Abstract
Most clinically approved biomarkers of cancer are glycoproteins, and those residing on the cell surface are of particular interest in biotherapeutics. We report a method for selective labeling, affinity enrichment, and identification of cell-surface glycoproteins. PC-3 cells and primary human prostate cancer tissue were treated with peracetylated N-azidoacetylgalactosamine, resulting in metabolic labeling of cell surface glycans with the azidosugar. We used mass spectrometry to identify over 70 cell surface glycoproteins and biochemically validated CD146 and integrin beta-4, both of which are known to promote metastatic behavior. These results establish cell-surface glycoproteomics as an effective technique for discovery of cancer biomarkers.
Keywords: Metabolic labeling, Glycoproteomics, Prostate cancer, Azidosugar, Tissue slice cultures
Prostate cancer (PC) is the second leading cause of death from cancer in men, with over 25,000 succumbing to the disease each year.1 While prostate-specific antigen (PSA) screening detects a large number of cases, cancer detection in some men with low levels of PSA is missed. Conversely, PSA screening leads to overdetection of men with nonaggressive cancer.2 Several recent compelling examples support the notion that glycoproteins may be strong predictors of the progression of PC. PSA itself is a secreted glycoprotein, the glycan structures of which have been elucidated and shown to be altered on PSA derived from malignant cells.3,4 Additionally, the overproduction of mucin-type glycoproteins, which are characterized by dense clusters of serine or threonine-bound glycans bearing conserved core N-acetylgalactosamine (GalNAc) residues, has been shown to correlate with increased metastatic potential and decreased survival rates in many cancer types.5 For instance, transcript microarrays have revealed that of the ~26,000 genes surveyed in normal, benign and metastatic prostate tissue, the transcript encoding the mucin MUC1 is one of the most informative characteristics of clinically relevant PC subtypes.6,7 A study comparing MUC1 from cancerous and normal prostate tissue identified glycosylation changes that are indicative of disease; these changes are also mirrored in immortalized prostate cell lines (PC-3, DU145 and BPH-1), providing a basis for clinical translation of studies initiated in cell culture.8 Lastly, glycoproteins are preeminent as an information-rich class of biomarkers for cancer, and likewise, virtually all biomarkers currently in use in the clinic are glycoproteins (e.g., CEA, CA125, PSA and HER2, among others).9,10
The evolving field of proteomics holds considerable promise for the identification of new PC biomarkers.11 However, one significant hurdle remains to be overcome before proteomics technology can realize its full potential for clinical utility—the difficulty inherent in identifying proteins of interest amongst the highly abundant, steady-state proteins that are not related to disease. Recent studies have attempted to overcome this issue through selective targeting of a subclass of glycoproteins, the N-linked type.12 Several groups have employed N-glycan-specific lectins as reagents for enriching N-glycoproteins prior to mass spectrometry analysis.13,14 Others have enriched the N-glycoproteome from serum or tissue samples by periodate oxidation of the glycan chains and selective capture on a hydrazide affinity matrix.14–16 This method has been recently employed in the search for biomarkers from lung cancer patients.15 Methods for enriching O-glycoproteins are less well developed, though periodate oxidation/capture/release16 and lectin-based17,18 approaches have been recently reported.
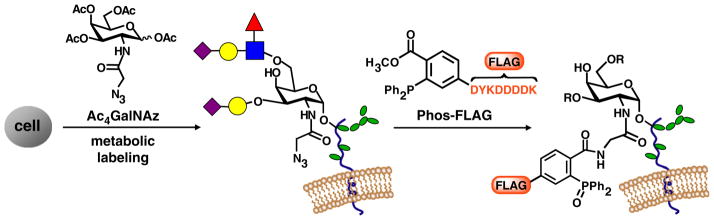
Metabolic labeling with bioorthogonal chemical reporters is an emerging strategy for glycoprotein profiling. The technique introduces unnatural sugars bearing either azide or alkyne groups into cellular glycans, thereby arming them for covalent reaction with affinity probes via Staudinger ligation, copper-catalyzed alkyne-azide cycloaddition, or copper-free click chemistry.19 Toward the development of glycoproteomics methods that target O-glycoproteins, we previously reported their metabolic labeling with the azidosugar N-azidoacetylgalactosamine (GalNAz, Fig. 1A), which replaces the conserved core GalNAc residue in both cultured cells and murine tissues.20,21 Staudinger ligation of the azides on GalNAz residues with phosphine-conjugated affinity reagents such as Phos-FLAG (Fig. 1) enabled selective detection of O-glycoproteins from cell and tissue lysates.22 More recently, Wong and coworkers and Yang and coworkers reported a similar approach to enrich sialylated glycoproteins from prostate cancer-derived PC-3 cells23 and Lemoine and coworkers employed metabolic labeling to profile O-N-acetylglucosamine (O-GlcNAc)-modified cytosolic proteins from a breast cancer cell line.24
Figure 1.
