Abstract
An emerging picture of the nature of immune systems across animal phyla reveals both conservatism of some features and the appearance among and within phyla of novel, lineage-specific defense solutions. The latter collectively represent a major and underappreciated form of animal diversity. Factors influencing this macroevolutionary (above the species level) pattern of novelty are considered and include adoption of different life styles, life histories, and body plans; a general advantage of being distinctive with respect to immune defenses; and the responses required to cope with parasites, many of which afflict hosts in a lineage-specific manner. This large-scale pattern of novelty implies that immunological phenomena can affect microevolutionary processes (at the population level within species) that can eventually lead to macroevolutionary events such as speciation, radiations, or extinctions. Immunologically based phenomena play a role in favoring intraspecific diversification, specialization and host specificity of parasites, and mechanisms are discussed whereby this could lead to parasite speciation. Host switching – the acquisition of new host species by parasites – is a major mechanism that drives parasite diversity and is frequently involved in disease emergence. It is also one that can be favored by reductions in immune competence of new hosts. Mechanisms involving immune phenomena favoring intraspecific diversification and speciation of host species are also discussed. A macroevolutionary perspective on immunology is invaluable in today’s world, including the need to study a broader range of species with distinctive immune systems. Many of these species are faced with extinction, another macroevolutionary process influenced by immune phenomena.
Keywords: macroevolution, immunology, host–parasite interactions, evolutionary immunology, host shifting, biodiversity
Introduction
Recent years have witnessed a dramatic increase in our understanding of the diversity of immune systems across animal phyla (Flajnik and Kasahara, 2010; Messier-Solek et al., 2010; Rast and Litman, 2010; this volume). Availability of genome sequences from a broad variety of animals coupled with an increased appreciation for the diversity of their defenses has given the study of immunity a much stronger evolutionary foundation, one that has been further enriched by studies of plant immunity and responses of bacteria and archaea to threats to their genomes (Horvath and Barrangou, 2010). The increasing depth and breadth of immunological studies is also bringing to light a greater awareness of the impact that immunity has had on all forms of life, especially parasites. Here “parasite” is used inclusively, referring to infectious agents ranging from viruses to bacteria to protists to multicellular helminths. The features uniting parasites are that they infect hosts, provoke some degree of fitness-diminishing harm, prompt the deployment of immune responses, and undertake immune evasive actions. “Immune systems” are referred to as those molecules, cells, tissues, and organs that protect hosts from parasites (see caveats below). This discussion excludes a broad range of behavioral defenses like preening (Bush and Clayton, 2006) or avoidance (e.g., Mooring et al., 2003; Garnick et al., 2010).
Here I attempt to draw together ideas that begin to put immunological phenomena into a broader macroevolutionary context. Macroevolution is the study of patterns, and the evolutionary processes that have generated them, at or above the species level (Stanley, 1998; Levinton, 2001). It is the study of how and why life has diversified, and attempts to document how and why lineages of organisms have come into being and either given rise to additional lineages or gone extinct. The process of speciation is germane to macroevolutionary studies because it is the process responsible for increasing the diversity of life forms. Extinction and its causes are also an essential part of such studies.
The attributes of immune systems across the spectrum of animal diversity provide a new way to view and reinterpret the diversity of animals. Immune systems exhibit unforeseen novelty and thus offer new insights into major selective forces influencing animal life. Also, phenomena that are fundamentally immunological provide fertile ground for investigating the impact of immunity as a driver of biodiversity. The role of immune systems in macroevolutionary processes is one that deserves recognition and more study.
In considering what is to follow, several caveats should be borne in mind:
-
(1)
We are just beginning to view molecular components of immune systems from a broad sampling of animal phyla. Detailed analyses are still few for how these presumptive immune components actually function in defense, and how critical their roles might be in protecting the organisms in question.
-
(2)
Also poorly known are the specific parasite threats faced by the more obscure groups of animals serving as hosts.
-
(3)
Many of the examples of immunological novelty presented below emphasize differences at the phylum level. Some phyla such as the Arthropoda, Mollusca, and Chordata are immense in species numbers and undoubtedly collectively employ as yet many undiscovered immune capabilities. Also, some of the smaller animal phyla are essentially unexplored with respect to their immune systems. Once understood, these additional examples will only add to the overall diversity of immune responses.
-
(4)
It is not always easy to circumscribe “the immune system” or an “immune response.” This is particularly so in cases where potent defenses for parasites result from selection for variant alleles for genes like hemoglobin B or apolipoprotein L-1 that otherwise might not be considered a core part of the immune system (Anstee, 2010; Barreiro and Quintana-Murci, 2010; Genovese et al., 2010; Wheeler, 2010).
Immunological Novelty Among Animal Phyla: An Underappreciated form of Diversity
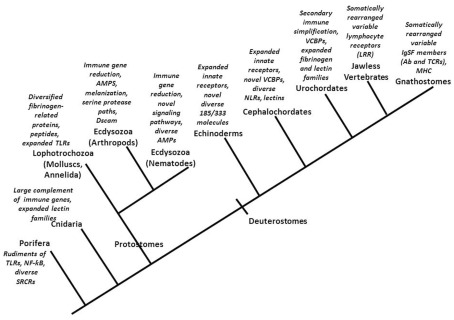
Discoveries relating to the innate immune systems of plants, flies, and mammals have tended to accentuate the similarities among them, implying a grand conservatism even across kingdoms with respect to basic immune system design and function. Indeed, there are intriguing similarities between the membrane-associated and intracytoplasmic receptors of plants and animals suggestive that some basic solutions to recognition and response to parasites have been conserved since at least the time animal and plant lineages diverged. However, particularly given that some of these similarities are a likely result of convergent evolution rather than indicative of a common origin (Ausubel, 2005), conserved immune features are not the emphasis here. Rather, this overview accentuates the emergence of immunological novelty among and within animal phyla (Figure 1; Table 1).
Figure 1.
An overview of some of the novel features associated with immune responses of representatives of major animal lineages (see text for details). TLR, Toll-like receptor; AMP, antimicrobial peptide; Dscam, Down syndrome cell adhesion protein; VCBPs, variable region-containing chitin-binding proteins; NLRs, intracellular NOD-like receptors; LRR, leucine-rich repeat; IgSF, immunoglobulin superfamily; Ab, antibodies; TCR, T cell receptor; MHC, major histocompatibility complex.
Table 1.
An overview of animal immune systems emphasizing distinctive immune features and diversified defense molecules.
