Summary
The prevalence of obesity and type 2-diabetes is increasing worldwide and threatens to shorten lifespan. Impaired insulin action in peripheral tissues is a major pathogenic factor. Insulin stimulates glucose uptake in adipose tissue through the Glut4-glucose transporter and alterations in adipose-Glut4 expression or function regulate systemic insulin sensitivity. Downregulation of adipose tissue-Glut4 occurs early in diabetes development. Here we report that adipose tissue-Glut4 regulates the expression of carbohydrate responsive-element binding protein (ChREBP), a transcriptional regulator of lipogenic and glycolytic genes. Furthermore, adipose-ChREBP is a major determinant of adipose tissue fatty acid synthesis and systemic insulin sensitivity. We discovered a new mechanism for glucose-regulation of ChREBP: Glucose-mediated activation of the canonical ChREBP isoform (ChREBPα) induces expression of a novel, potent isoform (ChREBPβ) that is transcribed from an alternative promoter. ChREBPβ expression in human adipose tissue predicts insulin sensitivity indicating that it may be an effective target for treating diabetes.
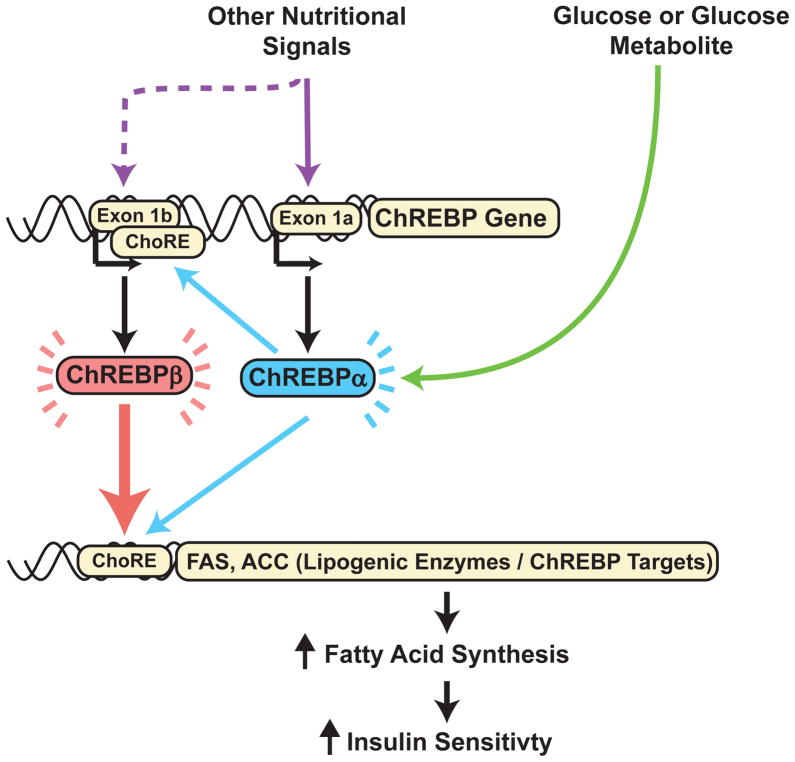
Insulin resistance is a common complication of obesity and a major factor in the pathogenesis of type 2 diabetes (T2D) and cardiovascular disease1. Adipose tissue contributes to the development of obesity-related insulin resistance through increased release of fatty acids, altered adipokine secretion, and/or macrophage infiltration and cytokine release2,3. Altered adipose tissue glucose metabolism is also an important cause of insulin resistance, and adipose tissue Glut4 (Slc2a4), the major insulin-responsive glucose transporter, plays a central role in systemic glucose metabolism1,4,5. In insulin-resistant states, Glut4 is down-regulated in adipose tissue, but not in muscle1, the major site of insulin-stimulated glucose uptake. In addition, mice with adipose-specific Glut4 overexpression (AG4OX) have improved glucose homeostasis5 while adipose-specific Glut4 knockout mice (AG4KO) have insulin resistance and T2D4. We investigated how altering adipose tissue glucose flux regulates glucose homeostasis. Here we show that carbohydrate responsive-element binding protein (ChREBP, also known as Mlxipl), a glucose-responsive transcription factor that regulates fatty acid synthesis and glycolysis,6 is highly regulated by Glut4 in adipose tissue and is a key determinant of systemic insulin sensitivity and glucose homeostasis. Also, ChREBP in adipose tissue is required for the improved glucose homeostasis resulting from increased adipose-Glut4 expression. Glut4-mediated glucose uptake induces ChREBP which activates adipose tissue de novo lipogenesis (DNL). The latter is associated with enhanced insulin sensitivity7–10. In obese humans adipose-ChREBP gene expression correlates with insulin sensitivity, suggesting that ChREBP protects against obesity-associated insulin resistance. In addition, we discovered a novel mechanism for glucose-regulated ChREBP expression involving a new isoform, ChREBPβ that is expressed from an alternative promoter in a glucose- and ChREBP-dependent manner. In contrast, expression of the canonical ChREBPα isoform is not regulated by glucose flux. However, glucose-induced ChREBPα transcriptional activity increases ChREBPβ expression. Furthermore, expression of ChREBPβ is more highly regulated than ChREBPα in adipose tissue in insulin resistant states. Thus, activation of adipose-ChREBP, and particularly ChREBPβ, may be a novel strategy for preventing and treating obesity-related metabolic dysfunction and T2D (Supplementary Fig. 1).