Metabolic labeling of glycoproteins via treatment of cells with Ac4GalNAz allows subsequent covalent tagging with phosphine probes such as Phos-FLAG. Colored shapes: other monosaccharides present in glycan structure.
In this study, we sought to evaluate metabolic labeling with GalNAz as a means to identify cell surface prostate cancer biomarkers. We reasoned that glycoproteins on the cell surface could be selectively tagged through reaction of live GalNAz-labeled cells with Phos-FLAG, a membrane-impermeant probe. This strategy would minimize contamination of enriched samples with intracellular glycoproteins, which are of lesser interest as biomarkers. Furthermore, we recently discovered that GalNAz labels cytosolic and nuclear O-GlcNAc-modified proteins after conversion to the intermediate UDP-N-azidoacetylglucosamine (UDP-GlcNAz).22 Selective Staudinger ligation on live, intact cells precludes reaction of the Phos-FLAG probe with this inaccessible class of glycoproteins, thereby focusing the enrichment on the cell surface glycoproteome. Since GalNAz should have access to all mucin-type O-glycans as well as some N-glycans and proteoglycans, this method can potentially select for a broad distribution of glycoproteins that is distinct from those enriched by other methods.
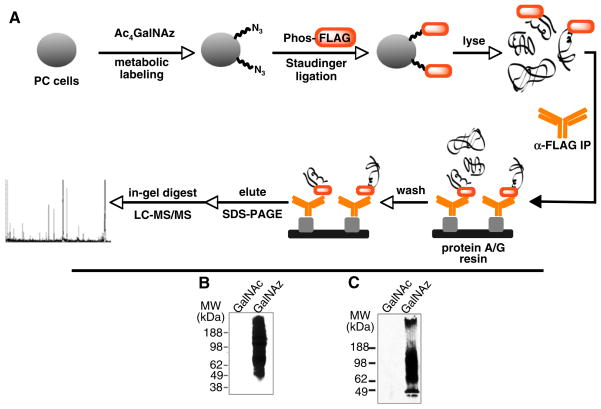
The PC-derived cell line PC-3 was chosen as a model of PC based on prior reports of aberrant mucin expression reminiscent of PC tissue (vide supra). Cells were cultured for three days in the presence of peracetylated GalNAz (Ac4GalNAz, a precursor that is converted to GalNAz in situ by cytosolic esterases21) (Fig. 2, panel A). The cells were washed and then treated with Phos-FLAG in standard cell culture media. After four hours, the cells were lysed and analyzed by Western blot probing with an anti-FLAG antibody (Fig. 2, panel B). The Staudinger ligation of GalNAz-labeled glycoproteins occurred with very high specificity, as determined by comparison to the immunoreactivity of that of control lysates from cells treated with peracetylated GalNAc (Ac4GalNAc). To determine whether the metabolic properties of PC-3 cells reflect those of human prostate cancer tissue, we metabolically labeled precision-cut, thin tissue slice cultures of primary human prostate adenocarcinoma25,26 under virtually identical conditions. Staudinger ligation of intact tissue slice cultures with Phos-FLAG produced selective labeling of glycoproteins with no detectable background labeling as determined by Western blot (Fig. 2, panel C). Although further biochemical analysis was not possible due to limited material, this result supports the utility of PC-3 cells as models of human PC.
Figure 2.
(A) Selective enrichment and identification of cell surface azidoglycoproteins by live-cell Staudinger ligation with Phosphine-FLAG. Labeled proteins are captured by immunoprecipitation of the FLAG tag, followed by SDS–PAGE. Gel bands are then excised and analyzed by LC–MS/MS. (B) PC-3 cells and (C) PC tissue slice cultures were labeled with GalNAz or GalNAc. The lysates were treated with Phos-FLAG and analyzed by Western blotting with anti-FLAG antibody. Very little background labeling was apparent in the control cells (treated with GalNAc).
For glycoproteomic analysis, we immunoprecipitated the FLAG-labeled glycoproteins from PC-3 cell lysates and resolved them by one-dimensional SDS–PAGE. This step was critical to separate the anti-FLAG IgG heavy and light chains from the captured glycoproteins prior to further analysis. Gel bands corresponding to high molecular weight (>70 kDa) labeled protein were then cut out and processed for mass spectrometry (MS) analysis using standard ingel digest procedures27 (Supplementary Fig. 1). In a control experiment, we treated PC-3 cells with Ac4GalNAc and subjected them to the same cell surface labeling and glycoprotein enrichment procedure. While no anti-FLAG immunoreactive species were observed, wenonetheless excised the region of the gel corresponding to the labeled species from Ac4GalNAz-treated cells (>70 kDa) for MS analysis (Supplementary Fig. 1, panel B). This process provided a means to determine which proteins identified by MS were retrieved in a Gal-NAz-specific manner rather than nonspecifically. Three biological replicates were prepared and analyzed independently.