| Phylum | Body plan | Specialized defense cells | Genome reduction | Distinctive immune features | Diversified defense molecules | Reference |
|---|---|---|---|---|---|---|
| Porifera (sponges) | Diploblastic | No | No | TIR domains, lack external LRR, MyD88 homolog, NF-κB rudiments, lack death domains, LPS-interacting proteins, perforin-like molecules, antiviral 2′-5′ oligoadenylate system | Scavenger receptor cysteine-rich molecules | Wiens et al. (2007), Schroder et al. (2008) |
| Cnidaria (anemones, corals, jellyfish, Hydra | Diploblastic with mesoglea | No | No | Bonafide TLR and NF-κB pathway, complement-3 component, multiple NACHT domains and NLRs likely, recognizable RAG1 homolog. Hydra lacks canonical TLR with both LRR and TIR, allorecognition molecules present including with IgSF domains | Diverse C-type lectins | Wood-Charlson and Weis (2009), Miller et al. (2007), Augustin et al. (2010), Hemmrich et al. (2007), Nicotra et al. (2009) |
| BILATERIA PROTOSTOMES ECDYSOZOA | ||||||
| Nematoda (Caenorhabditis and many others) | Triploblastic | No | Yes | One TLR that plays a role in defense against some bacteria, lacks canonical Toll pathway and NLRs, but can mount inducible defense responses and have several novel defense-related signaling pathways | Produces many caenopores and other antimicrobial peptides including 42 NLPs caenacins, diverse C-type lectins | Irazoqui et al. (2010), Roeder et al. (2010) |
| Arthropoda (insects, crustaceans, and many others | Triploblastic | Yes | Yes | NLR and complement-3 components lacking, have one or two TLRs functioning with NF-κB pathways, TLRs do not engage ligands directly, produce antimicrobial peptides, CLIP-protease cascades, melanization reactions | Multimeric fibrinogen-related molecules, the IgSF member Dscam with multiple isoforms | Lemaitre and Hoffmann (2007), Kanost et al. (2004), Dong and Dimopoulos (2009), Schmucker and Chen (2009), Dong et al. (2006), Brites et al. (2008), Smith et al. (2011) |
| LOPHOTROCHOZOA | ||||||
| Annelida (earthworms, leeches, polychaetes) | Triploblastic | Yes | No | Over 100 TLR genes, extensive involvement of coelomocytes from coelom in defense, cytotoxicity against allogeneic cells, hemolytic and clotting factors in body fluid, antimicrobial peptides, and protective body mucus | Expanded set of TLRs in polychaetes | Davidson et al. (2008), Salzet et al. (2006) |
| Mollusca (cephalopods, snails, bivalves chitons, others) | Triploblastic | Yes | No | Involvement of body mucus in protection, TLRs and Toll pathway present, little melanization, hemocytes working with lectins like galectins or fibrinogen-containing proteins (FREPs), mitogen-activated protein kinase pathways, complement-like factors antimicrobial peptides | Somatic diversification of FREPS by point mutation and gene conversion, large C-type lectin families, diversified myticin C in bivalves | Hanington et al. (2010a,b), Bayne (2009), Zhang et al. (2004), Yoshino et al. (2008), Costa et al. (2009), Loker (2010) |
| DEUTEROSTOMES | ||||||
| Echinodermata (sea urchins, starfish, brittle stars, crinoids, sea cucumbers) | Triploblastic | Yes | No | “Expanded” innate immune system with >220 TLRs, >200 NLRs, >200 SRCR genes, Toll pathway, lectin and alternative pathways, RAG1 and RAG2 homologs present | In addition to expanded sets of TLRs, NLRs, and SRCRs, they also have novel Sp 185/333 gene family producing diverse immune proteins | Messier-Solek et al. (2010), Hibino et al. (2006), Ghosh et al. (2010) |
| CHORDATA | ||||||
| Cephalochordata (amphioxus) | Triploblastic | Yes | No | “Expanded innate immune system with ∼72 TLRs, >92 NLRs, −270 SRCRs, >1200 C-type lectins, possesses distinctive variable region-containing chitin-binding proteins (VCBPs), have functioning complement, RAG1 and possibly RAG2 present | In addition to expanded sets of TLRs, NLRs, SRCRs, and lectins, they have polymorphic VCBPs arising from a variety of mechanisms | Huang et al. (2008), Messier-Solek et al. (2010), Dishaw et al. (2008, 2010), Cannon and Litman (2009) |
| Urochordata (tunicates) | Triploblastic | Yes | Yes | V-like and CI-like domains present, VCBPs present, have three TLRs, lack complement, or expansion of any gene family relevant to vertebrate immunity | Have expanded families of C-type lectins and fibrinogen-related proteins | Azumi et al. (2003), Cannon et al. (2004) |
| Agnathans (lampreys, hagfish) | Triploblastic | Yes | No | Lack RAG1 and RAG2 and do not produce TCRs or immunoglobulins, but do have two basic types of lymphocytes and produce variable lymphocyte receptors (VLRs) with LRRs | Produce diverse VLRs through somatic rearrangement of modules with leucine-rich repeats | Pancer et al. (2004), Rogozin et al. (2007) |
| Gnathostomes (fish, amphibians reptiles, birds, mammals) | Triploblastic | Yes | No | Modest numbers of TLRs (10–25) and NLRs (20–35), three complement pathways, somatic diversification of both Ig and TCR, involvement of MHC, memory, heightened secondary response, affinity maturation in some | Both Ig and TCRs diversified somatically | Messier-Solek et al. (2010), Litman et al. (2010) |
The most basal animal group is the phylum Porifera, the sponges (Srivastava et al., 2010). Sponges lack the complex tissue and organ structure found in other animal phyla, and lack cells specialized for protection from parasites. Although sponge immunobiology is in its infancy, one of the best-known sponges, Suberites domuncula, possesses membrane-spanning molecules that contain an intracellular Toll-interleukin 1 receptor (TIR) domain, though it lacks an external leucine-rich repeat pattern recognition receptor more typical of TLRs. On the basis of having a MyD88 homolog, S. domuncula has at least the rudiments of an NF-κB signaling pathway. Sponges also have molecules for attacking bacterial membranes, presumptive antiviral responses (Schroder et al., 2008), and diversified scavenger receptor cysteine-rich molecules of unknown function (Wiens et al., 2007).
Among basal animals, it is members of the phylum Cnidaria (jellyfish, Hydra, anemones, and corals) that have proven most surprising with respect to the large size and content of their genomes, including their immune systems. Cnidarians have distinct tissues but lack organs and are generally considered to be diploblastic, meaning they have recognizable ecto- and endoderm, but lack well-developed mesoderm tissue. Like sponges, they lack recognizable specialized immune cells. However, the starlet sea anemone Nematostella vectensis has at least one TLR, an NF-κB signaling pathway, a homolog of a complement 3-like molecule, the likely presence of functioning intracellular NOD-like receptors (NLRs), perforin-like molecules, diverse C-type lectins (Wood-Charlson and Weis, 2009), and even a recognizable homolog of the recombination activating gene, RAG1 (Miller et al., 2007; Augustin et al., 2010). Cnidarians often live in colonies and have to contend with encroaching competitors, including conspecifics. For this they have well-developed mechanisms to recognize self and non-self. One of the responsible molecules has been identified, and is surface expressed, polymorphic and possesses three external immunoglobulin superfamily (IgSF) domains (Nicotra et al., 2009).
The remaining animals, the Bilateria, are bilaterally symmetrical and triploblastic, with well-developed tissues and organs. They often have specialized immune cells. Most fall into two major lineages, the protostomes and deuterostomes. Among the protostomes, representatives of the molting clade (Ecdysozoa) have been most extensively studied in an immune context, as this clade includes nematodes and arthropods, both containing well-studied model organisms. Caenorhabditis elegans and other nematodes have reduced genomes and from an immunological perspective are surprising for what they do not have. Although C. elegans has one bonafide TLR that plays a role in defense against some bacteria (Tenor and Aballay, 2008), it lacks Myd88, NF-κB and several other components of the canonical Toll pathway. NLRs are also lacking. Nonetheless, C. elegans can mount inducible, parasite specific responses. It has several novel signaling pathways for defense (Irazoqui et al., 2010) and produces many distinctive antimicrobial peptides (AMPs) for protection from bacteria (Roeder et al., 2010). C-type lectins may serve as recognition molecules in C. elegans. The preoccupation with production of AMPs by gut cells reflects their diet of bacteria, which could include potential parasites (Roeder et al., 2010).
Another prominent model of ecdysozoan immunity is Drosophila (Lemaitre and Hoffmann, 2007), but increasingly other insect are studied as well, such as mosquitoes because of their role in transmitting human parasites (Bartholomay et al., 2010). Insects have dedicated immune cells such as plasmatocytes and lamellocytes that circulate through their open circulatory system and phagocytose or encapsulate foreign objects. The Drosophila immune system also shows evidence of gene loss: it lacks the C3-like complement component and NLR homologs found in cnidarians. Their TLRs are different from those of vertebrates in that they do not engage microbial ligands directly. Of their nine TLR genes, only one or two function in immunity, activating NF-κB signaling pathways in the fat body to produce AMPs (Lemaitre and Hoffmann, 2007). Insects are by no means immunologically bereft though. They have a number of other effective defense components not seen in many other organisms. They have elaborate cascades of CLIP-serine proteases that mediate and coordinate phagocytosis, nodule formation, encapsulation, and AMP formation, and they can deposit layers of melanin around foreign objects (Kanost et al., 2004). They engage multimeric fibrinogen-related proteins (FREPs) in parasite recognition (Dong and Dimopoulos, 2009) and employ Down syndrome cell adhesion molecule (Dscam), a member of the IgSF, in antigen recognition. Tens of thousands of Dscam isoforms can potentially be generated by alternative splicing (Schmucker and Chen, 2009) and parasite challenge-specific Dscam splice form repertoires can be produced (Dong et al., 2006).