Glucose Regulates Adipose Tissue ChREBP
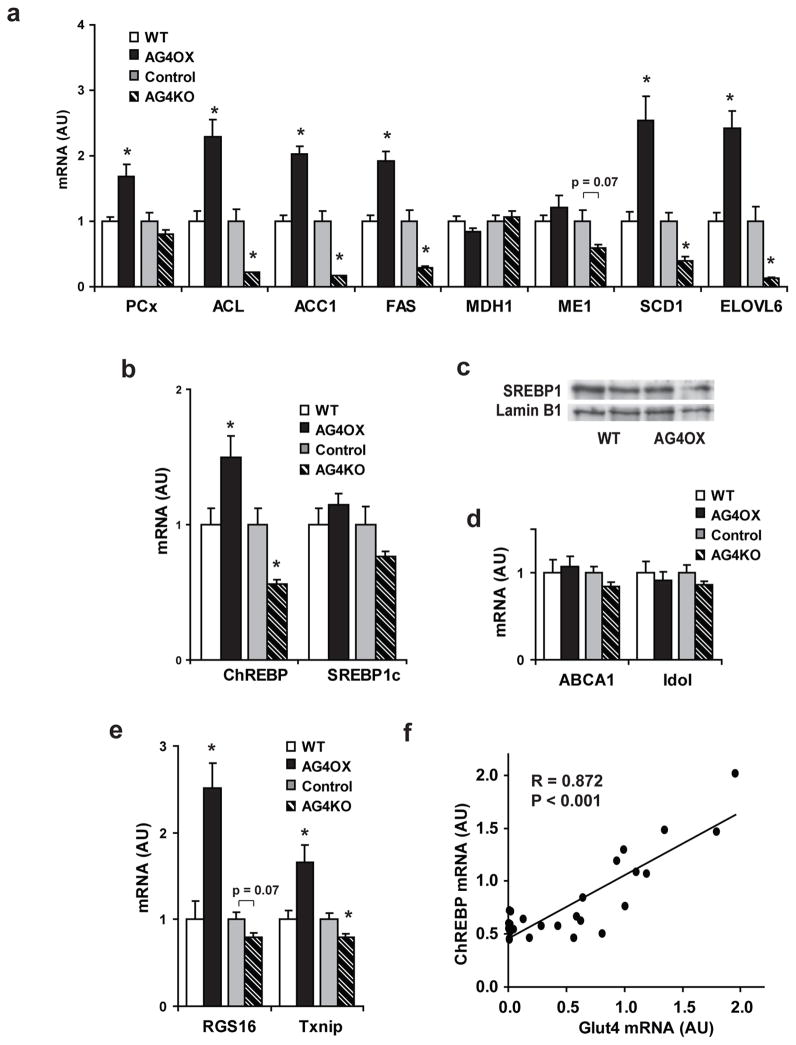
To understand the mechanisms by which adipocytes respond to changes in glucose flux, we analyzed global gene expression in adipose tissue from AG4OX and AG4KO mice. Gene-set enrichment analysis11 demonstrated coordinate up-regulation of DNL enzymes in AG4OX mice (Supplementary Table 1) which we confirmed (Fig 1a). DNL enzymes were down-regulated in AG4KO mice (Fig. 1a). Therefore we investigated the expression of transcription factors known to regulate DNL enzymes, e.g. sterol regulatory element binding protein 1c (SREBP-1c) and ChREBP6,12. ChREBP, but not SREBP-1c, expression is increased 50% in AG4OX and decreased 44% in AG4KO adipose tissue compared to controls (Fig. 1b). SREBP-1c transcriptional activity is primarily determined by the accumulation of mature SREBP-1c in the nucleus13. However, the nuclear abundance of SREBP-1c is not increased in AG4OX adipose tissue (Supplementary Fig. 2a). Liver X receptors (LXRα and LXRβ) can regulate the expression of both ChREBP and SREBP-1c 14,15, and DNL enzymes16. Expression of canonical LXR targets, do not change (Supplementary Fig. 2b) in AG4OX or AG4KO adipose tissue indicating that LXR activity is unchanged and is not driving the changes in ChREBP or DNL enzyme expression. In contrast, expression of RGS16 and Txnip, two ChREBP transcriptional targets17–19 not known to be regulated by other lipogenic transcription factors, were reciprocally regulated in AG4OX and AG4KO mice (Supplementary Fig. 2c). In AG4KO and control mice (Fig. 1c) and also in 30 different mouse strains (Supplementary Fig. 3), adipose-ChREBP expression strongly correlates with Glut4 expression. Expression of ChREBP transcriptional targets FAS and ACC1 also strongly correlate with ChREBP expression across these strains (Supplementary Fig. 3). Thus, adipose-ChREBP may mediate the effects of altered Glut4 expression and glucose flux on lipogenic enzyme expression.
Figure 1. Genetically altering adipose tissue glucose flux regulates the expression of ChREBP and its lipogenic targets.
a, mRNA expression of fatty acid synthetic enzymes, and b, lipogenic transcription factors in perigonadal fat from 6-week-old female mice (n=10–14 per group). *P<0.05 compared to respective controls. c, ChREBP and Glut4 mRNA correlate highly in PG WAT from control and AG4KO mice (n=27). Values are means ± S.E.
ChREBP Mediates Effects of Altered Adipose Glut4
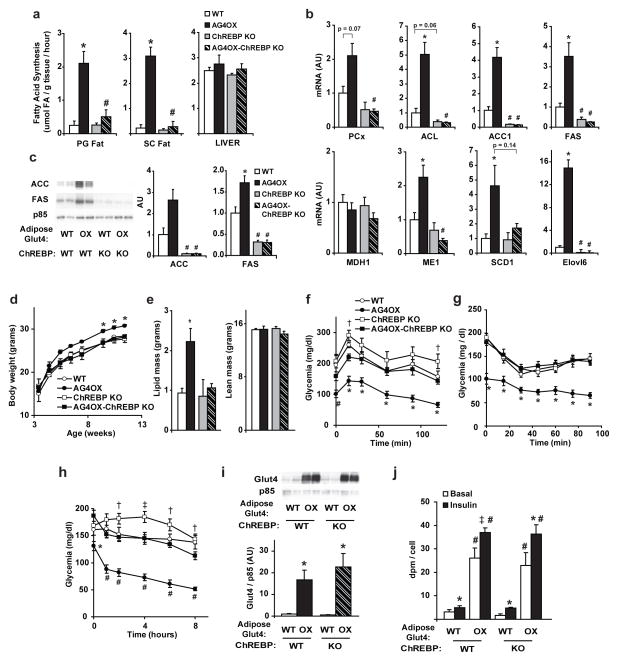
Increased DNL in adipose tissue is associated with enhanced glucose tolerance and insulin sensitivity20,21. Consistent with the increases in ChREBP and DNL enzyme expression (Fig. 1a–b), DNL in vivo is increased in perigonadal and subcutaneous white adipose tissue (WAT) in AG4OX mice compared to wild-type (WT) (Fig. 2a).
Figure 2. ChREBP is essential for the effects of adipose tissue Glut4 on adiposity, DNL, and glucose homeostasis.
a, DNL measured in vivo in fed, 4-month-old male mice PG=perigonadal, SC=subcutaneous, (n=5–6 per group). b, Quantification of western blots of FAS and ACC in SC fat from fed, 6-month-old females (n=9–10 per group). For a and b, *P<0.05 versus same ChREBP genotype, different AG4OX genotype. # p<0.05 versus same AG4OX genotype, different ChREBP genotype. c, Body weights in male mice on chow (n=5–7 per group). *P<0.05 versus all other groups at the indicated time. d, Body composition in 8-week-old, female mice (for d–g; n=10–12 per group). *P<0.01 versus all others. e, Glucose tolerance test and f, insulin tolerance test. *P<0.05 versus all others; #P<0.05 versus WT and ChREBP KO. g, Glycemia following food removal * P<0.05 versus all others; # P<0.05 versus ChREBP KO and AG4OX-ChREBP KO; †P<0.05 versus AG4OX; Values are means ± S.E.
To determine whether adipose-ChREBP is required for the increased adipose tissue DNL resulting from Glut4 overexpression, we crossbred AG4OX mice with whole-body ChREBP KO mice6. DNL normalizes in adipose tissue after deletion of ChREBP in AG4OX mice (Fig. 2a). Glucose incorporation into newly synthesized fatty acids is also decreased in WT and AG4OX adipose following ChREBP knockout (Supplementary Fig. 4). In liver, DNL is normal in AG4OX and AG4OX-ChREBP KO mice and is not decreased in ChREBP KO (Fig. 2a) contrasting with a prior report6. This difference may be attributable to significant differences in age, sex, genetic background and/or diet.
In parallel with reduction in DNL in AG4OX adipose tissue following ChREBP deletion, DNL enzyme mRNA and protein expression levels also normalize (Fig. 2b and Supplementary Fig. 5). Thus, ChREBP is a major regulator of DNL enzymes in adipose tissue and mediates the changes in DNL enzyme expression resulting from increased glucose flux.