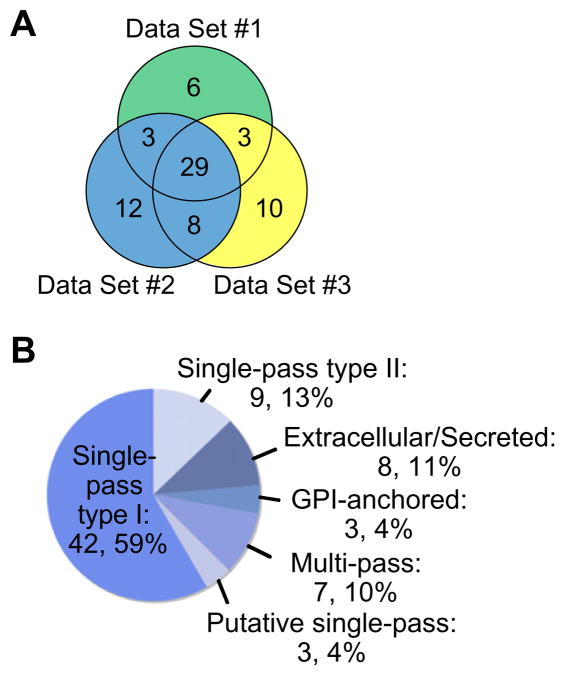
The digests were analyzed by liquid chromatography nanospray tandem MS (LC–MS/MS) and the data were generated with an LTQ XL MS. Using stringent filter criteria28 in our analysis of the data, we identified 71 nonredundant cell surface membrane, extracellular matrix, and secreted glycoproteins from the three data sets, with an average of 48 proteins per sample (Fig. 3, panel A). Over 40% of the proteins appeared in all three data sets (29 out of 71). Full tables of the identified proteins and individual data sets, including the control data derived from Ac4GalNAc-treated cells, are available in the Supporting Information (Supplementary Table 1). A variety of types of cell surface proteins was identified (Fig. 3, panel B), the majority of which are single-pass type I membrane proteins, in which the C-terminal domain is cytosolic and the N-terminal domain extracellular. Three glycosylphosphatidylinositol (GPI)-anchored proteins were identified as well, each of which is also putatively glycosylated. Approximately 10% were multi-pass membrane proteins, while another 10% were extracellular matrix or secreted proteins. Every cell surface protein identified in this study is putatively glycosylated according to the Swiss-Prot database.
Figure 3.
(A) A Venn diagram of PC-3 cell surface glycoproteins identified from three biological replicates, demonstrating a high degree of overlap in protein identification. (B) A pie chart of the diverse membrane classifications of the labeled cell surface proteins.
The selectivity of the method for cell surface proteins appears to be dependent on the total protein being processed in the enrichment step (Supplementary Table 2). The number of contaminating intracellular proteins identified in each data set scaled with increasing amounts of lysate subjected to immunoprecipitation. This observation likely results from incomplete removal of contaminating proteins during the anti-FLAG immunoenrichment, wherein the mild washing step becomes less effective with higher levels of protein. Further studies utilizing affinity tags that tolerate more stringent washing conditions are underway to address this issue. Importantly, of the proteins observed in the control data sets, fewer than 10% were membrane proteins; the majority were likely due to contamination during sample preparation (e.g., human keratin and related epidermal proteins).
The 29 glycoproteins identified in all three data sets in this study are listed in Table 1. A number of these proteins has been associated with disease progression. For example, CD44, a cell adhesion molecule known to possess chondroitin sulfate chains as well as both O- and N-glycans, participates in cancer dissemination and invasion and is highly prognostic of aggressive malignancy. 29 ICAM-1, another adhesion molecule, was recently identified using an antibody-based proteomics strategy as a marker of hormone-refractory prostate cancer cells.30 Notably, Wong and coworkers previously reported an N-glycan-specific glycoproteomic study of whole PC-3 cell lysates using alkynyl sialic acid precursor as a metabolic label for covalent capture and enrichment of sialylated glycoproteins23. Their identified species included both intracellular and cell surface molecules, including the three mentioned above. Although each study focused on a different subset of the glycoproteome in PC-3 cells (our study examines O-glycans and Wong’s focuses on N-glycans), a total of 29 glycoproteins were found in both studies, while the remainder were unique to one data set or the other. This further validates the utility of such methods for reliable identification of glycoproteins using metabolic oligosaccharide engineering.
Table 1.
