Abstract
Over 90 capsular serotypes of Streptococcus pneumoniae, a common nasopharyngeal colonizer and major cause of pneumonia, bacteremia, and meningitis, are known. It is unclear why some serotypes can persist at all: they are more easily cleared from carriage and compete poorly in vivo. Serotype-specific immune responses, which could promote diversity in principle, are weak enough to allow repeated colonizations by the same type. Here we show that weak serotype-specific immunity and an acquired response not specific to the capsule can together reproduce observed diversity. Serotype-specific immunity stabilizes competition, and acquired immunity to noncapsular antigens reduces fitness differences. Our model can be used to explain the effects of pneumococcal vaccination and indicates general factors that regulate the diversity of pathogens.
The growing use of multivalent vaccines against antigenically variable pathogens creates a need to understand the individual- and population-level forces that maintain the coexistence of pathogen strains so that the evolutionary effects of such vaccines can be anticipated. One of the oldest ideas in ecology is that species are able to coexist by occupying unique niches (1). Stable coexistence involving a shared limiting resource can result when individuals compete more strongly with members of their own species than other species, creating a negative feedback loop that limits growth at high abundance (a “stabilizing” effect) (2). Among closely related pathogen strains, this kind of niche separation often arises through antigenic variation. By differing in their appearance to the immune system, competing strains effectively subdivide the host population, with each host a “habitat” more favorable to strains not yet encountered and for which strain-specific immunity is lacking. In this way, antigenic types to which few hosts are immune maintain a relative advantage. Niche partitioning through antigenic variation can explain the dynamics of protozoal (3), viral (4), and bacterial (5) pathogen populations and is a core assumption of theoretical models of strain competition (6).
Streptococcus pneumoniae, or pneumococcus, is a ubiquitous pathogen and a leading cause of pneumonia, meningitis, and bacteremia in children worldwide (7). The serotypes of pneumococcus are distinguished by their polysaccharide capsules. Licensed pneumococcal vaccines are serotype-specific and, despite overall benefits, have led to an increase in disease from nonvaccine serotypes. Thus, understanding the forces shaping pneumococcal serotype dynamics is important to interpret the effects of vaccine on disease and predict trends. Serotype-specific (henceforth, “anticapsular”) immunity might in principle provide a mechanism to maintain the diversity of pneumococcal serotypes. To permit coexistence, niche differences must be large enough to overcome differences in fitness that would otherwise lead to competitive exclusion (2). Many biological and epidemiological properties of pneumococcal strains, including properties that contribute directly to fitness, strongly correlate with the thickness of the capsule, which depends on the serotype. Serotypes with thicker capsules tend to have longer durations of nasopharyngeal carriage in young children (8), a competitive advantage in the nasopharynx (9), and a greater ability to escape neutrophil-mediated phagocytosis (10) in vitro, for example, compared to serotypes with thinner capsules. These traits also predict the prevalence of serotypes in carriage (9, 10), which is relatively consistent across human populations (Section S1): serotypes with thicker capsules are relatively more common. Studies of anticapsular immunity have shown, however, that the serotype-specific immunity elicited by natural exposure to pneumococci is imperfect, frequently allowing repeated colonizations with the same serotype. Three separate studies have found evidence of strong immunity for one serotype (individuals who have carried type 14 are ~90% less likely to carry it again); however, they suggest that such immunity for all other serotypes, if it exists, is much less protective (e.g., point estimates of ≤50% protection) (11–13). We sought to understand how such relatively weak type-specific immune responses could support the coexistence of serotypes with substantial fitness differences.