AG4OX mice are obese (Fig. 2c–d) but have enhanced glucose tolerance (Fig. 2e)5. Increased body weight (Fig. 2c), adiposity (Fig. 2d) and a tendency toward increased food intake normalize with ChREBP deletion; energy expenditure was normal (Supplementary Fig. 6). In spite of obesity, AG4OX mice are more insulin-sensitive than WT mice (Supplementary Table 2). Decreased fed glycemia (Supplementary Table 2), markedly enhanced glucose tolerance (Fig. 2e), and lower glycemia during insulin tolerance testing (Fig. 2f) in AG4OX mice normalized after ChREBP deletion. Genetically ablating ChREBP in WT and AG4OX mice impairs insulin sensitivity (Supplementary Table 2). ChREBP KO mice have modest glucose intolerance (Fig. 2e) as reported6. Glycemia in ChREBP KO mice remains elevated following food removal compared to WT mice (Fig. 2g). ChREBP KO prevents the hypoglycemia which develops in AG4OX mice after food removal (Fig. 2g). ChREBP deletion did not decrease Glut4 overexpression or glucose uptake (Supplementary Fig. 7) in AG4OX adipocytes indicating that the mechanism by which ChREBP regulates glucose homeostasis is not by altering adipose-glucose uptake. The decreased insulin sensitivity and glucose tolerance in AG4OX-ChREBP KO mice is not due to elevated serum triglycerides or non-esterified fatty acids since both metabolites decrease compared to AG4OX mice (Supplementary Table 2). The effects of ChREBP KO on metabolism and gene expression in AG4OX mice appear to result from the absence of ChREBP specifically in adipose tissue because ChREBP expression is increased in adipose tissue (Fig. 1b) but not in liver of AG4OX mice (Supplementary Fig. 8). Reversal of enhanced glucose tolerance and insulin sensitivity with normalization of DNL is consistent with a role for adipose tissue DNL in improving systemic glucose metabolism.
Obesity Regulates Adipose Tissue ChREBP
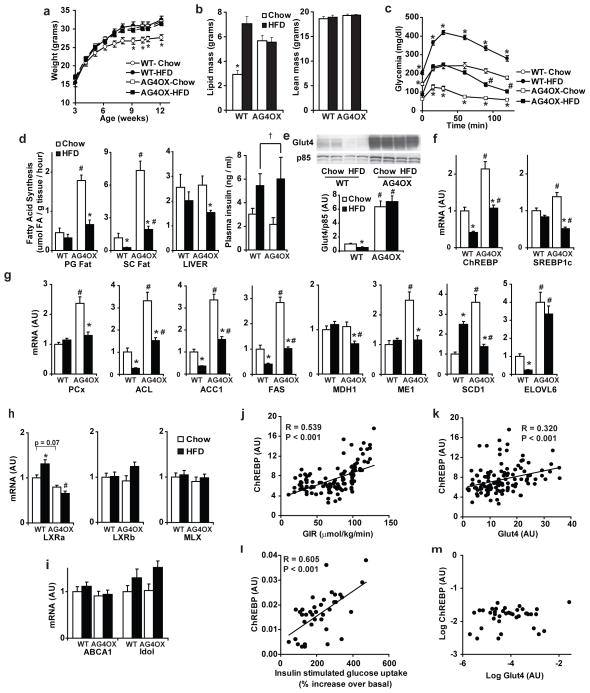
High-fat diet (HFD) causes obesity and insulin resistance and down-regulates Glut4 expression selectively in adipose tissue1. To investigate whether adipose ChREBP expression plays a role in the insulin resistance resulting from adipose-Glut4 down-regulation, we subjected mice to HFD. On chow, AG4OX mice are obese (Fig. 2c–d and 3a). AG4OX do not gain more weight on HFD and their degree of obesity on both diets is similar to HFD-fed WT (Fig. 3a and Supplementary Fig. 9). HFD induces insulin resistance in both genotypes (Supplementary Table 3). In WT, HFD causes a diabetic GTT (Fig. 3b) but not in AG4OX mice. The enhanced glucose tolerance in AG4OX mice does not result from changes in serum metabolites typically associated with insulin sensitivity. AG4OX mice have higher non-esterified fatty acids and triglycerides, and lower leptin levels on both diets (Supplementary Table 3), and lower adiponectin levels22 on chow compared to WT. This pattern is thought to contribute to insulin resistance2,3,23 and not to enhanced insulin sensitivity.
Figure 3. ChREBP is regulated in mouse and human adipose tissue in pathologic conditions.
a, Body weight in chow- versus HFD-WT and AG4OX mice (9-week-old, n=8 per group), *P<0.05 WT-Chow compared to all others at the same time point. b, Glucose tolerance test in 7-week-old males (n=8 per group). *P<0.001 versus all others; c, Fatty acid synthesis measured in vivo in 5–6 month-old males (n=7 per group). Last panel, plasma insulin: HFD increases insulin (†P=0.01 by 2-way ANOVA). mRNA expression of d, ChREBP and SREBP-1c and e, ACC and FAS in SC fat from 4-month-old male mice (n=8–14 per group). For panels c–e, *P<0.05 compared to same diet, different genotype. #P<0.05 compared to same genotype, different diet. f, SC fat ChREBP mRNA correlates with insulin sensitivity in non-diabetic, normal glucose-tolerant humans (n=123). g, ChREBP and Glut4 mRNA expression in SC fat correlate in this group. h, SC fat ChREBP mRNA correlates highly with insulin sensitivity in obese, non-diabetic, BMI-matched humans (n=38). i, ChREBP and Glut4 mRNA expression do not correlate in this group. Values are means ± S.E.
HFD reduces expression of adipose tissue DNL enzymes24,25 which correlates with insulin resistance7–10. Therefore, we examined whether downregulation of adipose-ChREBP and DNL contributes to HFD-induced insulin resistance. HFD in WT mice reduces DNL in subcutaneous but not perigonadal adipose tissue, (Fig. 3c). In AG4OX, HFD markedly reduces DNL in WAT, but DNL remains 2–7-fold higher than in WT on HFD. This intermediate rate of DNL parallels the change in glucose tolerance in AG4OX mice on HFD (Fig. 3b). Changes in glucose incorporation into newly synthesized fatty acids (Supplementary Fig. 10) are similar to the changes in total (from all substrates) DNL (Fig. 3c) in WAT. In liver, DNL is not higher in AG4OX on either diet (Fig. 3c). HFD inhibits adipose-DNL independently of adipose tissue glucose uptake in AG4OX mice since Glut4 protein (Supplementary Fig. 11) and glucose transport remain elevated26.
To investigate the mechanisms underlying HFD-induced changes in adipose-DNL, we examined the expression of lipogenic transcription factors and DNL enzymes. In WT, HFD diminishes ChREBP but not SREBP-1c expression in WAT (Fig. 3d) as reported25,27. In AG4OX WAT, HFD markedly decreases ChREBP and also SREBP-1c expression but ChREBP remains elevated compared to WT on HFD. Nevertheless, HFD reduces the degree of ChREBP induction in AG4OX adipose tissue despite persistent Glut4 overexpression (Supplementary Fig. 11). Thus, HFD may regulate ChREBP expression in part independently of Glut4 expression and glucose flux, consistent with HFD effects on DNL (Fig. 3c).
Many DNL enzymes that are up-regulated in WAT of AG4OX on chow are down-regulated in HFD WT and AG4OX mice (Fig. 3e and Supplementary Fig. 12) but remain higher in AG4OX WAT compared to WT in a pattern paralleling ChREBP expression (Fig. 3d) and DNL (Fig. 3c). In WT WAT, ChREBP down-regulation most likely accounts for HFD-induced down-regulation of DNL gene expression since WAT SREBP-1c expression is not altered. Furthermore, whole body genetic ablation of SREBP-1c does not diminish expression of these genes in WAT28.
In contrast, both ChREBP and SREBP-1c expression are down-regulated in WAT in AG4OX mice on HFD compared to chow (Fig. 3d). Persistent modest elevation of ChREBP in WAT of AG4OX on HFD compared to WT on HFD (Fig. 3d) is likely responsible for the increased DNL which contributes to improved glucose tolerance in AG4OX (Fig. 3b).