Twenty-nine cell surface glycoproteiuns are found from all three biological replicates of GalNAz-treated PC-3 cells after live-cell labeling with Phosphine-FLAG
| Protein | Accession # | MW (kDa) | Protein probability | Membrane localization | Protein function |
|---|---|---|---|---|---|
| CD109 antigen | Q6YHK3 | 162 | 9.2 e-014 | GPI-anchor | Signal modulator |
| 5′-nucleotidase | P21589 | 63 | 2.2 e-011 | GPI-anchor | Nucleotide hydrolysis |
| CD97 antigen | P48960 | 92 | 1.8 e-006 | Multi-pass Putative | Cell adhesion |
| Transmembrane protein 2 | Q9UHN6 | 154 | 6.4 e-009 | Single-pass Putative | Cell signaling |
| CUB domain-containing protein 1 | Q9H5V8 | 93 | 2.7 e-006 | Single-pass | Adhesion/ECM association |
| Cell surface glycoprotein CD146 | P43121 | 72 | 1.1 e-015 | Single-pass type I | Cell adhesion |
| Integrin alpha-2 | P17301 | 129 | 1.0 e-030 | Single-pass type I | Laminin/collagen receptor |
| Integrin beta-1 | P05556 | 88 | 1.0 e-030 | Single-pass type I | Laminin receptor |
| Integrin alpha-6 | P23229 | 127 | 4.4 e-015 | Single-pass type I | ECM protein receptor |
| Integrin alpha-3 | P26006 | 119 | 1.0 e-013 | Single-pass type I | Cell adhesion |
| CD166 antigen | Q13740 | 65 | 5.6 e-016 | Single-pass type I | Hyaluronic acid receptor |
| CD44 antigen | P16070 | 82 | 2.0 e-010 | Single-pass type I | Laminin receptor |
| Integrin beta-4 | P16144 | 202 | 3.5 e-011 | Single-pass type I | Receptor, cell migration |
| Plexin-B2 | O15031 | 205 | 2.3 e-011 | Single-pass type I | Cell adhesion |
| Intercellular adhesion molecule 1 | P05362 | 58 | 2.8 e-009 | Single-pass type I | Laminin receptor |
| C-type mannose receptor 2 | Q9UBG0 | 167 | 6.5 e-009 | Single-pass type I | Endocytosis |
| Integrin alpha-1 | P56199 | 131 | 1.0 e-030 | Single-pass type I | Laminin/collagen receptor |
| Syndecan-1 | P18827 | 32 | 4.1 e-011 | Single-pass type I | Cytoskeleton/ECM link |
| Receptor-type tyrosine-protein phosphatase eta | Q12913 | 146 | 3.6 e-012 | type I | Phosphotyrosine hydrolysis |
| Integrin beta-5 | P18084Q | 88 | 6.3 e-009 | Single-pass type I | Cell adhesion |
| Integrin alpha-V | P06756 | 116 | 8.5 e-013 | Single-pass type I | Cell adhesion |
| Podocalyxin-like protein 1 | O00592 | 59 | 8.6 e-009 | Single-pass type I | Anti-adhesion |
| Tyrosine-protein kinase-like 7 | Q13308 | 118 | 2.3 e-010 | Single-pass type I | Cell adhesion |
| Epidermal growth factor receptor | P00533 | 134 | 3.8 e-010 | Single-pass type I | Proliferation/adhesion |
| Poliovirus receptor | P15151 | 45 | 3.8 e-006 | Single-pass type I | Cell adhesion |
| 4F2 cell surface antigen heavy chain | P08195 | 68 | 3.6 e-012 | Single-pass type II | Amino acid transporter |
| Ectonucleotide pyrophosphatase/phosphodiesterase family member 1 | P22413 | 105 | 1.0 e-011 | Single-pass type II | Pyrophosphate regulation |
| Aminopeptidase N | P15144 | 109 | 1.1 e-014 | Single-pass type II | Metalloprotease |
| Transferrin receptor protein 1 | P02786 | 85 | 9.9 e-013 | Single-pass type II | Endocytosis |
Several cell surface glycoproteins that were uniquely identified using GalNAz as a metabolic label stood out as particularly relevant to cancer progression. We were intrigued by two uniquely identified proteins: CD146 and integrin beta-4, both of which were identified in all three of our biological replicates with 28% and 33% total amino acid sequence coverage, respectively (Supplementary Table 3). CD146 was first characterized for its function in melanoma cell adhesion31,32, and is also known as MCAM, Mel-CAM or MUC18 (although the latter nomenclature may be misleading as CD146/MUC18 is not a member of the mucin (MUC) family of glycoproteins). Although CD146 has been implicated and characterized for its role in metastasis in several tissue types33, CD146 has also been characterized as a marker of metastatic disease in PC. High levels of CD146 protein and also hypermethylation of the CD146 gene were observed in PC tissues and cell lines, including PC-3, but not in healthy prostate tissue, suggesting that CD146 expression may serve as a marker of aggressive prostatic disease34,35. Furthermore, expression of CD146 in PC cells was found to increase metastatic ability in xenograft and transgenic mouse models of PC33,36. CD146 is a transmembrane cell adhesion molecule belonging to the immunoglobulin (Ig) super family. Although CD146’s glycosylation profile has not been extensively studied, one N-glycan has been annotated37 and 8 more are predicted. Analysis of the protein sequence using the NetOGlyc 3.1 algorithm38 suggests a high likelihood of O-glycosylation as well.