Insect studies provide additional examples of immunological novelty, at the ordinal, family, or even genus level. One example is provided by Drosophila and Anopheles, both in the same order (Diptera), but representing very different life styles and having been separate lineages for 250 million years. Gene families involved in immunity have evolved rapidly and divergently in the two dipterans. For example, with respect to thioester containing proteins (TEPs), Anopheles has 10 genes and Drosophila only four, with only one orthologous pair between the two. Anopheles has 58 fibrinogen-like immunogenes whereas Drosophila has only 14, with only two shared orthologous pairs (Dong and Dimopoulos, 2009).
At the family level, a comparison of three different mosquito genera (Aedes, Anopheles, and Culex, all in the Culicidae) has revealed prominent genus specific expansion of some immune gene families (Bartholomay et al., 2010). Comparative studies of Drosophila species are particularly revealing, showing that novel immune genes and immune gene families have originated relatively recently, suggestive of a role of parasites in driving adaptive evolution in flies (Sackton et al., 2007). Furthermore, for particular immune proteins, the amino acids under positive selection vary between Drosophila species groups, suggesting different fly species experience different parasite pressures (Morales-Hojas et al., 2009). Insects with very different life styles, such as the social honey bees (Evans et al., 2006) and ants (Smith et al., 2011), and symbiont-dependent aphids (Pennisi, 2009) likewise have immune systems that are surprisingly divergent from the Drosophila immune system.
The other major lineage of protostomes, the Lophotrochozoa, includes prominent groups such as the flatworms, annelids, and mollusks. In the polychete annelid Capitella capitata, TLRs have undergone an expansion to over 100 genes, most of which are similar, suggestive of recent duplication. Another annelid, the leech Helobdella robusta, has only 16 TLR homologs which are not only highly divergent from one another but also are not orthologous to any of the polychete sequences (Davidson et al., 2008). In the freshwater snail Biomphalaria glabrata, FREPs are encoded by an expanded gene family, and are implicated in defense against gastropod parasites such as digenetic trematodes (Hanington et al., 2010a,b). In B. glabrata, FREPs are particularly noteworthy for being comprised of juxtaposed IgSF and fibrinogen domains, and for the fact they are somatically diversified during the production of hemocytes by the snails (Zhang et al., 2004). Expanded families of C-type lectins are present in other mollusks and the bivalve Mytilus edulis is capable of generating diversified forms of the AMP myticin C both within and among individuals (Costa et al., 2009).
Among invertebrate deuterostomes, the sea urchin has proven surprising in featuring dramatic echinoderm-specific expansion of several recognition molecules (Hibino et al., 2006). Sea urchins possess >220 TLR genes (vertebrates usually have 21–25), >200 NLRs (mammals have 20–35), >200 SRCR genes (humans have 16; Messier-Solek et al., 2010), and a novel Sp 185/333 gene family. The latter gene family produces a repertoire of defense proteins more diverse than the sequence diversity encoded in the genes, indicative of the presence of another mechanism to generate diversity (Ghosh et al., 2010). Sea urchins also possess an NF-κB pathway, lectin and alternative complement pathways and homologs of RAG1 and RAG2, but do not produce immunoglobulins (Ig), T cell receptors (TCRs), or have a major histocompatibility complex (MHC; Hibino et al., 2006).
With respect to our own phylum, the Chordata, the cephalochordate Branchiostoma (better known as Amphioxus or the lancelet), is novel in having expanded families of TLRs, NLRs, and SRCRs (Huang et al., 2008), over 1,200 C-type lectins, and an extraordinary diversity in adaptors/facilitators and signaling/effector domains functioning downstream from their NLRs (Huang et al., 2008; Messier-Solek et al., 2010). Amphioxus also possesses distinctive variable region-containing chitin-binding proteins (VCBPs; Dishaw et al., 2008; Cannon and Litman, 2009) which are further distinguished by high levels of polymorphism, resulting in yet another distinct “hyper-diversified,” multigene immune receptors family (Dishaw et al., 2010). Cephalochordates have a functioning complement system operating via the alternative and lectin pathways, including with a distinctive expanded number of C1q-like genes (Huang et al., 2008; Messier-Solek et al., 2010). A RAG1 gene is present, and possibly a RAG2 gene as well (Dong et al., 2005).
The urochordates, or tunicates, the sister group to the vertebrates, in the same immunological vein as nematodes and flies, are surprising for what they do not have. None of the genes playing a pivotal role in adaptive immunity in the jawed vertebrates are present. MHC, TCRs, Ig, RAG, and activation-induced cytidine deaminase (AID) genes are all lacking. V-like and C1-like domains are present and VCBPs have been identified (Cannon et al., 2004), and they do have complement components, three TLRs, an expanded family of C-type lectins and FREPs. However, urochordates lack obvious expansions of any gene family highly relevant to vertebrate immunity (Azumi et al., 2003). Based on what we know thus far, genome reduction is the hallmark of urochordate immunobiology.
Even closer to home are the agnathans or jawless vertebrates, lampreys and hagfish, the sister group to the jawed vertebrates or gnathostomes. We now know they lack RAG1 and RAG2 and do not produce TCRs or Ig, however, they have a remarkable ability to make highly diverse variable lymphocyte receptors (VLRs) that consist of somatically re-arranged modules containing leucine-rich repeats (Pancer et al., 2004). It is striking that agnathans and gnathostomes have adopted divergent solutions to the same problem of generation of recognition capability, both of which involve rearrangements of germ-line encoded genes, but in entirely different ways with different starting sets of molecules.
The basic gnathostome immune system, the one most familiar to immunologists, features a close collaboration between innate and adaptive arms. As noted above, relative to some of the invertebrate deuterostomes such as echinoderms or cephalochordates, gnathostome innate immune components are modest in numbers, typically possessing 10–25 TLRs and 20–35 NLRs (Messier-Solek et al., 2010). The gnathostome adaptive immune system features somatic diversification of both TCRs and Ig, requiring for this process RAG1 and RAG2, the former likely derived and modified from a transib-like transposon (Fugmann, 2010). The gnathostome immune system works in conjunction with a unique antigen processing and presentation system, the MHC, to limit self-reactivity. It is notable for its specificity, its emphasis on expansion of relevant clones of lymphocytes, and for its memory and capacity to produce heightened secondary responses long after primary stimulation (Litman et al., 2010). The ongoing discovery of new types of immune cells (Neill et al., 2010; Saenz et al., 2010) and novel receptors (Parra et al., 2007) strongly suggests there are more fundamental insights to come with respect to gnathostome immunology. Furthermore, and a point relevant for the general discussion here, there is considerable variability among gnathostomes in how their immune systems function (Flajnik and Kasahara, 2010).
To conclude this overview, it is indeed remarkable that organisms as diverse as cnidarians and humans have some immune architecture such as TLRs (and associated pathways) and NLRs in common. However, it is argued from the examples provided above that at least as compelling are the differences among and within phyla, even among species in a genus. Surprises abound, such as in the unexpectedly complete set of immune genes found in basal cnidarians and the immune genome reductions exhibited by nematodes, arthropods, and urochordates. Even the more familiar examples of conservatism such as TLRs and NLRs in arthropods and vertebrates may have been derived independently (Hughes, 1998; Hughes and Piontkivska, 2008; Zhang et al., 2010). Large lineage-specific gene expansions such as noted for echinoderms, and domain reshuffling such as for invertebrate NLRs (Zhang et al., 2010) have occurred, creating remarkable heterogeneities among and within phyla. Layered on top of this are other forms of innovation such as elaboration of novel signaling pathways and production of associated AMPs in nematodes, distinct antigen recognition and melanin-deposition systems in arthropods, and the emergence of several distinct mechanisms for generating diverse antigen receptors in mollusks, arthropods, echinoderms, cephalochordates, agnathans, and gnathostomes. From this it is concluded that immune systems across and within phyla have a remarkable propensity to generate novelty and distinctiveness. As we learn about the immune systems of more animals, this diversity is bound to increase.
Drivers of Immunological Novelty
It is hardly surprising that immune systems are so variable. Animals have been extant and diversifying for up to 800 million years (Erwin et al., 2011). They have adopted a diversity of life styles: sessile, colonial predators; inhabitants of extreme environments dependent on chemosynthetic symbionts; animals that are at times net producers of energy due to their photosynthetic symbionts; pelagic species that live in enormous schools; inhabitants of arid terrestrial environments; social species living in large colonies; and endoparasites that are so modified morphologically as to belie their origins, to name just a few. These different life styles will impose very different exposures to potential parasites.