LXRs are unlikely to contribute to down-regulation of ChREBP expression, DNL, or DNL enzyme expression in WT mice on HFD because expression of LXRs and canonical LXR targets are either unchanged or modestly increased (Supplementary Fig. 13). Mlx (Max like protein X) is an obligate dimerization partner for ChREBP transcriptional activity 29. Neither HFD nor Glut4 overexpression alters Mlx expression in WAT (Supplementary Fig. 13).
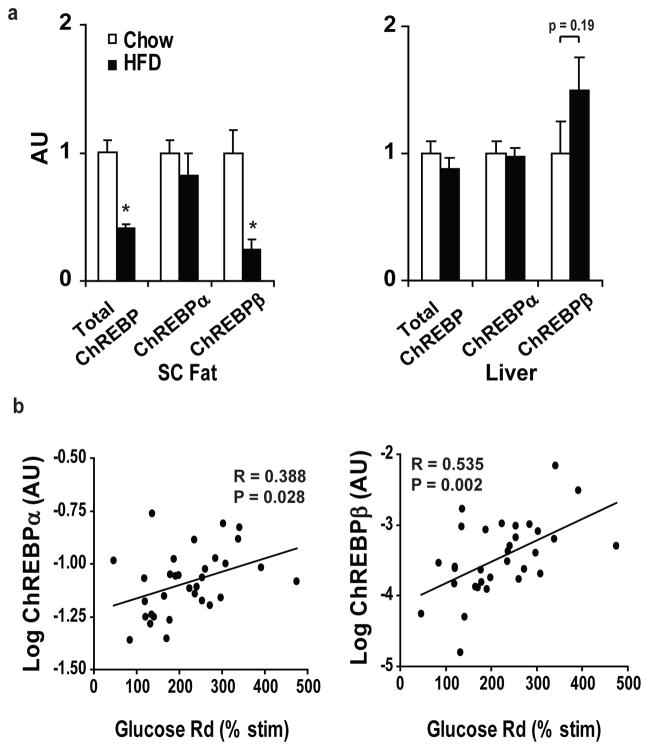
We next sought to determine whether adipose-ChREBP might contribute to regulating insulin sensitivity and glucose homeostasis in humans. In 123 non-diabetic individuals with normal glucose tolerance and widely ranging body mass index (Supplementary Table 4), ChREBP expression in subcutaneous WAT correlates strongly with insulin sensitivity measured during a euglycemic-hyperinsulinemic clamp procedure (Fig. 3f). Adipose-ChREBP correlates with Glut4 (Fig. 3g), consistent with a role for ChREBP in the beneficial effects of adipose-Glut4 expression on glucose homeostasis.
Most, but not all, obese people are insulin resistant. To determine whether adipose-ChREBP expression could have a role in regulating insulin sensitivity in obese people, we also investigated adipose-ChREBP expression in non-diabetic obese individuals with widely ranging insulin-sensitivity (Supplementary Table 4). Adipose-ChREBP expression was directly associated with insulin-stimulated glucose uptake during a clamp, independent of BMI (Fig. 3h). Surprisingly, ChREBP and Glut4 expression did not correlate in these obese individuals (Fig. 3i). Thus, adipose-ChREBP expression may have beneficial effects on insulin sensitivity, even among obese subjects, and this can be independent of Glut4 expression.
Identification of a Novel ChREBP Isoform
The mechanism by which glucose activates ChREBP transcriptional activity is complex and controversial. It has been attributed to xylulose-5-phosphate induced dephosphorylation of specific ChREBP residues30. Alternatively, glucose or a metabolite may prevent the interaction of an N-terminal inhibitory domain with a glucose-activating domain within the ChREBP protein31. Glucose and other carbohydrates can also induce ChREBP expression27 but whether this contributes to ChREBP activity is unknown. The mechanism for carbohydrate-mediated ChREBP induction may involve “feed-forward” auto-regulation since a dominant negative Mlx mutant which inhibits ChREBP activity reduces endogenous ChREBP expression19,32.
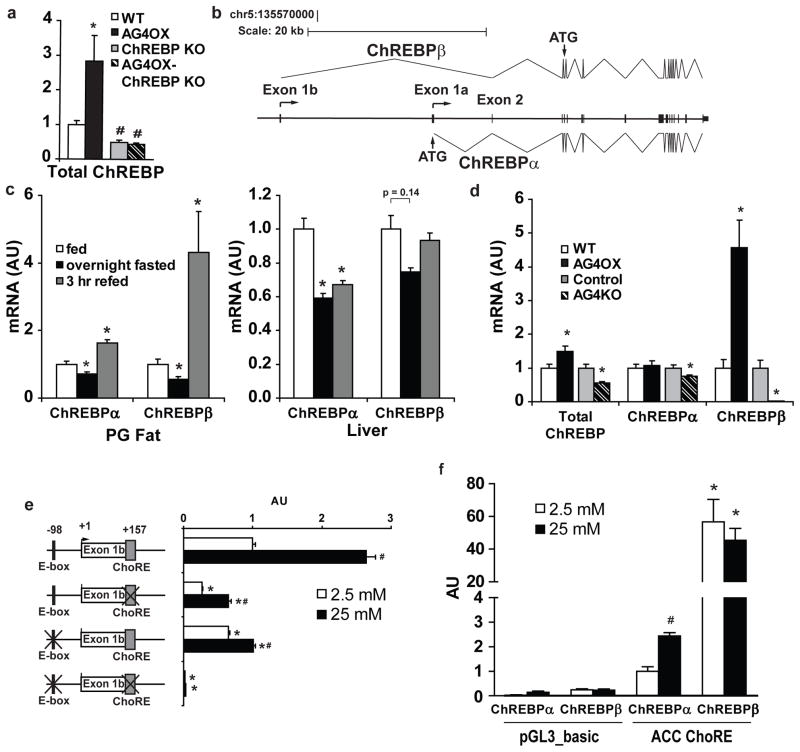
To investigate the molecular mechanisms by which ChREBP might transactivate its own expression, we searched the genomic sequence around the ChREBP transcriptional start site for a carbohydrate response element (ChoRE) which is defined by two E-boxes (CACGTG) separated by 5 nucleotides33. We identified a single candidate ChoRE 17 kb upstream of the mouse ChREBP transcriptional start site (Supplementary Fig. 15 and 16). Additionally, a separate E-box was identified 255 base pairs proximal to the putative ChoRE (Supplementary Fig. 16). In this region, the genomic sequence including the ChoRE and 5′ E-box is highly conserved (Supplementary Fig. 16 and 17). H3K4me3 and H3K4me1 histone methylation marks aligned with this conserved genomic region (Supplementary Fig. 18) suggesting the presence of an alternative promoter34 and potentially an alternative first exon. By 5′ rapid amplification of cDNA ends, we confirmed an alternative first exon, exon 1b (Fig. 4a and Supplementary Figs. 16–18) and cloned an mRNA species transcribed from this alternative promoter in which exon 1b is spliced to exon 2 bypassing exon 1a and retaining the remainder of exons present in the canonical ChREBP mRNA species (Fig. 4a).