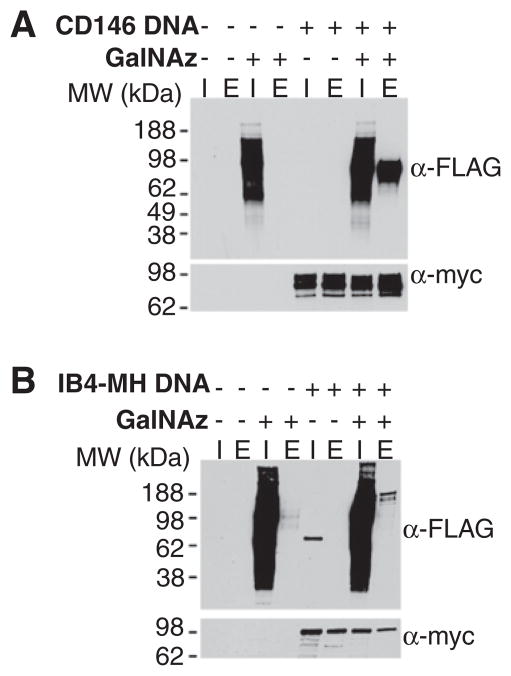
To validate CD146 as a bona fide metabolically labeled cell surface glycoprotein, we transiently expressed human CD146 bearing C-terminal myc and His6 tags in PC-3 cells. The cells were incubated with Ac4GalNAz or vehicle only, and cell surface labeled with Phos-FLAG as in the above proteomics experiments. Cell lysates were purified on Ni-NTA-agarose and eluted proteins were separated by SDS–PAGE and probed by Western blot with anti-FLAG and anti-myc antibodies (Fig. 4, panel A). CD146 showed strong GalNAz-dependent labeling, validating the approach as a means to identify cell surface glycoproteins relevant to disease.
Figure 4.
Validation of azide-specific labeling of Myc-His tagged CD146 (A) and integrin beta-4 (B). PC-3 cells were transfected with the indicated DNA (+) or mock transfected (−) and, at the same time, Ac4GalNAz (+) or DMSO (−, vehicle) was added to the culture media. After 24 h, the cells were labeled with Phos-FLAG and then lysed, and the lysates were purified on Ni-NTA agarose. Samples of the input (I, 1% of the total) and imidazole eluate (E, 5% of the total) were analysed by Western blot probing with an anti-FLAG antibody (top) or an anti-myc antibody (bottom).
Integrin beta-4 has been associated with angiogenesis39 and aggressive cancer progression40. In a report last year, the integrin α6β4 complex was linked to breast cancer cell invasion41. While these studies correlate protein expression with disease, none has examined the role of glycosylation of integrin beta-4 with malignant transformation. Out of 5 putative N-glycosylation sites, one has been annotated42, and no reports have yet been published regarding O-glycosylation, if any, of integrin beta-4. A C-terminal myc and His6-tagged construct for integrin beta-4 was acquired from the Addgene repository. The protein was expressed in PC-3 cells and analyzed as performed above with CD146. Again, GalNAz-specific Phos-FLAG labeling of the protein was observed (Fig. 4, panel B). Differences in the glycan heterogeneity or the number of available epitopes for antibody recognition may explain the banding patterns observed in the anti-FLAG versus anti-myc blots.
These results validate the strategy of metabolic labeling and identification of potential cell surface glycoprotein markers of disease. Metabolic labeling methods are distinct from and complementary to lectin and periodate-based enrichment techniques. While the latter methods capture the steady-state glycoprotein repertoire, metabolic labeling selects for glycoproteins with high rates of de novo biosynthesis; consequently, metabolic labeling may be more sensitive to changes in biosynthetic flux that accompany disease progression. In addition, the azide is a versatile handle for attachment of a wide variety of probes tailored for affinity capture or quantitation purposes. Further studies to elucidate the structure of the glycan moieties present on these proteins are underway. Metabolic labeling with GalNAz may in the future facilitate glycan characterization, quantitation, and site mapping – all particularly obstinate challenges in the study of mucin-type O-glycosylation.