Similarly, life histories vary radically from wind-dispersed organisms like tardigrades or rotifers that live in ephemeral habitats or that have life spans measured in hours, to sessile filter feeders like marine bivalves that routinely live for over 100 years in the same location. The role of life history traits such as survival rates and reproductive output are predicted to strongly influence the extent and kinds of particular immune responses that might be expected both among and within particular host species (Lee, 2006).
Another factor likely to have influenced immune capability is the nature and extent of commitment to mutualistic symbionts. Some animals have established mutualistic associations with what are essentially monocultures of specialized bacteria (Nyholm and Nishiguchi, 2008; Pais et al., 2008). Others, like ourselves, are dependent on a diversity of both archaeal and bacterial mutualists to which our immune system has made extensive accommodation. Third party symbionts have long played a role in educating, augmenting, and modulating animal immune systems (Turnbaugh et al., 2007). The outcome of host–parasite interactions is often influenced by third party symbionts which probably play a far great role in host defense in many animal groups than customarily realized (Loker, 1994; Welchman et al., 2009; Gross et al., 2009; Eberl, 2010).
The adoption of body plans differing in complexity and mass has also influenced immune system structure and function. It has been argued that evolution of the vertebrate jaw and an accompanying predatory life style introduced parasites into the gut and required a more elaborate adaptive immune system that now typifies gnathostomes (Matsunaga and Rahman, 1998). It has also been argued that the complexities of adaptive immunity could not have evolved in animals with limited numbers of cells or with small size or simplified body architecture (Hauton and Smith, 2007). With respect to body mass, for vertebrates, it has been suggested that the number of B and likely T cells in a clone scale with body mass as does the B cell repertoire (Wiegel and Perelson, 2004). The general point is that the adoption of different habitats, life histories, symbionts, and bodies of differing body mass and complexity are all factors that will influence immune system design and mode of action.
A general advantage of immunological novelty – being different from your neighbors
In addition to the above considerations, all organisms have to contend with another category of symbionts, namely parasites. Because viruses, bacteria, and protists were present before animals arose, all animals from their inception would have had to contend with these parasites. Several modes of transmission of such parasites among early animals of disparate lineages were available, including: intimate proximity of many different kinds of animals (such as on a coral reef), predation, presence of vectors imbibing blood or plant juices containing parasites, and even one parasite serving as a vector for another, as for example a trematode vectoring a bacterium into new animal hosts.
In such a situation, where frequent transfer of parasites was possible among hosts from even disparate phyla, if all emerging animal lineages had the same defenses, it would be possible for an effective parasite that had overcome the defenses of one host group to simply spread into another host phylum. Consequently, having an immune system with distinctive means of antigen recognition and/or novel effector mechanisms would have been a distinct advantage when inevitably confronted with parasites that had evolved in other host groups (Figure 2). Being immunologically different increases the odds that parasites from other inhabitants of the same coral reef are not as easily acquired. The notion that a parasite can track and exploit a common host genotype creating an advantage of rareness has been predicted and observed in specific host–parasite systems (Trachtenberg et al., 2003; Wolinska and Spaak, 2009), further suggestive of a similar dynamic favoring distinctiveness or “avoidance of commonness” among members of different host lineages.
Figure 2.
One scenario for early in the divergence of animals is that different lineages (A–D) have fundamentally similar immune systems such that they all are colonized by the same parasites. In the case shown at the top, an immunological innovation occurred in lineage A, that allowed it to resist these parasites. This may have permitted a subsequent radiation in “parasite-free space” in this host lineage. At the bottom, lineage D has acquired a lineage-specific parasite different from those previously experienced. This requires an immunological accommodation that causes the immune system of lineage D1 to diverge. Both the host lineage and the lineage-specific parasites along with them may subsequently diverge.
Lineage-specific animal parasites as additional drivers of novelty
Once animals began to diversify, a major trend was for some animals to parasitize others. Some animal parasites became wholly or largely committed to particular lineages of animal hosts, in which they subsequently diversified. Such lineage-specific parasites (some examples in Table 2) are another general factor expected to drive immunological novelty. These parasites often establish prolonged, intimate, and extensive infections in their chosen hosts that have profound fitness consequences such as castration or death (Lafferty and Kuris, 2009a). Furthermore, given the phylogenetic diversity represented among these parasites, it is not surprising they would evolve novel methods of infectivity. For example, ichneumonid and braconid hymenopteran parasitoids have acquired mutualistic polydnaviruses that function to suppress the immune responsiveness of their hosts and facilitate parasitoid infection (Webb et al., 2009). In contrast, without the aid of viral symbionts, larvae of digenetic trematodes secrete both anti-oxidants and immunosuppressive factors that down-regulate snail host immune components for a period sufficient to enable them to complete their lengthy period of larval development (Hanington et al., 2010a). Therefore, the immune response devised by a particular host lineage afflicted with its own phylogenetically distinct, host specific, and harmful parasites would likely be divergent from the responses mounted by a different host group experiencing its own lineage-specific parasites (Figure 2). This is not to imply that only animal parasites have developed lineage-specific associations with hosts, but merely serves to show that specialized parasites can help us understand the origins of immunological novelties.
Table 2.
Examples of parasites that are “lineage-specific” in particular host groups during at least part of their life cycles.
| Parasites | Species (n) | Hosts |
|---|---|---|
| Digenetic trematodes (as larvae) | 18,000 | Mollusks (usually gastropods) |
| Unionid bivalves | 1,000 | Fish |
| Rhizocephalans | 260 | Decapod crustaceans |
| Poecilostome copepods | 400 | Cnidarians |
| Sisyridid sponge flies | 50 | FW sponges and some bryozoans |
| Hydracarina water mites | 5,000 | Aquatic insects |
| Tantulocaridans | 30 | Crustaceans |
| Acroceridae | >500 | Spiders |
| Pipunculids | 1,388 | Leafhoppers and planthoppers |
| Tetracneminae chalcidoid wasps | 815 | Pseudococcid insects |
| Banchinid ichneumonid wasps | 1,500 | Lepidopteran insects |
| Ichneumoninae ichneumonid wasps | 350 genera | Lepidopteran insects |
| Aphidiinae braconid wasps | 400 | Aphids |
| Conopidae | 800 | Mostly wasps and bees |
This list is not exhaustive and merely serves to illustrate the concept that particular host lineages acquire unique parasites that are likely to have distinctive methods of infectivity that could influence how their host’s immune systems are shaped by selection. Although cases where members of the parasite groups identified colonize hosts outside the indicated host lineage certainly occur, they do not negate the idea that the host groups indicated above have been far more affected in aggregate than a host lineage containing an isolated member harboring a peculiar outlying parasite. Also, for some huge groups, such as the ichneumonid wasps, although when viewed more inclusively they infect much broader groups of hosts (such as insects or terrestrial arthropods), the point remains they have had relatively little impact on other major host lineages beyond the insects.
A Role for Immunity in Generating Parasite Biodiversity
The macroevolutionary patterns noted above with respect to novelty in defense strategies among animal lineages could not occur if there were not microevolutionary processes ultimately involved in generating them. These microevolutionary processes occur below the species level, within and among populations of either host or parasite species, and might culminate in speciation of either participant. Speciation may be accompanied by colonization of new habitats, and further divergence to create major new lineages. Starting with the process of parasite diversification, the sections below discuss how the involvement of immunology in microevolutionary processes could lead to events that can help explain the macroevolutionary patterns discussed above.
The following quotes outline some of the key ideas for how immunity can play a role in generation of parasite diversity.
“For a pathogen, the selective pressures arising from the host immune system are a major influence on the evolution of mechanisms of infectivity and of immune-recognition avoidance” (Acevedo-Whitehouse and Cunningham, 2006).
“In parasitism an essential factor in survival is immune escape, which allows a parasite to resist host attack. Immune escape is a mechanism for reducing gene flow at the level of the compatibility filter because its result is assortative survival (Combes and Théron, 2000) as opposed to assortative mating.” (Combes, 2001, p. 154).
“The stepwise coevolutionary process results in extreme specialization and complex defense mechanisms…specialization is likely to increase the rate of speciation that may occur in both host and parasite” (Price, 1980)
“Host specificity thus is an ideal prerequisite for rapid speciation” (Mayr, 1963).