Figure 4. Expression of the novel ChREBPβ isoform is regulated in a glucose- and ChREBP-dependent manner.
a, Model of ChREBPα and ChREBPβ gene structure with indication of splice sites and translational start sites (ATG). b, Regulation of ChREBPα and β mRNA expression in perigonadal fat and liver of 10-week-old, female mice with fasting and refeeding (n=6/group). *P<0.05 compared to fed group. c, ChREBPα and β mRNA expression in perigonadal fat from 6-week-old female AG4OX and AG4KO compared to littermate controls (n=10–14 per group). *P<0.05 versus respective control. d, Glucose regulation of exon 1b promoter-luciferase reporter and indicated ChREBPβ mutants, co-transfected with ChREBPα and Mlx (n=3/group). *P<0.05 compared to non-mutated exon 1b-luciferase construct in the same glucose. #P<0.05 compared to the same construct in low glucose. e, ChREBPα and β induce an ACC ChoRE-luciferase reporter compared to pGL3_basic control vector in both low and high glucose. *P<0.05 compared to ChREBPα in the same glucose, #P<0.05 compared to ChREBPα, low glucose. Values are means ± S.E.
Physiologic Regulation of ChREBP Isoforms
To determine whether the two distinct ChREBP mRNA species are physiologically regulated, we performed qPCR with primers specific for exon 1a (ChREBPα) or exon 1b (ChREBPβ). In WAT, ChREBPα expression declines modestly with overnight fasting, and increases 64% after 3 hours of refeeding a chow diet compared to ad libitum fed mice (Fig. 4b). In contrast, adipose-ChREBPβ declines 45% after an overnight fast and increases 4.3-fold with refeeding compared to ad libitum feeding. Thus, ChREBPβ expression is more highly regulated in WAT than ChREBPα with fasting and refeeding. In liver, ChREBPα declines 40% and ChREBPβ expression tends to decline (25%) with overnight fasting. Following 3 hours of refeeding, ChREBPα remains suppressed while ChREBPβ returns to baseline. After 6 hours of refeeding, liver ChREBPα levels return to normal (data not shown).Thus, in liver, the time course of ChREBPα and ChREBPβ regulation differ. Furthermore, the refeeding effects on ChREBPβ are more rapid in WAT than liver.
To determine whether alterations in glucose flux selectively regulate ChREBPα or β, we examined the expression of the two isoforms in AG4OX and AG4KO WAT. Total ChREBP expression increases 50% in AG4OX and decreases 50% in AG4KO WAT (Fig. 4d). Adipose-ChREBPα expression remains unchanged in AG4OX and declines modestly in AG4KO. In contrast, in WAT, ChREBPβ increases 4.6-fold in AG4OX mice, and decreases 97% in AG4KO (Fig. 4c). Considering the difference in magnitude of change of total ChREBP compared to ChREBPβ, ChREBPβ is likely much less abundant than ChREBPα. While expression of both ChREBPα and β are nutritionally regulated (Fig. 4b), ChREBPβ alone responds robustly to Glut4-mediated changes in glucose flux (Fig 4c).
Glucose-activated ChREBPα Induces ChREBPβ
We investigated the molecular mechanism by which ChREBPβ expression is regulated in response to changes in adipose-Glut4 expression. Expression of a ChREBPβ-promoter-luciferase construct is markedly increased in a glucose-dependent manner with co-transfection of ChREBPα and Mlx (Fig. 4d). Mlx, the dimerization partner for ChREBP, is required for ChREBP transcriptional activity29. Co-transfection of both ChREBPα and Mlx were required for induction of the ChREBPβ-promoter-luciferase activity in high glucose (Supplementary Fig. 19) indicating that ChREBPβ requires transactivation by ChREBPα for expression. Basal and glucose-stimulated expression is attenuated with deletion of either the ChoRE or the upstream E-box and abolished entirely with deletion of both (Fig. 4d). Glucose and ChREBPα/Mlx had no effect on the expression of a luciferase reporter containing 5 kb of the ChREBPα promoter indicating that ChREBPα does not regulate its own expression (data not shown). Thus, ChREBPα specifically regulates expression of ChREBPβ.
The translational start site for ChREBPα resides in exon 1a and translation from this site produces an 864 amino acid protein. No translational start site is present in exon 1b and translation beginning at the next start site, in exon 4, produces a 687 amino acid protein in which two nuclear export signals, a nuclear localization signal, and a domain that inhibits ChREBP transcriptional activity in low glucose conditions are deleted (Supplementary Fig. 20)31,35. N-terminally-deleted ChREBP mutants demonstrate increased nuclear localization and enhanced transcriptional activity in both low and high glucose31. Using an ACC-ChoRE-luciferase reporter construct36, we compared the transcriptional activity of ChREBPα and ChREBPβ (Fig. 4e). High glucose increased ChREBPα transcriptional activity 2.4-fold over low glucose. ChREBPβ activity in either low or high glucose is increased ~20-fold compared to ChREBPα activity in high glucose, and shows no glucose regulation. The elevated ChREBPβ transcriptional activity may result in part from absence of the N-terminal inhibitory domain or its constitutive nuclear localization in low and high glucose conditions (Supplementary Fig. 21). These results suggest a feed-forward mechanism in which glucose-stimulated ChREBPα transactivates expression of ChREBPβ (Fig. 4d and Supplementary Fig. 22), a more potent activator of ChREBP transcriptional targets (Fig. 4e).
Pathophysiologic Regulation of ChREBPα/β
On HFD, ChREBPβ expression declines in WAT while ChREBPα expression remains unchanged (Fig. 5a), consistent with the possibility that downregulation of adipose ChREBPβ contributes to insulin resistance.
Figure 5. Adipose tissue ChREBPβ expression predicts insulin sensitivity.
a, ChREBPα and β mRNA expression in SC fat of 4-month-old male mice on chow or HFD (n=10–14 per group). *P<0.05 compared to Chow-fed. b, mRNA expression of ChREBPβ in SC fat correlates more highly with insulin sensitivity (% increase in insulin-stimulated glucose uptake over basal measured during a euglycemic-hyperinsulinemic clamp procedure) than ChREBPα in obese, non-diabetic, BMI-matched humans (n=38). Values are means ± S.E.
In human WAT, expression of both ChREBPα and β correlate with insulin sensitivity (Fig. 5b). Multiple linear regression analysis demonstrates that expression of ChREBPβ (β=0.454, P=0.016) and not ChREBPα (β=0.136, P=0.356) predicts insulin sensitivity (Supplementary Table 5).
Significance
The adipocyte is a key regulator of whole-body energy homeostasis and metabolic function2,4. Our prior work demonstrating that adipose-Glut4 expression regulates systemic insulin sensitivity4,5 indicated that adipocytes are capable of sensing and coordinating responses to changes in glucose availability. The elucidation here of the molecular mechanism by which glucose-mediated ChREBP activation transactivates expression of a novel, potent ChREBPβ (Supplementary Fig. 22) isoform reveals the complexity underlying cellular responses to nutrient availability. Our data demonstrate that adipose tissue-ChREBP plays a key role in integrating adipocyte and whole-body metabolic function and this may be mediated by transcriptional regulation of the potent ChREBPβ isoform.
The beneficial effects of adipose tissue-ChREBP on glucose homeostasis may result from upregulation of adipose tissue-DNL and/or other ChREBP-dependent actions such as changing the metabolic fate of glucose after it enters adipocytes and/or regulating a novel adipokine. The strong, direct correlation between adipose-ChREBP expression and insulin sensitivity in humans suggests that adipose-ChREBP may be involved in regulating whole-body insulin action in people. The beneficial metabolic effects of increased DNL in adipose tissue contrast with the adverse effects in the liver which increases intrahepatic triglyceride content and may contribute to insulin resistance and the metabolic syndrome37,38. In summary, our data support the importance of adipose-ChREBP in regulating insulin action and glucose homeostasis in human obesity and diabetes. These data suggest that selective activation of adipose tissue-ChREBP, and specifically ChREBPβ, could be an effective therapeutic strategy for preventing and treating T2D and obesity-related metabolic diseases.