Materials and methods
Metabolic labeling of cell surface glycoproteins
Conditions for primary human PC tissue slice culture are available in the Supplementary data. PC-3 cells were plated at approximately 100,000 cells mL−1 in Ham’s F12 media containing L-glutamine, 10% v/v FBS, 100 units mL−1 penicillin, and 100 g mL−1 streptomycin and 50 μM Ac4GalNAz, or the control sugar, Ac4Gal- NAc, and incubated at 37 °C under 5% CO2 atmosphere. After three days, the media were removed, and the cells were washed twice with PBS. To label the cell surfaces, 5 mL of fresh media containing 1 mM Phos-FLAG was then added and the plates incubated at 37 °C for 4 h. The Phos-FLAG media was then aspirated and the cells washed twice with cold PBS. To lift the cells, 1 mM sterile EDTA (Invitrogen) in PBS was added, and the cells were scraped from the plates and collected. The cells were pelleted and washed with PBS and then resuspended in lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% v/v NP-40, and protease inhibitors (Roche Diagnostics). The cells were fully lysed by sonication (Misonex) while on ice. This protocol was followed using two additional biological replicates to generate two additional sets of samples for further analysis.
Cell surface glycoprotein capture and MS analysis
Lysates were first pre-cleared with Ultralink Protein A/G resin (ThermoFisher) for one hour with end-over-end inversion and then centrifuged at 10,000g to remove the resin and insoluble protein. The protein concentrations for the supernatants were then determined by DC assay. Approximately 2 mg of each sample from set #1, 5 mg from set #2, and 10 mg from set #3, were then diluted to a final concentration of 0.2 mg mL−1 with additional lysis buffer (Data Sets #1, #2, and #3, respectively). M2 anti-FLAG antibody (Sigma) was then added to both samples for a final antibody concentration of 15 μgmL−1, and the solutions were incubated overnight at 4 °C with mixing. Protein A/G resin was then equilibrated in lysis buffer and added to the lysate/antibody mixture to precipitate the labeled protein. The solutions were mixed for 1.5 h before centrifugation at 750g to pellet the resin and bound protein. The resin was then washed four times with lysis buffer and three times with lysis buffer containing 0.1% v/v SDS. A minimal volume of 4× SDS protein loading buffer with reducing agent was added, and the samples were boiled for 10 min at 100 °C to elute the protein. Approximately 10% of each elution was resolved by SDS–PAGE using 4–12% bis–tris Criterion gels, followed by transfer to nitrocellulose. The blots were blocked overnight in 5% w/v dry non-fat milk in phosphate-buffered saline containing 0.1% v/v Tween-20 (PBST). The blots were then incubated with a fresh milk solution containing M2 anti-FLAG antibody conjugated with horseradish peroxidase (Sigma, 1:5000) for one hour and washed with PBST prior to incubation with SuperSignal Pico West Chemiluminescent substrate and exposure to film and development to verify azide-specific signal. The remaining 90% of the elutions were resolved on 4–12% Bis–Tris Criterion gels and then stained with Simply Blue Safe Stain (Invitrogen) according to the manufacturer’s instructions. Bands of approximately 1 mm width were then cut from the control and azide-treated gels and analyzed by mass spectrometry using conditions described in the literature and the Supplementary data.27
Validation of azidosugar incorporation into PC-3 cell surfaces
The pcDNA3.1/Myc-His beta 4 (IB4-MH) construct was obtained from Addgene.43 Methods for generating the CD146-Myc-His6 (CD146-MH) construct are described in the Supplementary data. PC-3 cells were seeded in eight 10 cm plates at 150,000 cells mL−1 and incubated for 24 h prior to transfection with mock (four plates), IB4-MH (two plates), or CD146-MH DNA (two plates) using the TransIT-Prostate Transfection Kit (Mirus) according to the manufacturer’s instructions. At the same time, 50 μM Ac4GalNAz was added to half of the transfections and DMSO to the other four plates. The plates were then incubated for 24 h. The cell surfaces were then labeled with Phos-FLAG as described above, followed by lysis into buffer containing 8 M urea, 15 mM imidazole, and 1% Triton X-100 in PBS. Protein concentration of the supernatant was determined by BCA assay (Thermo Scientific). Equal amounts of protein were then incubated with Ni-NTA agarose (Qiagen) with end-over-end rotation for 45 min. The resin was washed three times with buffer, followed by three incubations with buffer containing 250 mM imidazole to elute the bound protein. Approximately 1% of the inputs and 5% of the elutions were then resolved by SDS–PAGE and transferred to nitrocellulose membrane. The blots were blocked with 5% w/v bovine serum albumin (BSA) or dry non-fat milk in PBST for one hour. The BSA-blocked blots were incubated with α-c-myc 9E10 antibody (1:1000, Santa Cruz Biotechnology), followed by two washes with PBST. The blots were then incubated with a mouse light chain-specific secondary antibody conjugated to HRP (1:5000, Southern Biotech), and the milk-blocked blots were incubated with M2 α-FLAG conjugated to HRP (1:1000). The blots were then washed with PBST prior to development as described above.