Immunity is often responsible for the specialization to particular host species shown by parasites
Parasites are often cited as examples of specialists because they have limited ranges of host species, often with restricted ranges of habitats within their chosen hosts. For example, the lineage-specific parasites mentioned above, often show considerable specificity to particular species or genera within their adopted host lineages. Most animal parasites are host specific (Poulin and Keeney, 2008; Agosta et al., 2010), but this by no means is to suggest that generalists do not occur: parasites like Schistosoma japonicum, Toxoplasma gondii, Borrelia burgdorferi, or the rabies virus routinely infect a remarkably broad range of host species. Using molecular techniques to identify parasites, species formerly considered to be generalists have in some cases been shown to be complexes of cryptic, host specific species (Poulin and Keeney, 2008).
As eloquently documented by Combes (2001), both encounter and compatibility filters operate to restrict the spectrum of host usage. Encounter filters pertain in situations where host and parasite live in different geographic localities, have different ecological circumstances, or where host or parasite behavioral tendencies preclude contact. The compatibility filter refers to barriers imposed by the host that prevent infection once contact has occurred. The compatibility filter includes both physiological and biochemical suitability of the host to support the parasite, and the active defense provided by the immune system.
Encounter filters are undeniably important in restricting parasite host range. Many examples of emerging infections (Goss et al., 2009; Gray et al., 2009; Pfeffer and Dobler, 2010) owe their emergence to a change in the encounter filter such that a new combination of parasite (often including its vector) and host are juxtaposed (Daszak et al., 2000; Parrish et al., 2008; Weissenbock et al., 2010). Emerging diseases are an indication that parasite infectivity is not always dependent on a long accommodation to a particular host species: lack of contact may have prevented prior infections. Similarly, experimental infections of new hosts with parasites essentially bypass the encounter filter, and are sometimes successful (Poulin and Keeney, 2008), affirming the reality and importance of the encounter filter.
With respect to the compatibility filter, a role for unsuitability should not be discounted and could be manifested in several ways, such as a lack of receptors needed for efficient viral entry into cells (Parrish et al., 2008), lack of appropriate structures for parasite attachment (Tompkins and Clayton, 1999), or by a general failure to provide the biochemical environment needed for the parasite to survive (Sullivan and Richards, 1981).
Although unsuitability is probably underappreciated with respect to preventing infections, relying on the possibility of being unsuitable is not a cogent defense strategy. The importance of active immunity to the compatibility filter is illustrated by several lines of evidence.
-
(1)
As illustrated by HIV, when the immune system is compromised, the door is opened to opportunists that themselves can become life-threatening (Holmes et al., 2003).
-
(2)
Genetic defects in the immune system, such as with TLRs, are associated with increased susceptibility to several different pathogens (Qureshi and Medzhitov, 2003).
-
(3)
Experimental exposures of hosts to parasites they have not previously encountered often fail (Bowen, 1976; Bozeman et al., 1981; Vidal-Martinez et al., 1994; Philips and Clarkson, 1998; Sapp and Loker, 2000; Duke, 2004; de Vienne et al., 2009; Giraud et al., 2010) or the parasite replicates poorly or is inefficiently transmitted in a new host (Komar et al., 2003; Parrish et al., 2008). Table 3 provides examples of parasites in novel hosts that are engaged and killed by immune responses.
-
(4)
Host defense genes are under strong selection and are conspicuous for evolving quickly (Sackton et al., 2007; Viljakainen et al., 2009; Barreiro and Quintana-Murci, 2010; Schulte et al., 2010).
-
(5)
The extraordinary diversity of strategies undertaken by parasites to evade, manipulate, or suppress the immune system is testament to the impact of immunity on their success (Schmid-Hempel, 2008). These evasive strategies have been shown in some cases to be specific with respect to the particular hosts involved (Table 4), providing a mechanistic basis for the connection between immunity and parasite specialization.
Table 3.
Examples of colonizing parasites, or parasites placed in novel hosts, that are killed or limited by immune responses.
| Infection of the crab Pachygrapsus marmoratus with the rhizocephalan barnacle Sacculina carcini results in melanization of larvae in thoracic ganglia (Kuris et al., 2007) |
| Antibody/factor that activates complement in serum of the non-host Raja radiata kills the tapeworm Acanthobothrium quadripartitum whereas larvae survive in serum of the normal host, Raja naevus (McVicar and Fletcher, 1970) |
| Destruction of cercariae of avian schistosomes in the skin of mammals associated with a mixed Th1/Th2 lymphocyte cytokine response followed by more polarized Th2 response upon repeated exposures (Horak and Kolarova, 2005) |
| Encapsulation of hymenopteran parasitoids by hemocytes of non-permissive insect hosts (Schmidt et al., 2001) |
| Lysis of the trypanosome Trypanosoma brucei brucei by apolipoprotein L-1 in serum of humans who are refractory to this subspecies (Wheeler, 2010). |
| Disruption of the Erk-STAT1 signaling pathway allows cross species transmission of the normally rabbit-specific myxoma virus to mice (Wang et al., 2004) |
| Animal handlers who were exposed to a new coronavirus developed antibodies to the new virus and did not develop clinical infections (Guan et al., 2003) |
| Species specific forms of APOBEC3G and other innate, intracellular defense components, can prevent cross species transfer of lentiviruses (Mangeat et al., 2004; VandeWoude et al., 2010) |
Table 4.
Examples of parasite immune evasive factors that are host specific in their action.
| A staphylococcal complement inhibitor that specifically blocks human C4b2a and C3bBb, interfering with additional C3b deposition through classical, lectin or alternative pathways (Rooijakkers et al., 2005). Sung et al. (2008) found several genes conserved in all Staphylococcus aureus isolates from humans were variable or missing in one or more animal isolates, including fnbA, fnbB, and coa. |
| Human and murine chlamydial infections depend on different virulence factor genes that coevolved to counter host species specific IFN-γ-mediated effector responses mounted by the particular host species (Nelson et al., 2005). |
| Orf virus encodes a secreted protein GIF that binds to and inhibits GM-CSF and IL-2 of ovines but not humans or murines, consistent with the idea that Orf virus is evolutionarily adapted to sheep as its primary host (Seet et al., 2003). |
| Different strains of influenza A virus likely have NS1 genes adapted to antagonize the IFNα/β antiviral system of their specific host species (Garcia-Sastre, 2006). |
| In a review of the interactions between monogenean parasites and their fish hosts, Buchmann and Lindenstrøm (2002) concluded that “immune evasion mechanisms are probably a main factor in host specificity.” |
| Rosengard et al. (2002) noted that the smallpox inhibitor of complement enzymes (SPICE) is nearly 100-fold more potent than the vaccinia homolog in inactivating human C3b and sixfold more potent at inactivating C4b, providing evidence for how variola proteins are particularly adept at overcoming human immunity relative to vaccinia. |
| The host specificity of three species of Bacillus (B. cereus, B. thuringiensis, and B. anthracis) is determined by the presence of virulence plasmids that determine the type of particular virulence factors produced (Gohar et al., 2005). |
Given that the consequences to a parasite for engaging the compatibility filter of an atypical host could be disastrous and result in its death, strong selection to avoid colonization of such hosts would be expected in some cases. Similar considerations may also apply to the host as well, as mounting an immune response can be costly and detrimental (Graham et al., 2011). This means that some avoidance behaviors attributed to the encounter filter may actually be a consequence of the operation of a strong host immune response (Kuris et al., 2007; Keesing et al., 2009).
To conclude this section, specialization and attendant host specificity is a central, emergent property of parasitism and has multiple underlying determinants, involving both encounter and compatibility filters. The ubiquity of host defenses and the evidence that they often eliminate novel parasites argue that host immune systems play a critical role in limiting parasite host ranges and thereby at least in part dictate the specialization so characteristic of parasitic organisms. Also in support of this claim is that some patterns of parasite host specificity can be attributed to the operation of specific immune evasion strategies, and that such strategies are pervasive among parasites.
Does specialization dictated by immunity actually lead to parasite speciation?
As exemplified by the parasites indicated in Table 2, in addition to being specialized to exploit particular host groups, they are remarkably diverse in species, as are the lineages of many parasites. One of the potential consequences of specialization, including in an immunological context, is diversification in species, of both parasite and host lineages. The mechanisms involved in promoting speciation remain a matter of active investigation and for the discussion below, the purpose is to indicate that immunological phenomena may play a role in this process that deserves further attention. One prominent mechanism of parasite speciation is switching to a new host species, and the role of accommodation to the immune system of new hosts to permit such switches is discussed in a separate section below.