Methods Summary
AG4OX, AG4KO, and ChREBP KO mice were described previously4–6 except that AG4KO mice were generated using adiponectin-Cre expressing mice39 rather than aP2-Cre expressing mice. Phenotypic analyses were performed as described4,5,40. All mouse studies were conducted in accordance with federal guidelines and were approved by the BIDMC Institutional Animal Care and Use Committee. For microarray analysis, RNA was isolated from perigonadal adipose tissue and the cDNA was analyzed on the Affymetrix MG-U74-A.v2 Genechip. Quantitative real-time PCR was performed on the Applied Biosystems 7900 HT using SYBR Green PCR Master Mix. Cloning, mutagenesis, and luciferase assays were performed by standard methods. Human subjects were recruited as described37,41. Human studies were approved by the Human Studies Committee of Washington University School of Medicine or the Ethics Committee of the University of Leipzig.
Methods
Animal Studies
Generation and initial metabolic characterization of the adipose-specific Glut4-overexpressing mice (AG4OX), adipose-specific Glut4 knock-out mice (AG4KO), and ChREBP KO mice were previously described4–6 except that AG4KO mice were generated using adiponectin-Cre expressing mice39 rather than aP2-Cre expressing mice. WT and adiponectin-Cre littermates were used as controls for AG4KO mice. Mice were housed at Beth Israel Deaconess Medical Center with a 14/10 light-dark cycle and were fed standard chow (Formulab 5008) or HFD (Harlan-Teklad TD.93075) ad libitum. All studies were performed on age- and sex-matched littermates. Blood collections were performed by submandibular vein or tail vein bleeding. Body composition was measured by NMR (Echo Medical Systems). Glucose tolerance tests were performed by injection of 1 mg glucose per kg body weight i.p. after 5 hours food removal. For the fasting glycemic time course experiment, food was removed at 8 AM. Mice were sacrificed by CO2 euthanasia or decapitation and serum was collected, and tissues were harvested, snap frozen in liquid nitrogen, and stored at −80C for processing. Mouse studies were conducted in accordance with federal guidelines and were approved by the BIDMC Institutional Animal Care and Use Committee.
Human Studies
For the cross-sectional cohort, adipose tissue samples were obtained from 123 men (n = 64) and women (n = 59) who underwent open abdominal surgery for gastric banding, cholecystectomy, appendectomy, weight reduction surgery, abdominal injuries or explorative laparotomy. These patients represent a normal glucose tolerant subset of patients previously described41. All subjects had a stable weight, defined as the absence of fluctuations of >2% of body weight for at least 3 months before surgery. Patients with malignant diseases or any acute or chronic inflammatory disease, as determined by a leukocyte count of >7,000 Gpt/l, C-reactive protein levels of >50 mg/l or clinical signs of infection, were excluded from the study. After resection, samples of visceral and subcutaneous adipose tissue were immediately frozen in liquid nitrogen. Euglycemic-hyperinsulinemic clamp studies were performed prior to surgery in subjects undergoing bariatric surgery or 3 months after surgery in subjects who underwent non-elective surgeries. The clamp study was performed with an insulin infusion rate of 20 mU/kg/min, as described and the glucose disposal rate was defined as the glucose infusion rate during the last 30 min of the study41. The study was approved by the ethics committee of the University of Leipzig. All participants gave written informed consent before taking part in the study.
For the obese cohort, 38 obese subjects (10 men, 28 women; 41±11 years old) who had normal oral glucose tolerance were studied. Subjects were sedentary (i.e., participated in regular exercise <1 hour/wk and ≤1 time/wk) and weight stable. All subjects provided written, informed consent before participating in the study, which was approved by the Human Studies Committee of Washington University School of Medicine in St. Louis.
Subjects were admitted to the Clinical Research Unit at Washington University School of Medicine in the evening before the clamp procedure. The next morning, after subjects fasted for 12 h overnight, a hyperinsulinemic-euglycemic clamp procedure in conjunction with [6,6-2H2]glucose tracer infusion was performed, as described previously37. Euglycemia was maintained at a blood glucose concentration of approximately 5.6 mmol/L (100 mg/dL) by infusing 20% dextrose enriched to 2.5% with [6,6-2H2]glucose. Skeletal muscle insulin sensitivity was determined as the relative increase in the rate of glucose uptake during insulin infusion (50 mU/m2/min).
Microarray Analysis
Total RNA from epididymal adipose tissue was extracted using the RNeasy Mini Kit from Qiagen from three mice from each of four genotypes: aP2-Cre transgenic littermates (controls for AG4KO mice), AG4KO mice; FVB littermates (controls for AG4OX) and AG4OX. RNA from each mouse was hybridized on an Affymetrix MG-U74-A.v2 Genechip microarray. Affymetrix gene chip hybridization and analysis were performed at the Genomics Core Facility of the Beth Israel Deaconess Medical Center. Genome-wide expression analysis of the microarray data were performed using gene set enrichment analysis (GSEA)11 and the s2.mgu74av2.gmt gene set database available through the Broad Institute.
Gene Expression analysis
Mouse tissues were harvested following CO2 euthanasia or decapitation, snap frozen in liquid nitrogen, and stored at −80C for processing. Total RNA was extracted from frozen tissue with TRI Reagent (Molecular Research Center, Inc.). Reverse transcription was performed using the Advantage RT-for-PCR kit (Clontech). Quantitative PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) in a 7900 HT thermocycler (Applied Biosystems). SDS 2.3 (Applied Biosystems) was used for calculation of cycle thresholds. Relative expression levels were determined using the standard curve method with normalization of target gene expression levels to 18s or Tbp. See Supplementary Table 6 for primer sequences.
For the cross-sectional cohort, total RNA was isolated from subcutaneous adipose tissue samples using TRIzol (Life Technologies) and cDNA was synthesized withstandard reagents (Life Technologies). Quantitative PCR was performed using Brilliant SYBR Green QPCR Core Reagent Kit (Stratagene) in a PRISM 7900 thermocycler (Applied Biosystems). Relative expression levels were determined using the standard curve method with normalization of target gene expression levels to 18s.
For the obese cohort, total RNA was isolated from subcutaneous adipose tissue samples using TRIzol (Invitrogen) and cDNA was synthesized using Taqman Reverse Transcription (Applied Biosystems). Quantitative PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) on the ABI 7500 Real-Time PCR System (Applied Biosystems). Relative expression levels were determined using the 2−ΔCt method with normalization of target gene expression levels to the 36B4 gene.
Western Blotting
For adipose tissue lysates, aliquots of frozen adipose tissue were homogenized on ice in RIPA buffer supplemented with phospho-preserving and anti-protease agents (sodium fluoride, sodium pyrophosphate, sodium orthovanadate, PMSF, aprotinin, and leupeptin). For adipose tissue nuclear lysates, an adipose tissue sample was ground under liquid nitrogen, then dounce homogenized in low salt buffer [potassium chloride, 10 mM; Hepes pH 7.9, 10 mM; EDTA 0.1mM; EGTA 0.1 mM; DTT 1 mM] with protease inhibitors. Homogenates were centrifuged at 1000 RCF for 10 minutes at 4C and nuclear pellets were collected and washed with low salt buffer. The nuclear pellets were resuspended in RIPA buffer and vortexed vigorously. Nuclear lysates from 3 to 4 animals from the same genotype were pooled for further processing. Protein concentrations were assayed using the BCA assay. Equal amounts of protein were loaded and transferred to nitrocellulose membranes. Membranes were probed with antibodies against Acetyl-CoA Carboxylase (Cell Signaling), Fatty Acid Synthase (Cell Signaling), Glut4 (provided by H. Haspel), SREBP1 (Novus Biologicals), and Lamin B (Santa Cruz) and PI3 kinase p85 (Upstate). Film images were scanned (Epson Expression 10000 XL) and results were quantified with ImageQuant TL software (GE).