Supplementary Material
Acknowledgments
The authors thank B. Smart for his assistance with mass spectrometry analysis. C.R.B. acknowledges support from the National Institutes of Health (GM66047). D.M.P. acknowledges support from the National Institutes of Health (U01CA128416). C.R.B. and D.M.P. acknowledge joint support from the Department of Defense (PC080659). S.C.H. was supported by a pre-doctoral fellowship from the National Science Foundation. M.B. was supported by Howard Hughes Medical Institute and is a fellow of the Life Sciences Research Foundation.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmcl.2011.05.045.
References and notes
- 1.American Cancer Society. Cancer Facts & Figures. 2009. [Google Scholar]
- 2.Albertsen PC. Urology. 2010;75:399. doi: 10.1016/j.urology.2009.08.078. [DOI] [PubMed] [Google Scholar]
- 3.Tabares G, Radcliffe CM, Barrabes S, Ramirez M, Aleixandre RN, Hoesel W, Dwek RA, Rudd PM, Peracaula R, de Llorens R. Glycobiology. 2006;16:132. doi: 10.1093/glycob/cwj042. [DOI] [PubMed] [Google Scholar]
- 4.Tajiri M, Ohyama C, Wada Y. Glycobiology. 2008;18:2. doi: 10.1093/glycob/cwm117. [DOI] [PubMed] [Google Scholar]
- 5.Hollingsworth MA, Swanson BJ. Nat Rev Cancer. 2004;4:45. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 6.Andrianifahanana M, Moniaux N, Batra SK. Biochim Biophys Acta, Rev Cancer. 2006;1765:189. doi: 10.1016/j.bbcan.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Proc Natl Acad Sci USA. 2004;101:811. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schut IC, Waterfall PM, Ross M, O’Sullivan C, Miller WR, Habib FK, Bayne CW. BJU Int. 2003;91:278. doi: 10.1046/j.1464-410x.2003.03062.x. [DOI] [PubMed] [Google Scholar]
- 9.Dube DH, Bertozzi CR. Nat Rev Drug Discovery. 2005;4:477. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 10.Fuster MM, Esko JD. Nat Rev Cancer. 2005;5:526. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 11.Ornstein DK, Rayford W, Fusaro VA, Conrads TP, Ross SJ, Hitt BA, Wiggins WW, Veenstra TD, Liotta LA, Petricoin EFJ. Urology. 2004;172:1302. doi: 10.1097/01.ju.0000139572.88463.39. [DOI] [PubMed] [Google Scholar]
- 12.Schiess R, Wollscheid B, Aebersold R. Mol Oncol. 2009;3:33. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung KY, Cho WR, Regnier FE. J Proteome Res. 2009;8:643. doi: 10.1021/pr8007495. [DOI] [PubMed] [Google Scholar]
- 14.Madera M, Mann B, Mechref Y, Novotny MV. J Sep Sci. 2008;31:2722. doi: 10.1002/jssc.200800094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soltermann A, Ossola R, Kilgus-Hawelski S, von Eckardstein A, Suter T, Aebersold R, Moch H. Cancer Cytopathology. 2008;144:124. doi: 10.1002/cncr.23349. [DOI] [PubMed] [Google Scholar]
- 16.Hanisch FG, Teitz S, Schwientek T, Muller S. Proteomics. 2009;9:710. doi: 10.1002/pmic.200800492. [DOI] [PubMed] [Google Scholar]
- 17.Schwientek T, Mandel U, Roth U, Muler S, Hanisch FG. Proteomics. 2007;7:3264. doi: 10.1002/pmic.200600793. [DOI] [PubMed] [Google Scholar]
- 18.Durham M, Regnier FE. J Chromatogr, A. 2006;1132:165. doi: 10.1016/j.chroma.2006.07.070. [DOI] [PubMed] [Google Scholar]
