss et al, 2000). Reduced mtDNA copy number has been found in 72% of breast cancers with mtDNA D-loop mutations (Tseng et al, 2006). The decrease in mtDNA content in breast cancer may consequently increase mitochondrial genomic instability, causing alterations in energy metabolism and promoting tumour development (Lu et al, 2009). It is therefore plausible that D310 mutations promote the early development of breast cancer by depleting mtDNA content in cells, thus altering energy metabolism and promoting genetic instability. Further studies are needed to confirm the role of D310 mutation in carcinogenesis.
Finally, field cancerisation has been observed in breast cancer and it is a postulated explanation for the occurrence of D310 mutations in normal epithelium adjacent to tumours. First introduced by Slaughter et al (1953) ‘field cancerisation’ is now commonly used to describe the occurrence of genetic and epigenetic alterations in normal tissues surrounding an area of cancer. Field cancerisation has been implicated in the lung, colon, cervix, bladder, skin, and breast cancers (Heaphy et al, 2006). Previous studies have found telomere DNA alterations and allelic imbalance in histologically normal ductal epithelial tissues. These alterations decreased as a function of distance from tumour (Heaphy et al, 2006). Similarly, in this study, 50% (5/10) of patients with matched DCIS, IDC, and adjacent normal tissue have normal tissue that shared at least one allele with the tumour tissues (Table 3). When measured under the microscope, histologically normal, but D310 mutant containing, tissues were found at an average distance of 1.08±1.0 mm from tumours, a significantly shorter distance in comparison with normal tissues that did not display D310 mutations (2.64±0.93 mm). An explanation for this finding is that there are genetically altered, premalignant cells within histologically normal breast tissue ‘field cancerised’. This interpretation is corroborated by the fact that in these cases, all of the mutant alleles in adjacent histologically normal tissues are conserved in the matched tumours, but not in another set of normal tissues taken this time at least 2 cm away from tumour. Similarly, in a study of terminal ductal–lobular units (TDLUs) adjacent to cancer, LOH was observed in 26% (8 out of 30) of normal TDLUs adjacent to tumour sites, but not in normal TDLU samples taken two blocks away from the tumour (Deng et al, 1996). In gallbladder carcinoma, and head and neck carcinoma, identical D310 sequences to tumour tissues have been found in 86% (19/22) and 37.5% (3/8) of microdissected epithelial tissues adjacent to tumour tissue, respectively (Ha et al, 2002; Tang et al, 2004). Thus, as demonstrated in this study and others, D310 mutations in mtDNA can function as a marker of clonality and be used to define a field of genetic abnormality. As it has been suggested that local normal remnants of a field may develop into cancer after removal of the primary tumour site, further investigation is warranted to examine whether D310 mutations in adjacent normal tissue is associated with tumour recurrence.
In conclusion, our study has showed that mitochondrial D310 mutations are an early genetic event in the carcinogenesis of breast cancer, and probably occur before DCIS transitioning to IDC. Tumours relative to normal tissues are enriched with mutant alleles C6, C7, C10, and C11. Although D310 alterations do not seem to be related to DCIS progression, they were found in histologically normal cells adjacent to tumour. This suggests a field of genetically altered cells, thus D310 mutations could represent a potential marker for the clonal expansion of premalignant breast cancer cells. Given the abundance of mtDNA in each cell, and the success of identifying mtDNA mutations in fine-needle aspirates (Parrella et al, 2001) and bodily fluids of cancer patients (Fliss et al, 2000), D310 mutation assessment may become a convenient biomarker to apply in clinical settings.