A second mechanism is co-speciation. For some parasite groups closely tied to their hosts and with limited options for colonization of new hosts, such as sucking lice on burrowing mammals (Light and Hafner, 2008), speciation may occur if the hosts upon which they are found themselves speciate, often following a physical separation of populations of the host species. In such cases, persistence of new daughter parasite species should be favored by the fact that the parental species had already achieved successful accommodation to the parental host species. Although the actual role of specific immune phenomena in influencing the persistence of incipient parasite species in co-speciating systems is not known, an important underlying role for a preexisting immunological accommodation between parental host and parasite species that also favors persistence of the new parasite species should not be discounted.
Another important way in which specialization dictated by immunological phenomena can increase the probability of formation of new parasite species is by promoting intraspecific diversification. The interactions between a particular host and parasite species can be expected to be variable across space (Wood et al., 2007). Parasite abundance will vary across local scales, possibly because of the variable presence of other hosts needed to complete its life cycle (Byers et al., 2008). Other parasite species impacting the same host may be present or absent, such that the host experiences different overall parasite pressure in different locations within its range. Furthermore, the host itself will be variable across its range owing to its responses to other local circumstances. All of these factors conspire to create heterogeneities with respect to how the host potentially mounts immune responses to the parasite (Kraaijeveld and Godfray, 1999; Thomas et al., 2000; Lindstrom et al., 2004; Kalbe and Kurtz, 2006; Blais et al., 2007; Bryan-Walker et al., 2007; Scharsack et al., 2007; Matthews et al., 2010). Variability within parasite species with respect to infectivity to their hosts is a pervasive phenomenon (Carius et al., 2001; Schulenburg and Ewbank, 2004; Seppala et al., 2007; Vorburger et al., 2009) and this is likely driven in part by variations in immune evasive measures taken by parasites (Hammerschmidt and Kurtz, 2005; Cornet et al., 2009; Vorburger et al., 2009). These dynamics are compatible with general theoretical predictions that parasite variation is driven by immunity, and hosts themselves are variable with respect to immunity due to pressure posed by parasitism (Frank, 2002). Immune responses are drivers for parasite diversification (Summers et al., 2003; Lazzaro and Little, 2009; McKeever, 2009). An overall increase in intraspecific genetic variability, with that variation partitioned into regionally differentiated parasite populations accommodated to local host populations provide rich opportunities for further divergence.
One possibility for further divergence is that local adaptation to host immunity could potentially lead to “assortative survival” (Combes and Théron, 2000; Combes, 2001, p. 154), meaning that the only options for mating (parasites frequently seek mates and undergo sexual reproduction within their hosts) occur between individuals able to survive in hosts with similar immune capability that are vulnerable to the same parasite immune evasive capacity (Giraud et al., 2010). This would further accentuate local differentiation of parasites, potentially leading to ecological speciation of the parasite, particularly if subsequent gene flow is prevented by failure of immune adapted parasites to thrive in hosts (from other localities) with different immune capacities.
It must be noted that fluctuations in local patterns of abundance of hosts and parasites may diminish the strength of local adaptation and promote gene flow such that speciation is precluded (Lazzaro and Little, 2009), and that in general, evidence that parasite speciation is effected by underlying immune mechanisms is sparse. However, given the need for parasites to accommodate to a host’s internal environment and that a host species is likely to be confronted with varying parasite pressure, it seems host immune responses will favor diversification in parasite lineages. To add an additional dimension to the concept that spatially variable relationships favor parasite diversification, it has recently been argued a general underlying mechanism favoring biological diversification is the existence of localized parasite-coevolutionary races that select hosts to prefer immunologically similar conspecifics and to avoid out-group individuals, thereby minimizing the risk of exotic disease acquisition (Fincher and Thornhill, 2008). By promoting strong intraspecific diversification within host species based on avoidance of contagion, this mechanism has also been predicted to lead to parasite diversification (Fincher and Thornhill, 2008).
To conclude this section, all of these observations fit into the more generalized geographic mosaic theory of coevolved relationships (Thompson, 2005): in this particular case, local adaptations based on immunological accommodation of host and parasite can lead to diversification of parasites and potentially speciation.
A Role for Immunity in Host Switching and Parasite Diversification
An important way diversity in parasite lineages is generated, one that has increasingly come to light from molecular phylogenetic studies and the study of emerging diseases, is via switching to new hosts (Table 5). Although successful host switching cannot be a ubiquitous process, otherwise we might expect to find only a few species of generalist parasites instead of a predominance of host specific parasites, clearly it has been an important factor historically and examples continue to be regularly documented. A priori, it seems logical that most successful switches would be to hosts not phylogenetically distant from the original host species. Such close range switches are likely favored by a degree of phenotypic plasticity and preadaptation (exaptation) of the parasite and its use of phylogenetically conserved resources in the new host species such that new attributes are not needed to overcome a new host’s immune system (de Vienne et al., 2009; Agosta et al., 2010). For example, in a study of host switches in bats involving the fast-evolving RNA virus causing rabies, the success of cross species transfers diminished as the phylogenetic distances among the hosts involved increased (Streicker et al., 2010).
Table 5.
Examples of parasite groups exhibiting hosts switches likely to have played a major role in diversification of that group.
| With respect to Plasmodium and related genera of blood parasites, major clades are associated with shifts into different families of dipteran vectors, and the Plasmodium species of birds and squamate reptiles show evidence of repeated switching back and forth (Martinsen et al., 2008). |
| Major lineages within the blood fluke genus Schistosoma are defined by acquisition of different genera of even families of snail intermediate hosts, by host switching (Morgan et al., 2003). Long-range host shifts involving acquisition of both new snail and vertebrate hosts appear to have occurred during the history of schistosomes (Brant and Loker, 2005). |
| Zietara and Lumme (2002) note that as many as 20,000 species of the monogenean genus Gyrodactylus may exist, and note that in a study of one subgenus (Limnonephrotus) that several host switch events were statistically confirmed, including into new host families, supporting the idea that host switching is a means to drive innovation and adaptive radiation in these ectoparasites. |
| It appears that host switching has been common in trypanorhynch tapeworms, one of the most diverse and abundant groups of metazoan parasites of elasmobranchs (Olson et al., 2010) |
| Coronaviruses have likely undergone several host switches, between mouse and rat, chicken and turkey, birds and mammals, and between humans and other mammals (Rest and Mindell, 2003). |
| Braconid wasps of the subfamily Euphorinae have undergone extensive host switching among phylogenetically distantly related insect host groups, often followed by adaptive radiations of the parasitoids within a particular host lineage (Shaw, 1988). |
| “Infection of a novel host is the most frequent cause of fungal emerging disease” (Stukenbrock and McDonald, 2008; Giraud et al., 2010) |
It is also possible for parasite switches to occur when the original and new host species are not closely related (Brant and Loker, 2005). This has been observed for emerging human parasites for which ungulates and carnivores were more likely originating host species than primates, and it was concluded that an already broad host range as opposed to the phylogenetic relatedness of the new and old host species was the more important factor dictating success in interspecific parasite jumps (Woolhouse et al., 2005).
In any case, a host switch can lead to a speciation event if the parasite in the new host becomes isolated from the founding stock, or can have even more profound effects if the switch is into a new host lineage and leads to the founding of a diverse new parasite lineage (Agosta, 2006; Janz et al., 2006; Hoberg and Brooks, 2008; Martinsen et al., 2008; Refrégier et al., 2008; Winkler et al., 2009; Giraud et al., 2010; Nyman, 2010). The isolation of the switching parasite from the founding stock is reinforced because even a single individual may be able to establish a new population and because differing ecological circumstances of the new host may preclude mixing of parasite progeny with the source population: the new parasites may never get back into the original host and thus mate only with other parasites in the new host. Assortative survival and mating would again be factors favoring isolation of the founding parasites. Host switching is also relevant to the idea stated above that if a host lineage acquires a new parasite, it may then have its immune system substantially altered. Particularly if the parasite is successful and radiates, then the immune system of the new host lineage may be forced to diverge to adjust to the new challenge.