To do so, we developed an individual-based model (Sections S2–S5) constructed to avoid an intrinsic bias toward coexistence (14). We simulated the transmission dynamics of 25 serotypes that differed only in their intrinsic durations of carriage and their in vivo competitive abilities (15, 16). These two components of fitness were positively correlated: serotypes with longer intrinsic durations of carriage were also better at excluding other serotypes in vivo (9). This competitive ability was modeled as a reduction in a host’s susceptibility to a colonizing serotype, with the size of the reduction set by the fitness of any resident serotypes. Anticapsular immunity reduced a host’s susceptibility to colonization by a fraction σ if the host had ever previously cleared that serotype (11). This fraction was the same for all serotypes but was varied across simulations. Because anticapsular immunity is thought to result from antibodies, once acquired, it was assumed to persist for the duration of a host’s life. To facilitate comparisons of simulations involving different parameter values, we fitted transmission rates so that the total carriage prevalence of pneumococcus in children <5 y old was 40%. Simulations were run for 150 y, and serotype frequencies from the last 20 y were analyzed.
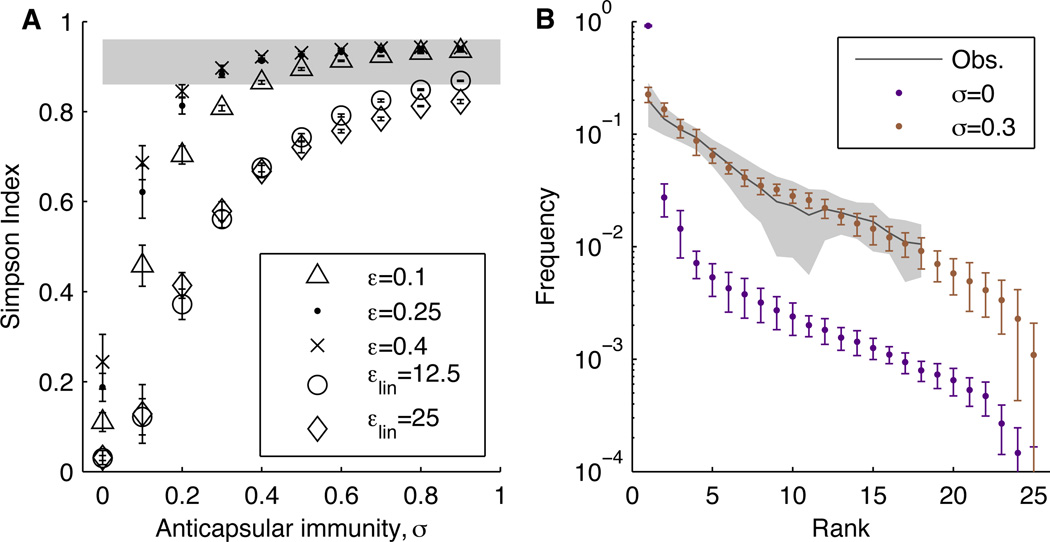
We first considered models with anticapsular immunity and acquired non-serotype-specific immunity (henceforth “nonspecific”) that reduced each serotype’s duration of carriage (17) by a fixed amount for every past exposure to any serotype (Fig. S4). In these cases, the presence of anticapsular immunity, a stabilizing mechanism, promoted serotype diversity (Fig. 1A). However, anticapsular immunity alone was insufficient to overcome differences in serotypes’ fitnesses: even implausibly high levels of immunity (e.g., a 90% reduction in susceptibility from prior homologous carriage for each serotype) could not generate the high diversity observed in studies (Section S6). Furthermore, high levels of anticapsular immunity in this scenario created an unrealistically low carriage prevalence and diversity in adults (Section S6).
Fig. 1.
Serotype diversity is sensitive to the form of acquisition of nonspecific immunity and the strength of anticapsular immunity. (A) Diversity, measured by the Simpson Index, as a function of the strength of anticapsular immunity for different forms of acquired nonspecific immunity (durations that decay linearly to 0, εlin = 12.5 and εlin = 25; durations that decay exponentially to a nonzero value, ε = 0.10, ε = 0.25, and ε = 0.40; Section S4). The Simpson Index equals the probability of randomly picking two serotypes from the population with replacement; it was calculated as 1 - D, where D = Σpi2, and pi denotes the frequency of the ith species. Measures were obtained by averaging annual estimates of the Simpson Index for each of the last 20 y of each simulation. The shaded area shows the range of diversity observed in carriage studies. (B) Observed rank-frequency distributions from carriage studies and the default model (ε = 0.25) without (σ = 0) and with (σ = 0.3) anticapsular immunity. The gray line shows the mean distribution from carriage studies, and the shading indicates the S.D. (Section S1). Error bars show S.D. based on ten simulations.