- 19.Agard NJ, Bertozzi CR. Acc Chem Res. 2009;42:788. doi: 10.1021/ar800267j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dube DH, Prescher JA, Quang CN, Bertozzi CR. Proc Natl Acad Sci USA. 2006;103:4819. doi: 10.1073/pnas.0506855103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hang HC, Yu C, Kato DL, Bertozzi CR. Proc Natl Acad Sci USA. 2003;100:14846. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyce M, Carrico IS, Ganguli AS, Yu S, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Proc Natl Acad Sci USA. 2011;108:3141. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Hanson SR, Hsu TL, Weerapana E, Kishikawa K, Simon GM, Cravatt BF, Wong CH. J Am Chem Soc. 2007;129:7266. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yang L, Nyalwidhe JO, Guo S, Drake RR, Semmes O. J Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M110.007294. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gurcel C, Vercoutter-Edouart AS, Fonbonne C, Mortuaire M, Salvador A, Michalski JC, Lemoine J. Anal Bioanal Chem. 2008;390:2089. doi: 10.1007/s00216-008-1950-y. [DOI] [PubMed] [Google Scholar]
- 25.Parrish AR, Gandolfi AJ, Brendel K. Life Sciences. 1995;57:1901. doi: 10.1016/0024-3205(95)02176-j. [DOI] [PubMed] [Google Scholar]
- 26.Hallstrom T, Jaamaa S, Monkkonen M, Peltonen K, Andersson LC, Medema RH, Peehl DM, Laiho M. Proc Natl Acad Sci USA. 2007;104:7211. doi: 10.1073/pnas.0609299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowley A, Choudhary JS, Marzioch M, Ward MA, Weir M, Solari RCE, Blackstock WP. Methods. 2000;20:383. doi: 10.1006/meth.2000.0951. [DOI] [PubMed] [Google Scholar]
- 28.Tabb DL, McDonald WH, Yates JR. J Proteome Res. 2002;1:21. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naor D, Sionov RV, IshShalom D. CD44: Structure, function, and association with the malignant process. Advances in Cancer Research. 1997;71:241. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 30.Conrad F, Zhu XD, Zhang X, Chalkley RJ, Burlingame AL, Marks JD, Liu B. J Mol Med. 2009;87:507. doi: 10.1007/s00109-009-0446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann JM, Holzmann B, Breitbart EW, Schmiegelow P, Riethmuller G, Johnson JP. Cancer Res. 1987;47:841. [PubMed] [Google Scholar]
- 32.Ouhtit A, Gaur RL, Elmageed ZYA, Fernando A, Thouta R, Trappey AK, Abdraboh ME, El-Sayyad HI, Rao P, Raj MGH. Biochim Biophys Acta, Rev Cancer. 2009;1795:130. doi: 10.1016/j.bbcan.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Wu GJ. Curr Genomics. 2005;6:333. [Google Scholar]
- 34.Wu GJ, Varma VA, Wu MWH, Wang SW, Qu PP, Yang HC, Petros JA, Lim SD, Amin MB. Prostate. 2001;48:305. doi: 10.1002/pros.1111. [DOI] [PubMed] [Google Scholar]
- 35.Liu JW, Nagpal JK, Jeronimo C, Lee JE, Henrique R, Kim MS, Ostrow KL, Yamashita K, van Criekinge V, Wu GJ, Moon CS, Trink B, Sidransky D. Prostate. 2008;68:418. doi: 10.1002/pros.20709. [DOI] [PubMed] [Google Scholar]
- 36.Wu GJ, Fu PP, Chiang CF, Huss WJ, Greenberg NM, Wu MWH. J Urology. 2005;173:1778. doi: 10.1097/01.ju.0000154643.30048.2c. [DOI] [PubMed] [Google Scholar]
- 37.Chen R, Jiang XN, Sun DG, Han GH, Wang FJ, Ye ML, Wang LM, Zou HF. J Proteome Res. 2009;8:651. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 38.Julenius K, Molgaard A, Gupta R, Brunak S. Glycobiology. 2005;15:153. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 39.Ohira T, Akutagawa S, Usuda J, Nakamura T, Hirano T, Tsuboi M, Nishio K, Taguchi F, Ikeda N, Nakamura H, Konaka C, Saijo N, Kato H. Oncol Rep. 2002;9:723. [PubMed] [Google Scholar]
- 40.Kitajiri S, Hosaka N, Hiraumi H, Hirose T, Ikehara S. Pathol Int. 2002;2:438. doi: 10.1046/j.1440-1827.2002.01379.x. [DOI] [PubMed] [Google Scholar]
- 41.Kim TH, Kim HI, Soung YH, Shaw LA, Chung J. Mol Cancer Res. 2009;7:1605. doi: 10.1158/1541-7786.MCR-09-0102. [DOI] [PubMed] [Google Scholar]
- 42.Liu T, Qian WJ, Gritsenko MA, Camp DG, Monroe ME, Moore RJ, Smith RD. J Proteome Res. 2005;4:2070. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dans M, Gagnoux-Palacios L, Blaikie P, Klein S, Mariotti A, Giancotti FG. J Biol Chem. 2001;276:1494. doi: 10.1074/jbc.M008663200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.