Several examples from the literature of emerging infectious disease indicate that switches are often favored by changes in the encounter and not the compatibility filter (Woolhouse et al., 2005). Ecological circumstances have exposed humans to a parasite they previously did not encounter. Such examples of host switches, particularly if the new host is distantly related to the original host, would seem to argue against the points made in the preceding sections regarding the importance of parasite accommodation to the idiosyncrasies of their host’s immune system. If immunity is important in restricting parasite host ranges, how can such switches occur?
First, these conspicuous successes need to be weighed against all the encounters between novel parasite and host combinations that fail and therefore go unnoticed, which is likely a far more frequent outcome (de Vienne et al., 2009; Tunaz and Stanley, 2009; Giraud et al., 2010; see also Table 3). In cross species transfers of rabies into bats, the vast majority are dead ends: they did not establish sustained infections (Streicker et al., 2010). Although some of the failures could be explicable because of less frequent contact among more distantly related bats (the encounter filter), increasingly divergent defense systems leading to higher levels of innate resistance were also invoked as an explanation (Daszak, 2010; Streicker et al., 2010). The role of immune systems in preventing such infections would be easy to underestimate because the result is a failed experiment that in all probability we never even knew had happened. In a similar vein, a survey of field-trapped insects in turkey revealed that 98% exhibited some kind of melanotic hemocyte nodule (Tunaz and Stanley, 2009). Such host reactions provide a convenient historical record of previous parasite encounters (Kuris et al., 2007). It was concluded that insects are regularly challenged by infections from which they recover. The action of innate immunity in routinely preventing acquisition of new parasites is probably considerable and easy to underestimate.
Secondly, host switches would be favored if the new parasite, as exemplified by HIV, directly attacks the host’s immune system and compromises it, or if the new host is immunocompromised by some other means. Diminished levels of immune competence can occur for several reasons, including ones likely to have been in operation throughout animal evolution. One possible means is that the host’s indigenous parasites might use immunosuppression to favor initiation and persistence of their own long-term infections (Table 6) and thereby facilitate colonization of that host by other parasites (Krasnov et al., 2005). An intriguing possibility is that the successful colonization of a host species by one or more immunosuppressive parasites might then favor colonization by opportunist parasites, resulting in an unusually diverse parasite fauna supported by that host. The large number of species of larval digenetic trematodes known to be supported by some snail species (Loker et al., 1981; Lafferty and Kuris, 2009b) might exemplify this possibility.
Table 6.
Examples of immunosuppression by one parasite that could favor acquisition of new parasites, and potentially an eventual speciation event.
| Varroa mites (Varroa destructor) in honey bees (Apis mellifera) suppress the activity of several immune-related genes (encoding both antimicrobial peptides and enzymes) and favor higher infection titers with the deformed wing virus (Yang and Cox-Foster, 2005). |
| Drosophila simulans infected with Wolbachia have reduced ability to encapsulate eggs of the parasitoid Leptopilina heterotoma (Fytrou et al., 2006). |
| The malaria parasite Plasmodium gallinaceum suppresses the encapsulation response of the mosquito Aedes aegypti (Boëte et al., 2004). |
| Two acanthocephalan parasites Pomphorhynchus laevis and Polymorphus minutus both have the effect of decreasing the standing level of immune defense (as measured by reduced phenoloxidase enzyme activity) in their local gammarid hosts, Gammarus pulex, but not in their more recently introduced host Gammarus roeseli (Rigaud and Moret, 2003). |
| Hymenopteran parasitoids induce immunosuppression in their host insects in part by the injection of polydnaviruses which target and inhibit both cellular and humoral components of the host response (e.g., Labropoulou et al., 2008) and the parasitized hosts become increasingly susceptible to opportunistic infections by viruses (Rivkin et al., 2006), bacteria (Shelby et al., 1998), and other parasitoids (Guzo and Stoltz, 1985). |
| As noted by Lie (1982), interference by trematode larvae with gastropod defense responses appears to be a common consequence of infection (Hanington et al., 2010a), and the presence of one trematode infection can facilitate the colonization of an infected snail by trematodes that would not ordinarily be able to infect that species of host (Walker, 1979; Southgate et al., 1989). |
| HIV in people was associated with parasites that rarely if ever had been implicated in causing human disease including microsporidia, cryptosporidia, JC virus, and Mycobacterium avium (Kovacs and Masur, 2008). |
| Studies of parasite communities suggest that taxonomic distinctiveness of ectoparasites and endoparasite richness are positively correlated across species of rodent hosts, indicative of immune responses to some parasites depleting energy reserves and facilitating colonization by others (Krasnov et al., 2005). |
High host density, stressful thermal (Bruno et al., 2007) or oxygen regimes (Aeby and Santavy, 2006), and even mating (Rolff and Siva-Jothy, 2002) are some natural situations that can also lower immune competence. To this list can be added a number of human-imposed immune stressors including crowded aquaculture conditions (Flemming et al., 2007), use of harmful chemicals on fields or roads (Rohr et al., 2008; Karraker and Ruthig, 2009), altered diets (Sahu et al., 2008), elevated or altered environmental nutrient conditions (McKenzie and Townsend, 2007; Wedekind et al., 2010), and deliberate implementation of immunosuppressive therapies. Not only can these situations lower host immune competence, they may also increase parasite virulence and thereby alter probabilities of successful colonization in a new host species (Wedekind et al., 2010).
A switch into an immunocompromised individual of a new host species is likely to be temporary and not lead to speciation unless the new parasite can better adapt to its new host, at the same time with minimal gene flow occurring with conspecifics from its ancestral host. Availability of populations of similarly immunocompromised new hosts that allow continued transmission and adaptation of the new parasite host could favor divergence from the founding parasite and speciation.
A Role for Immunity in Driving Host Diversity?
How might immunological phenomena influence the degree of diversity shown among the hosts contending with parasites? As highlighted below, parasite pressure clearly favors immunological diversification at microevolutionary scales. Whether this diversification contributes in a meaningful way to host speciation remains controversial, but has attracted considerable attention and is gaining some support, as discussed below.
As early as Haldane (1949), and as more recently underscored (Frank, 2002; Lazzaro and Little, 2009), it has been recognized that parasites drive polymorphisms in host immune competence, particularly in variable environments. This can occur by balancing (Wegner, 2008) or disruptive selection (Duffy et al., 2008; Matthews et al., 2010). High levels of polymorphism are found in several genes of both the innate and adaptive immune systems (Hill, 2001; Trowsdale and Parham, 2004; Acevedo-Whitehouse and Cunningham, 2006). MHC genes show a predominance of non-synonymous over synonymous mutations in their peptide-binding regions, and have extensive allelic diversity, indicative of strong role of selection, generally considered to be mediated by parasites. For example, parasite pressure is believed to have promoted maintenance of high MHC diversity in sticklebacks (Wegner et al., 2003) and Atlantic salmon (Dionne et al., 2007). In humans, regional differences in HLA class I diversity has been associated with intracellular parasite richness (Prugnolle et al., 2005). Across species comparisons of rodents have associated helminth species richness with increased MHC class II polymorphisms (de Bellocq et al., 2008). MHC genes are also known for their role in mediating mate choice through olfactory systems in humans (McClintock et al., 2002), rodents (Sommer, 2005), and fishes (Landry et al., 2001; Milinski et al., 2005).
The evidence supporting the idea that variability in immune response driven by parasites can be a factor favoring speciation of host lineages is mostly correlational, but is supported by a growing body of literature. One general factor favoring this is the development of strong local immunological accommodations of particular host populations to the distinctive parasites they encounter, such that across a broader host range different host populations differ significantly with respect to the nature of their responses (Wheatley, 1980; Blais et al., 2007; Scharsack et al., 2007; Matthews et al., 2010). For example, malaria might be encountered in some but not all parts of the host range where appropriate mosquitoes were present, or different suites of parasites might be encountered in different foraging habitats such as rivers or lakes (Eizaguirre et al., 2011). Differences among populations of the same host species with respect to their immune defenses have been noted in Drosophila against parasitoids (Kraaijeveld and Godfray, 1999), Darwin’s finches (Lindstrom et al., 2004), sticklebacks coping with eye flukes (Kalbe and Kurtz, 2006), and marine amphipods infected with trematodes (Thomas et al., 2000; Bryan-Walker et al., 2007). Differentiation resulting from spatially variable antagonistic interactions with parasites would in this case provide the substrate for further diversification of host lineages (Thompson, 2005, 2009).