In Vivo Fatty Acid Synthesis
Conscious, freely moving mice in the fed state were injected intraperitoneally with 5 mCi of 3H2O (MP Biomedicals) and 10 μCi of [U-14C]-glucose (Perkin Elmer) and sacrificed 1 hour later using an overdose of ketamine and xylazine (160 and 24 mg/kg i.p., respectively). Samples of plasma were obtained serially at 5, 10, 30, and 60 minutes after injection for measurement of the plasma glucose levels and the specific activity of plasma water and glucose. At sacrifice, tissues were rapidly removed, frozen in liquid N2 and stored at −80C for processing. Lipids were extracted by the Folch method with chloroform/methanol (2:1). Fatty acids were obtained by saponification and extraction with petroleum ether. Incorporation of 3H and 14C into saponified fatty acids were measured. The rate of synthesis was calculated as a molar rate based on the estimate that 13.3 mol of H2O are incorporated into each newly synthesized C16 fatty acid42. The rate of glucose incorporation into newly synthesized fatty acids was calculated as nanomoles of 14C-glucose incorporated into fatty acids/gram tissue per minute, using 14C counts in saponified fatty acids extracted from tissue normalized by the time averaged specific activity of glucose in plasma over the course of the hour-long experiment.
Glucose Uptake in Isolated Adipocytes
Adipocytes were isolated from perigonadal fat pads and glucose uptake was measured as previously described5. Briefly, perigonadal fat pads were digested with collagenase (1 mg/ml) and cells were incubated at 37 °C with constant shaking in Krebs-Ringer-Hepes (30 mM) buffer (pH 7.4) with 2% bovine serum albumin, 200 nM adenosine, and without (basal) or with (insulin-stimulated) 80 nM insulin. Following a 30-min incubation with or without insulin, U-14C glucose (3 μM) was added for 60 min and the reaction was terminated by separating cells from media by spinning the suspension through dinonyl-phthalate oil. A portion of isolated adipocytes from each sample were fixed with osmic acid and counted in a Coulter counter to normalize glucose uptake per cell.
Identification of ChoRE and ChREBP Exon 1b
A position weighted matrix representative of a consensus ChoRE was generated from sequences for 16 mouse, human, or rat ChoREs previously identified and experimentally validated (Supplementary Fig. 15). Genomic sequence 20 kb upstream and downstream of the mouse ChREBP transcriptional start site was obtained from the UCSC Genome Browser (Mouse July 2007 (NCBI37/mm9) Assembly)43,44 and was scanned using the Transcriptional Element Search System (www.cbil.upenn.edu/tess) and the positional weight described above. Genomic sequence in the exon 1b region was obtained through the UCSC genome browser and comparisons across species were performed with Megalign (Dnastar) using the Clustal W algorithm. Analysis of histone methylation marks was performed using the Encode browser and database45,46. ChREBP exon 1b was cloned from RNA prepared from AG4OX adipose tissue by 5′ rapid amplification of cDNA ends (GeneRacer, Invitrogen). A full-length ChREBPβ mRNA species was cloned using GeneRacer reagents. All cloned products were verified by sequencing.
Functional Analysis of ChoREs
PCR of bacterial artificial chromosome containing the mouse ChREBP gene (CH29-535J6, Children’s Hospital Oakland Research Institute) was used to generate the mouse ChREBPβ promoter sequence (−631 to +224, in reference to exon 1b transcriptional start site) which was cloned into the pGL3_basic reporter vector. The putative ChoRE sequence (+157 to +174) and E-box (−98 to −92) were deleted by site-directed mutagenesis (QuickChange II Site-Directed Mutagenesis Kit, Agilent). The exon 1b promoter_pGL3, mutant pGL3 constructs, or an empty pGL3_basic control were co-transfected using Lipofectamine LTX (Invitrogen) in HEK293T cells with Flag-tagged ChREBPα (ζ-isoform) and HA-tagged Mlx, both in the CMV4 vector 29, along with a renilla luciferase control. After 24 hours of transfection, cells were cultured in DMEM containing low (2.5 mM) versus high (25 mM) glucose. Cells were collected and firefly luciferase activity was measured 24 hours after the media change and normalized to renilla luciferase activity (Dual Luciferase Assay System, Promega). All experiments were repeated at least three times with comparable results.
Functional Analysis of ChREBPβ
Flag-tagged ChREBPβ was generated by deletion of N-terminal sequence in the mouse Flag-tagged ChREBPα (GenBank: AF245475) in the CMV4 vector 29 by site-directed mutagenesis (QuickChange II Site-Directed Mutagenesis Kit, Agilent). ChREBPβ sequence was confirmed by sequencing. Flag-tagged ChREBPα, Flag-tagged ChREBPβ, or empty pGL3_basic control were co-transfected with HA-tagged Mlx and a reporter plasmid containing a promoter region consisting of two copies of the ACC ChoRE fused to the firefly luciferase gene36. 24 hours after transfection, cells were cultured in DMEM containing low (2.5 mM) or high (25 mM) glucose. Cells were collected and firefly luciferase activity was measured 24 hours after the media change and normalized to renilla luciferase activity (Dual Luciferase Assay System, Promega). Similar results were also obtained when normalizing luciferase activity to ChREBPα or β protein levels, respectively (data not shown). All experiments were repeated at least three times with comparable results.
Statistics
All values are given as means ± S.E. Differences between two groups were assessed using unpaired two-tailed Student’s t-tests unless otherwise specified in figure legends. Comparisons between multiple groups were performed by ANOVA with pairwise comparisons and adjustment for multiple comparisons by the Tukey test unless otherwise specified in figure legends. For analyses with non-normal distributions or unequal variances, ANOVA and pairwise comparisons were performed on log-transformed values. All data presented in the figures are of non-transformed values. Due to the large variance and non-normal distribution of Glut4 expression in the obese human cohort (Fig. 3i), Log mRNA expression levels were analyzed and presented. Statistical Analyses including linear regressions were performed with SPSS 8.0.
Supplementary Material
Figure 6.
Acknowledgments
We thank H. Towle for providing reagents and discussion. We thank P. Pissios, I. Astapova, E. Rosen and the Rosen lab for discussion, K. Uyeda for providing ChREBP KO mice, and E. Shu for technical assistance. This work was supported by NIH R37 DK43051 (B.B.K), K08 DK076726 (M.A.H), BADERC DK057521 (B.B.K, M.A.H), BNORC DK046200 (M.A.H.), the Picower and JPB Foundations (B.B.K.), a Fellowship from the Radcliffe Institute for Advanced Study (B.B.K.), DK056341 (Nutrition and Obesity Research Unit) (S.K.), DK037948 (S.K.), and the Deutsche Forschungsgemeinschft DFG, KFO152, BL833/1 (M.B.)
Footnotes
The ChREBPβ sequence data has been deposited to NCBI Genbank under accession number JQ437838. The microarray data has been deposited to NCBI Gene Expression Omnibus under accession number GSE35378.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Author Contributions: M.A.H. and B.B.K. planned the experiments, interpreted the data, and wrote the paper. M.A.H. also performed the experiments except the human studies. O.D.P. and J.V. assisted with experimental design and provided technical support. N.A.A., M.R.S., M.B. and S.K. performed the human studies.