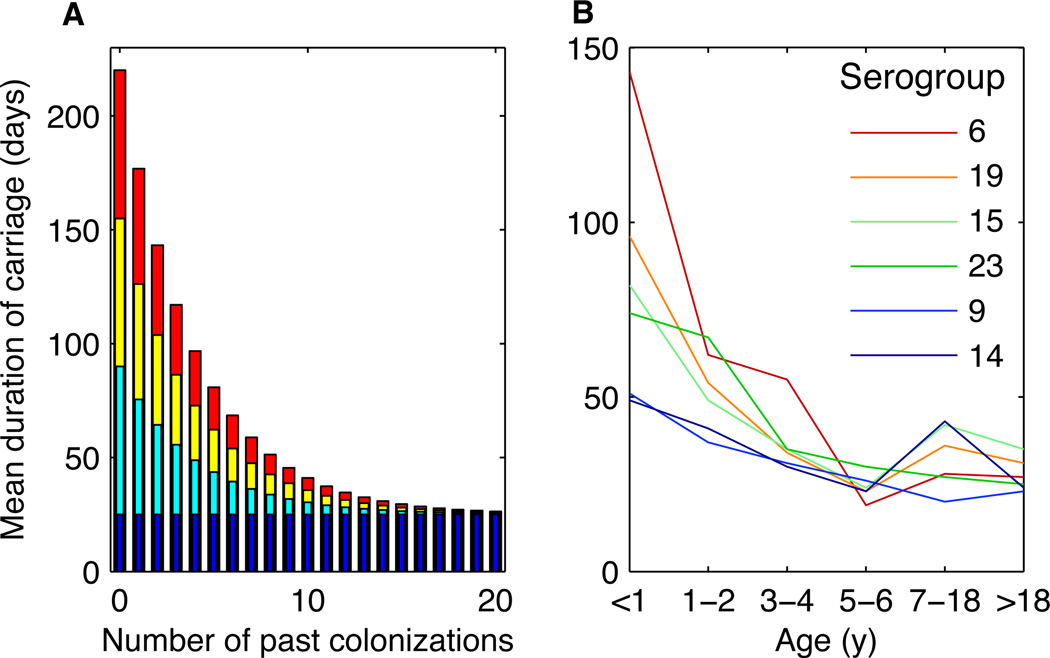
Epidemiologic studies of carriage duration (8) suggest that as individuals acquire nonspecific immunity to pneumococcus, the duration of carriage declines for all serotypes, approaching a fixed value of several weeks in individuals with multiple exposures, regardless of serotype (Fig. 2). This mechanism is thought to be largely driven by CD4+ Th17 cells (18, 19). Modeling this form of nonspecific immunity generated realistic patterns of diversity (Fig. 1 and Section S6). A consequence of this nonlinear decline in duration is that in highly exposed individuals, the fitness advantage of the “best” serotypes is reduced because their durations fall disproportionately (9). Acquired nonspecific immunity thus reduces fitness variation between serotypes, and this reduction is most pronounced in older individuals. This phenomenon might explain the higher diversity of serotypes (in both carriage and invasive disease) in older children and adults (20, 21). Some anticapsular immunity remains necessary in this scenario, underscoring the joint contribution of “niche” (stabilizing) and “neutral” (equalizing) mechanisms to coexistence (Fig. 1B) (2). In general, the simulated serotype distributions match observed distributions when anticapsular immunity confers a 30–60% reduction in susceptibility to future colonizations with that type (Figs. 1B and S9). This estimate is consistent with the finding that the confidence intervals of serotype-specific reductions in susceptibility from prior homologous carriage overlap in the range of 34–36% (11).
Fig. 2.