Acknowledgments
We thank N Kanwar for providing assistance in sequencing and optimising PCR conditions and T Cawthorn for help in statistical analysis. Tissue sectioning was performed by the Pathology Research Program Laboratory at the University Health Network. This work was supported by grants from the Canadian Breast Cancer Research Alliance.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG (2001) Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J 20: 1910–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ (1981) Sequence and organization of the human mitochondrial genome. Nature 290: 457–465 [DOI] [PubMed] [Google Scholar]
- Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS (1996) Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science 274: 2057–2059 [DOI] [PubMed] [Google Scholar]
- Done SJ, Eskandarian S, Bull S, Redston M, Andrulis IL (2001) P53 missense mutations in microdissected high-grade ductal carcinoma in situ of the breast. J Natl Cancer Inst 93: 700–704 [DOI] [PubMed] [Google Scholar]
- Erbas B, Provenzano E, Armes J, Gertig D (2006) The natural history of ductal carcinoma in situ of the breast: a review. Breast Cancer Res Treat 97: 135–144 [DOI] [PubMed] [Google Scholar]
- Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D (2000) Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 287: 2017–2019 [DOI] [PubMed] [Google Scholar]
- Ha PK, Tong BC, Westra WH, Sanchez-Cespedes M, Parrella P, Zahurak M, Sidransky D, Califano JA (2002) Mitochondrial C-tract alteration in premalignant lesions of the head and neck: a marker for progression and clonal proliferation. Clin Cancer Res 8: 2260–2265 [PubMed] [Google Scholar]
- Heaphy CM, Bisoffi M, Fordyce CA, Haaland CM, Hines WC, Joste NE, Griffith JK (2006) Telomere DNA content and allelic imbalance demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int J Cancer 119: 108–116 [DOI] [PubMed] [Google Scholar]
- Iakovlev VV, Arneson NC, Wong V, Wang C, Leung S, Iakovleva G, Warren K, Pintilie M, Done SJ (2008) Genomic differences between pure ductal carcinoma in situ of the breast and that associated with invasive disease: a calibrated aCGH study. Clin Cancer Res 14: 4446–4454 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Nomoto S, Caballero OL, Usadel H, Henrique R, Varzim G, Oliveira J, Lopes C, Fliss MS, Sidransky D (2001) Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene 20: 5195–5198 [DOI] [PubMed] [Google Scholar]
- Legras A, Lievre A, Bonaiti-Pellie C, Cottet V, Pariente A, Nalet B, Lafon J, Faivre J, Bonithon-Kopp C, Goasguen N, Penna C, Olschwang S (2008) Mitochondrial D310 mutations in colorectal adenomas: an early but not causative genetic event during colorectal carcinogenesis. Int J Cancer 122: 2242–2248 [DOI] [PubMed] [Google Scholar]
- Lievre A, Blons H, Houllier AM, Laccourreye O, Brasnu D, Beaune P, Laurent-Puig P (2006) Clinicopathological significance of mitochondrial D-Loop mutations in head and neck carcinoma. Br J Cancer 94: 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Sharma LK, Bai Y (2009) Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res 19: 802–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mambo E, Gao X, Cohen Y, Guo Z, Talalay P, Sidransky D (2003) Electrophile and oxidant damage of mitochondrial DNA leading to rapid evolution of homoplasmic mutations. Proc Natl Acad Sci USA 100: 1838–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono F, Schneider PM, Metzger R, Warnecke-Eberz U, Baldus SE, Dienes HP, Aikou T, Hoelscher AH (2002) Mutations in the mitochondrial DNA D-Loop region occur frequently in adenocarcinoma in Barrett's esophagus. Oncogene 21: 3780–3783 [DOI] [PubMed] [Google Scholar]
- Parrella P, Xiao Y, Fliss M, Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, Nicol T, Gabrielson E, Cuomo C, Cohen D, Pandit S, Spencer M (2001) Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res 61: 7623–7626 [PubMed] [Google Scholar]
- Slaughter DP, Southwick HW, Smejkal W (1953) Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 6: 963–968 [DOI] [PubMed] [Google Scholar]
- Tang M, Baez S, Pruyas M, Diaz A, Calvo A, Riquelme E, Wistuba II (2004) Mitochondrial DNA mutation at the D310 (displacement loop) mononucleotide sequence in the pathogenesis of gallbladder carcinoma. Clin Cancer Res 10: 1041–1046 [DOI] [PubMed] [Google Scholar]
- Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC (2006) Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 45: 629–638 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu VW, Tsang PC, Chiu PM, Cheung AN, Khoo US, Nagley P, Ngan HY (2006) Microsatellite instability in mitochondrial genome of common female cancers. Int J Gynecol Cancer 16(Suppl 1): 259–266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.