References
- 1.Shepherd PR, Kahn BB. Glucose transporters and insulin action--implications for insulin resistance and diabetes mellitus. N Engl J Med. 1999;341:248–257. doi: 10.1056/NEJM199907223410406. [DOI] [PubMed] [Google Scholar]
- 2.Attie AD, Scherer PE. Adipocyte metabolism and obesity. J Lipid Res. 2009;50 (Suppl):S395–399. doi: 10.1194/jlr.R800057-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boden G. Role of fatty acids in the pathogenesis of insulin resistance and NIDDM. Diabetes. 1997;46:3–10. [PubMed] [Google Scholar]
- 4.Abel ED, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 5.Shepherd PR, et al. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268:22243–22246. [PubMed] [Google Scholar]
- 6.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci U S A. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts R, et al. Markers of de novo lipogenesis in adipose tissue: associations with small adipocytes and insulin sensitivity in humans. Diabetologia. 2009;52:882–890. doi: 10.1007/s00125-009-1300-4. [DOI] [PubMed] [Google Scholar]
- 8.Hoffstedt J, Forster D, Lofgren P. Impaired subcutaneous adipocyte lipogenesis is associated with systemic insulin resistance and increased apolipoprotein B/AI ratio in men and women. J Intern Med. 2007;262:131–139. doi: 10.1111/j.1365-2796.2007.01811.x. [DOI] [PubMed] [Google Scholar]
- 9.Kursawe R, et al. Cellularity and adipogenic profile of the abdominal subcutaneous adipose tissue from obese adolescents: association with insulin resistance and hepatic steatosis. Diabetes. 2010;59:2288–2296. doi: 10.2337/db10-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranganathan G, et al. The lipogenic enzymes DGAT1, FAS, and LPL in adipose tissue: effects of obesity, insulin resistance, and TZD treatment. J Lipid Res. 2006;47:2444–2450. doi: 10.1194/jlr.M600248-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 14.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cha JY, Repa JJ. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J Biol Chem. 2007;282:743–751. doi: 10.1074/jbc.M605023200. [DOI] [PubMed] [Google Scholar]
- 16.Denechaud PD, Girard J, Postic C. Carbohydrate responsive element binding protein and lipid homeostasis. Curr Opin Lipidol. 2008;19:301–306. doi: 10.1097/MOL.0b013e3282ffafaa. [DOI] [PubMed] [Google Scholar]
- 17.Pashkov V, et al. Regulator of G protein signaling (RGS16) inhibits hepatic fatty acid oxidation in a carbohydrate response element-binding protein (ChREBP)-dependent manner. J Biol Chem. 286:15116–15125. doi: 10.1074/jbc.M110.216234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology. 2005;146:2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- 19.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281:28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 20.Kuriyama H, et al. Compensatory increase in fatty acid synthesis in adipose tissue of mice with conditional deficiency of SCAP in liver. Cell Metab. 2005;1:41–51. doi: 10.1016/j.cmet.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Cao H, et al. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho E, Kotani K, Peroni OD, Kahn BB. Adipose-specific overexpression of GLUT4 reverses insulin resistance and diabetes in mice lacking GLUT4 selectively in muscle. Am J Physiol Endocrinol Metab. 2005;289:E551–561. doi: 10.1152/ajpendo.00116.2005. [DOI] [PubMed] [Google Scholar]
- 23.Ahima RS, Flier JS. Leptin. Annu Rev Physiol. 2000;62:413–437. doi: 10.1146/annurev.physiol.62.1.413. [DOI] [PubMed] [Google Scholar]
- 24.Nadler ST, et al. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci U S A. 2000;97:11371–11376. doi: 10.1073/pnas.97.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caesar R, et al. A combined transcriptomics and lipidomics analysis of subcutaneous, epididymal and mesenteric adipose tissue reveals marked functional differences. PLoS One. 2010;5:e11525. doi: 10.1371/journal.pone.0011525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gnudi L, Tozzo E, Shepherd PR, Bliss JL, Kahn BB. High level overexpression of glucose transporter-4 driven by an adipose-specific promoter is maintained in transgenic mice on a high fat diet, but does not prevent impaired glucose tolerance. Endocrinology. 1995;136:995–1002. doi: 10.1210/endo.136.3.7867610. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez J, Palou A, Pico C. Response to carbohydrate and fat refeeding in the expression of genes involved in nutrient partitioning and metabolism: striking effects on fibroblast growth factor-21 induction. Endocrinology. 2009;150:5341–5350. doi: 10.1210/en.2009-0466. [DOI] [PubMed] [Google Scholar]
- 28.Shimano H, et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoeckman AK, Ma L, Towle HC. Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. J Biol Chem. 2004;279:15662–15669. doi: 10.1074/jbc.M311301200. [DOI] [PubMed] [Google Scholar]
- 30.Kabashima T, Kawaguchi T, Wadzinski BE, Uyeda K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc Natl Acad Sci U S A. 2003;100:5107–5112. doi: 10.1073/pnas.0730817100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li MV, Chang B, Imamura M, Poungvarin N, Chan L. Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes. 2006;55:1179–1189. doi: 10.2337/db05-0822. [DOI] [PubMed] [Google Scholar]
- 32.Iizuka K, Takeda J, Horikawa Y. Hepatic overexpression of dominant negative Mlx improves metabolic profile in diabetes-prone C57BL/6J mice. Biochem Biophys Res Commun. 2009;379:499–504. doi: 10.1016/j.bbrc.2008.12.100. [DOI] [PubMed] [Google Scholar]
- 33.Shih HM, Liu Z, Towle HC. Two CACGTG motifs with proper spacing dictate the carbohydrate regulation of hepatic gene transcription. J Biol Chem. 1995;270:21991–21997. doi: 10.1074/jbc.270.37.21991. [DOI] [PubMed] [Google Scholar]
- 34.Bernstein BE, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukasawa M, Ge Q, Wynn RM, Ishii S, Uyeda K. Coordinate regulation/localization of the carbohydrate responsive binding protein (ChREBP) by two nuclear export signal sites: discovery of a new leucine-rich nuclear export signal site. Biochem Biophys Res Commun. 391:1166–1169. doi: 10.1016/j.bbrc.2009.11.115. [DOI] [PubMed] [Google Scholar]
- 36.Tsatsos NG, Towle HC. Glucose activation of ChREBP in hepatocytes occurs via a two-step mechanism. Biochem Biophys Res Commun. 2006;340:449–456. doi: 10.1016/j.bbrc.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 37.Fabbrini E, et al. Intrahepatic fat, not visceral fat, is linked with metabolic complications of obesity. Proc Natl Acad Sci U S A. 2009;106:15430–15435. doi: 10.1073/pnas.0904944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donnelly KL, et al. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115:1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eguchi J, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotani K, Peroni OD, Minokoshi Y, Boss O, Kahn BB. GLUT4 glucose transporter deficiency increases hepatic lipid production and peripheral lipid utilization. J Clin Invest. 2004;114:1666–1675. doi: 10.1172/JCI21341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kloting N, et al. Serum retinol-binding protein is more highly expressed in visceral than in subcutaneous adipose tissue and is a marker of intra-abdominal fat mass. Cell Metab. 2007;6:79–87. doi: 10.1016/j.cmet.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Hems DA, Rath EA, Verrinder TR. Fatty acid synthesis in liver and adipose tissue of normal and genetically obese (ob/ob) mice during the 24-hour cycle. Biochem J. 1975;150:167–173. doi: 10.1042/bj1500167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39:D876–882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 38:D620–625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.