The rate of acquisition of nonspecific immunity was modeled as a linear or exponential function, with the latter supported by observations. (A) The duration of carriage of four serotypes modeled as an exponential function of the number of times a host has previously cleared any serotype of pneumococcus (ε = 0.25). The durations of the different serotypes are shown as superimposed bars with the color indicating the serotype’s fitness (red, most fit; blue, least fit). (B) Observed mean durations of carriage of different serogroups (serotypes or groups of antigenically related serotypes) as a function of host age from (8).
Models with intermediate levels of anticapsular immunity and a nonlinear reduction in the duration of carriage with exposure reproduced other key aspects of pneumococcal dynamics. These models produced relatively consistent rankings of serotype frequencies across populations, a realistic reduction in the duration of carriage with age, higher pneumococcal diversity in adults than in children, and realistic rates of co-colonization (Table 1). Large, multiannual fluctuations in the abundance of rarer serotypes were a common feature in models with anticapsular immunity, and similar fluctuations have been observed in time series of disease isolates (Section S6.6) (22).
Table 1.
Assumptions and constraints of the model and patterns reproduced by the calibrated model.
| Figure | Description/Reference | |
|---|---|---|
|
Epidemiologic observations used to define model assumptions | ||
| Weak to nonexistent anticapsular immunity | - | (11–13), Sections S2 |
| Reduction in the duration of carriage from nonspecific immunity | 1 | (17–19, 22), Sections S2 and S4.1 |
| In vivo competition strength correlating with serotype fitness (duration of carriage) | - | (9, 15), Section S2 |
|
Model outputs used to calibrate parameter values and functional forms | ||
| Total carriage prevalence in children <5 y | - | Table S1 |
| Diversity (Simpson Index) | 1A | Main text |
| Rank-frequency distribution | 1B, S9 | Sections S1 and S6.2 |
|
Observed patterns reproduced by the calibrated model | ||
| Stability of rank order | S8, S10 | Sections S1 and S6.2 |
| Decline in carriage duration with age | S11 | Section S6.3 |
| Increase in serotype diversity with age | S12 | Sections S1 and S6.4 |
| Frequency of co-colonizations | S13 | Section 6.5 |
| Epidemics of rarer serotypes | S14, S15 | Section 6.6 |
| Serotype replacement after vaccination | 3, A and B | Main text |
| Brief increase in diversity after vaccination | 3C | Main text |
The first section lists the major assumptions of the models and the empirical support for these assumptions. The second section lists the criteria by which the different models of immunity were judged. The third section lists additional patterns reproduced by the calibrated model (σ = 0.3, ε = 0.25). For each pattern, the supporting figure(s) and the sections in which the competing immune models are judged are listed.
The major results were robust to the size of the host population, the rate of immigration, and the rate of acquiring nonspecific immunity, and they were modestly affected by host population structure and the total prevalence of pneumococcus (Sections S6.7–S6.11). Quantitative validation of our model is limited, however, by the uncertainty in the actual intrinsic durations and transmissibilities of serotypes in nature. Our model assumed that serotypes were equally infectious per day (in the absence of evidence to the contrary) and had evenly distributed intrinsic durations of carriage. Consistent with theory (2), patterns of diversity were sensitive to the density of serotypes within this range (Fig. S16). For example, when we simulated 15 serotypes instead of 25 within the same range, more anticapsular immunity (higher σ) was required to overcome the larger differences in fitness between serotypes. With 35 serotypes, slightly less anticapsular immunity was needed. Our model used a relatively dense packing of fitnesses (Section S4.1), implying that the conditions we identify that promote diversity are conservative requirements.
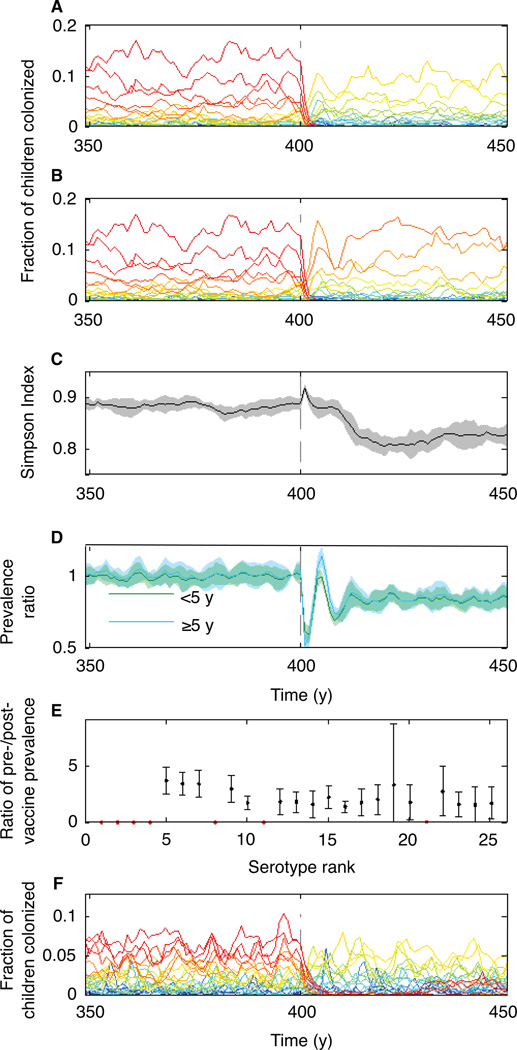
The dynamics of serotypes competing through naturally acquired immunity shed light on serotype replacement after vaccination. Unlike natural colonization, the pneumococcal conjugate vaccine induces anticapsular immunity that is strongly protective against future carriage (23). We expanded our model to include vaccination, assuming that vaccine-induced immunity, like naturally induced anticapsular immunity, permanently reduced susceptibility to future colonizations with specific serotypes. The strengths of natural and vaccine-acquired immunity were varied across simulations. Simulating vaccination with realistic parameters reproduced the observed rapid and sustained replacement of serotypes targeted by the vaccine (24) when the anticapsular immunity induced by the vaccine was stronger than the naturally occurring form (Fig. 3, A and B). In many scenarios, serotype diversity briefly increased in the few years following vaccination, a trend observed in human populations (Fig. 3C; Section S7) (24, 25). In addition, introduction of the simulated vaccine caused the prevalence of total carriage to fluctuate before approaching a new equilibrium (Fig. 3D). In human populations, changes in overall carriage post-vaccination have ranged from small increases to large decreases (Section S7.2). Whether the simulated serotype diversity or total carriage prevalence ultimately returned to pre-vaccine levels depended on the ranks of the targeted serotypes (Section S7.1). All non-vaccine serotypes increased in frequency following vaccine introduction in approximate proportion to their pre-vaccine frequencies (Fig. 3E). The precise effects of vaccination on carriage and invasive disease probably depend on the extent of natural versus vaccine-induced cross-immunity between potentially cross-reacting serotypes: certain serogroups (6, 19, and 23) contain several antigenically similar serotypes, of which only one was in the heptavalent conjugate vaccine (26) (Section S7.1). Effects might also depend on the vaccination schedule and rate of introduction, and the efficacy of the vaccine with respect to each serotype.
Fig. 3.
Vaccination causes at least short-term serotype replacement and can lower total carriage. (A, B) Sample dynamics involving vaccination assuming σ = 0.3. The prevalences (in children <5 y old) of individual serotypes, colored by their intrinsic fitness as in Fig. 2 are shown. The vaccine was given to all children six months old starting in year 400 (dashed line) and had a 60% efficacy against targeted serotypes: (A) Vaccination targeting the seven most intrinsically fit serotypes (ranks 1–7). (B) Vaccination targeting serotypes with ranks analogous to those included in the heptavalent pneumococcal conjugate vaccine (ranks 1–4, 8, 11, and 21; Section S7.1). (C) Simpson Index in children <5 y from the scenario in (B). (D) Total carriage prevalence (as a ratio of each year’s prevalence to the prevalence in year 399) over time in children <5 y and in individuals ≥5 y from the scenario in (B). (E) The ratio of each serotype’s post-vaccine prevalence (its mean prevalence in years 410–419) to its pre-vaccine prevalence (years 300–399) from the scenario in (B). Targeted serotypes are in red. In (C)-(E), shading or error bars show S.D. based on ten simulations. (F) As in (A), vaccination targeting the seven fittest serotypes but with natural immunity (σ = 0.7) stronger than vaccine-induced immunity.
A theoretically interesting prediction of the model is that sustained reductions in anticapsular and/or nonspecific immunity following vaccination can allow previously excluded serotypes to reinvade successfully if introduced through immigration. If vaccine-induced, serotype-specific immunity to carriage is weaker than that obtained through natural exposure, targeted serotypes can resurge after several years to decades (Fig. 3F). This transient period of low prevalence after vaccination is similar to the “honeymoon period” following vaccine introduction at a coverage just below what is required to eliminate transmission through herd immunity (27). Instead of arising from unvaccinated individuals, the resurgence here comes from incomplete immunity, particularly a reduction in anticapsular immunity. Even when vaccine-induced immunity is stronger than natural immunity, if total carriage drops precipitously from vaccination, the resulting decline in nonspecific immunity can also permit the return of previously eradicated serotypes (Fig. S18).
Previous efforts to model carriage of nasopharyngeal colonizing bacteria have focused on vaccine effects and have assumed, rather than tried to explain, coexistence of serotypes (28–30). This study has shown that the diversity of a pathogen can be maintained simply by the interaction of acquired specific and nonspecific immunity, despite fitness differences between serotypes. The sensitivity of the results to the distribution of simulated serotypes’ growth rates raises the intriguing question of what upper limits exist for the number of pathogen strains that can coexist stably in a host population and how this depends on the relation between their antigenicity and intrinsic fitness (31). More broadly, this study supports a growing perspective in ecology that recognizes the joint role of niche and neutral dynamics in shaping diversity (2, 32). As antigen-specific vaccines are introduced against pathogens such as human papillomavirus, meningococcus, malaria and other polymorphic infections, these findings suggest that attention to the details of immune responses may be required to explain prevaccine patterns and make predictions about the effects of vaccines on pathogen populations.
Supplementary Material
Acknowledgements
We thank Osman Abdullahi and Anthony Scott for sharing their data on the duration of carriage, Eugene Lee for help reviewing carriage studies, and Catherine Laine for sharing an unpublished earlier review of such studies. Caroline Buckee, William Hanage, Bruce Levin, Richard Malley, Virginia Pitzer, and Daniel Weinberger, and Jon Zelner gave helpful comments. Computations were run on the Odyssey cluster supported by the FAS Science Division Research Computing Group at Harvard University. Email scobey@hsph.harvard.edu for the location of the code repository. The project was supported by awards 5R01AI048935 from the NIAID and 1F32GM97997 and U54GM088558 from the NIGMS. The content is solely the responsibility of the authors and does not necessarily represent the views of the NIGMS or the NIH. M.L. has received consulting fees or honoraria from Pfizer, Novartis, AIR Worldwide, and the Avian/Pandemic Flu Registry (Outcome Sciences), supported by Roche.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Hutchinson GE. Quantitative Biology. 1957;22:415. [Google Scholar]
- 2.Chesson P. Annu Rev Ecol Syst. 2000;31:343. [Google Scholar]
- 3.Buckee CO, Bull PC, Gupta S. Proc Biol Sci. 2009 Feb 7;276:477. doi: 10.1098/rspb.2008.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koelle K, Cobey S, Grenfell B, Pascual M. Science. 2006;314:1898. doi: 10.1126/science.1132745. [DOI] [PubMed] [Google Scholar]
- 5.Buckee CO, Gupta S, Kriz P, Maiden MC, Jolley KA. Proc Biol Sci. 2010 Jun 7;277:1635. doi: 10.1098/rspb.2009.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta S, Ferguson NM, Anderson RM. Science. 1998;280:912. doi: 10.1126/science.280.5365.912. [DOI] [PubMed] [Google Scholar]
- 7.O'Brien KL, et al. Lancet. 2009 Sep 12;374:893. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 8.Hogberg L, et al. J Clin Microbiol. 2007 Mar;45:948. doi: 10.1128/JCM.01913-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipsitch M, et al. Epidemiology. 2012 Mar 21; [Google Scholar]
- 10.Weinberger DM, et al. PLoS Pathog. 2009 Jun;5:e1000476. doi: 10.1371/journal.ppat.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberger DM, et al. J Infect Dis. 2008 Jun 1;197:1511. doi: 10.1086/587941. [DOI] [PubMed] [Google Scholar]
- 12.Hill PC, et al. Clin Infect Dis. 2008 Mar 15;46:807. doi: 10.1086/528688. [DOI] [PubMed] [Google Scholar]
- 13.Goldblatt D, et al. J Infect Dis. 2005 Aug 1;192:387. doi: 10.1086/431524. [DOI] [PubMed] [Google Scholar]
- 14.Lipsitch M, Colijn C, Cohen T, Hanage WP, Fraser C. Epidemics. 2009 Mar;1:2. doi: 10.1016/j.epidem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipsitch M, et al. Vaccine. 2000 Jun 15;18:2895. doi: 10.1016/s0264-410x(00)00046-3. [DOI] [PubMed] [Google Scholar]
- 16.Lysenko ES, Lijek RS, Brown SP, Weiser JN. Curr Biol. 2010 Jul 13;20:1222. doi: 10.1016/j.cub.2010.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray BM, Converse GM, 3rd, Dillon HC., Jr J Infect Dis. 1980 Dec;142:923. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 18.Lu YJ, et al. PLoS Pathog. 2008;4:e1000159. doi: 10.1371/journal.ppat.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malley R, et al. Proc Natl Acad Sci U S A. 2005 Mar 29;102:4848. doi: 10.1073/pnas.0501254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdullahi O, Nyiro J, Lewa P, Slack M, Scott JA. Pediatr Infect Dis J. 2008 Jan;27:59. doi: 10.1097/INF.0b013e31814da70c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hausdorff WP, Bryant J, Kloek C, Paradiso PR, Siber GR. Clin Infect Dis. 2000 Jan;30:122. doi: 10.1086/313609. [DOI] [PubMed] [Google Scholar]
- 22.Harboe ZB, et al. Clin Infect Dis. 2010 Feb 1;50:329. doi: 10.1086/649872. [DOI] [PubMed] [Google Scholar]
- 23.Rinta-Kokko H, Dagan R, Givon-Lavi N, Auranen K. Vaccine. 2009 Jun 12;27:3831. doi: 10.1016/j.vaccine.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Hanage WP, et al. Epidemics. 2010 Jun;2:80. doi: 10.1016/j.epidem.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flasche S, et al. PLoS Med. 2011 Apr;8:e1001017. doi: 10.1371/journal.pmed.1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X, et al. J Infect Dis. 1999 Nov;180:1569. doi: 10.1086/315096. [DOI] [PubMed] [Google Scholar]
- 27.McLean AR. Br Med Bull. 1998;54:545. doi: 10.1093/oxfordjournals.bmb.a011709. [DOI] [PubMed] [Google Scholar]
- 28.Lipsitch M. Proc Natl Acad Sci U S A. 1997 Jun 10;94:6571. doi: 10.1073/pnas.94.12.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melegaro A, Choi Y, Pebody R, Gay N. Am J Epidemiol. 2007 Jul 15;166:228. doi: 10.1093/aje/kwm076. [DOI] [PubMed] [Google Scholar]
- 30.Trotter CL, Gay NJ, Edmunds WJ. Am J Epidemiol. 2005 Jul 1;162:89. doi: 10.1093/aje/kwi160. [DOI] [PubMed] [Google Scholar]
- 31.MacArthur R, Levins R. The American Naturalist. 1967;101:377. [Google Scholar]
- 32.Adler PB, Hillerislambers J, Levine JM. Ecol Lett. 2007 Feb;10:95. doi: 10.1111/j.1461-0248.2006.00996.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.