The impact of this local adaptation could be augmented by assortative mating mediated by sexual selection to favor further divergence (van Doorn et al., 2009). According to this line of thought, those locally adapted hosts that best resist parasites are able to elaborate ornaments that signal superior resistance to parasites, such that local mates preferentially breed with them. If these hosts were transplanted to other host populations with their own distinct parasite challenges, they would not be as resistant, their sexual ornamentation would suffer, and they would not be selected for mating. Thus a combination of natural and sexual selection could favor divergence of new host species.
Parasites can exert strong selection on traits known to affect mate choice (Hamilton and Zuk, 1982; Poulin and Thomas, 1999; MacColl, 2009) and in some cases the genes involved also have immune functions, such as genes of the MHC (Milinski et al., 2005; Milinski, 2006; Eizaguirre et al., 2009, 2011). MHC genes have been considered as possible “magic traits” in sticklebacks, influencing both defense and mate choice (Matthews et al., 2010). MHC divergence in a closely related and sympatric pair of cichlid species from Lake Malawi has been proposed to be a result of local host–parasite-coevolutionary associations, and to have influenced odor-mediated mate choice, and ultimately to have favored speciation (Blais et al., 2007).
Diverging cichlids of the genus Pundamilia in Lake Victoria provide another example of how local adaptation and assortative mating (both potentially influenced by immune traits) may work together to promote host speciation. In this system, females have a preference for conspicuously colored males. These bright colors seem to be reliable indicators of male fitness, including resistance to parasites. Conditions of light penetration favor blue males in shallow depths and red males in deeper waters. The parasites encountered at different depths also vary in density and composition such that habitat-specific defenses could occur. Females from the depths prefer brighter red males whereas those from shallow water prefer brighter blue males, potentially leading to divergence, with visual cues playing a key role. This example points out the immunology may work in concert with a number of other forces such as predator avoidance or dietary preferences which all conspire to reinforce divergence of the two incipient species by visual means (Maan and Seehausen, 2010). Mating among hosts between different locations would break down these differences, but might be disfavored if the progeny had reduced resistance to any local set of parasites.
“Infectious speciation” as exemplified by interactions between Drosophila species and inherited, endosymbiotic Wolbachia bacteria provides another possible mechanism for the involvement of immune processes in host speciation. For a group of six related species in the D. paulistorum complex, each species has its own distinct host specific obligatory and mutualistic Wolbachia with which it has achieved accommodation. This accommodation likely involves suppression of immune pathways involving apoptosis of infected cells. In hybrids, the Wolbachia over-replicate and cause embryonic inviability and male sterility, suggesting the unique host accommodation has been lost. In addition to such post-mating isolation, it has also been shown that females can detect and will reject males harboring the wrong symbiont, thus further reinforcing isolation (Miller et al., 2010).
Among animal hosts, hybrids are often more susceptible to parasites than parental species (see overviews provided by Fritz, 1999; Wolinska et al., 2008), potentially favoring isolation of the parental host species, but other outcomes have also been noted and the responsiveness of hybrids to infection recorded 1 year might differ from those reported the next. This implies that the interactions between hybrids and the parasites they experience exhibit complex temporal dynamics and that the parasites themselves have undergone complicated changes as a result of their hosts’ hybridization history that have not been sufficiently investigated (Detwiler and Criscione, 2010). Certainly some studies suggest that isolation of incipient parental species might be reinforced by a breakdown of co-adapted immune gene complexes among their hybrids. Also, in some cases, the act of hybridization contributes to the formation of new species by allopolyploidy, as has been postulated for anurans of the genus Xenopus. Hybrids in this case often have increased resistance for parasites, potentially providing a selective advantage to favor the persistence of new species of recent hybrid origin (Jackson and Tinsley, 2003).
Another hypothesized mechanism favoring diversification of host lineages is that localized interactions with particular parasites allows immunological accommodation to them, resulting in strong preference for interactions with individuals with similar immune accommodation and philopatry (limited host dispersal). This is coupled with avoidance of out-group individuals that might lead to introduction of exotic parasites (Fincher and Thornhill, 2008). Diverse parasite populations are thus hypothesized to drive diverse host populations, and ultimately speciation, leading to a general correlation between host and parasite biodiversity (Fincher and Thornhill, 2008).
The Role of Immunology in Extinction Events
Extinction too is a macroevolutionary process, including the major pulse of extinction events currently underway. Habitat destruction, human overpopulation, industrialization, threats from introduced species, emerging diseases, and global climate change, have lead to predictions that up to 50% of all species will be lost in the next 50 years (Pimm and Raven, 2000; Koh et al., 2004; Thomas et al., 2004). The study of immunology is relevant to extinction in at least three broad contexts outlined below.
First, as noted above, the interactions between parasite and host often lead the parasite to specialization and host specificity which may in part be dictated by interactions with the host immune system. It has long been argued that specialization leads to a greater probability of extinction because if the host on which the specialized parasite is dependent undergoes severe population fluctuations or itself goes extinct, the parasite will soon follow: a co-extinction event has occurred. Co-extinctions involving pairs of mutualists or host–parasite units may be the most common forms of loss of biodiversity (Koh et al., 2004; Dunn et al., 2009). By comparison, a generalist parasite able to exploit alternative hosts would have a greater likelihood of surviving under similar circumstances. The evolutionary trajectories taken by parasites have been much debated, and specialized parasites have been shown to give rise to large lineages of specialized, or even to more generalized descendents (Johnson et al., 2009). However, host specificity remains a salient feature affecting the odds of extinction (Koh et al., 2004; Poulin and Keeney, 2008).
Second, an inescapable feature of the modern world is the frequency with which invasions of exotic species occur (Torchin et al., 2002). Invasive species might be either hosts or parasites (possibly including their vectors), and all can present dire and often unpredictable consequences, including extinction, for indigenous organisms (Wyatt et al., 2008). A role for immunology in influencing invasions can occur in at least three ways:
-
(a)
Introduced host species often leave their native parasites behind and although they are likely to be colonized by parasites in their new environment, in some cases this colonization is slow to occur, such that they experience relatively low parasite burdens for a long time. Insofar as immune responses are costly to mount and the harmful effects of parasites are avoided, the invading species may enjoy a distinct advantage over its competitors in its relative freedom from parasites, particularly if they can adopt relatively low cost defense measures against their maladapted parasites (Lee and Klasing, 2004). The relative inability of parasites to colonize the new intruder is a testament to the specialization often required to achieve infectivity, as noted above.
-
(b)
If an introduced host is accompanied by some of its indigenous parasite fauna, then related hosts in the newly colonized area may then have to contend with a parasite to which they are not accustomed, a process that may take decades to achieve (Hedrick et al., 2003; Taraschewski, 2006). This is particularly likely to cause problems if the colonized area is an isolated habitat like an island where hosts have simply not encountered comparable parasites previously. The devastating impact of the introduction of mosquitoes followed by avian malaria and avian pox on the endemic honeycreepers of the Hawaiian islands, likely causing both extinctions and slowing recovery of still extant species, is an iconic example (Atkinson and Samuel, 2010). Islands often favor speciation yet the subsequent colonization of mainland habitats by island species is likely to be limited and unsuccessful due to active transmission of parasites there that a migrant is unable to handle immunologically.
-
(c)
In some cases, invasion of a parasite can occur even without the benefit of a simultaneous introduction of its indigenous host. An example is the eel swimbladder nematode Anguillicola crassus which has been introduced from the Orient into Europe where it provokes intense tissue reactions from European eel species (Taraschewski, 2006). By contrast, Oriental eels mount immune responses that prevent a high and robust population of worms from building up.
All of these examples centered on introductions have macroevolutionary implications as they might lead to expansion and radiation of hosts and their parasites into new habitats, or may directly cause extirpations of indigenous hosts (and possibly their co-adapted parasites as well). Immunological phenomena provide proximal causal explanations. Once again, as with the discussion above regarding host switching, there may be significance to what we do not see as well: many parasite introductions fail because the colonist is unable to breach indigenous host defenses and host introductions fail because they are ill-prepared for indigenous parasites. For example, the introduction of the American rainbow trout Oncorhynchus mykiss into Europe failed because they encountered the parasite Myxobolus cerebralis which is normally transmitted by the indigenous brown trout, Salmo trutta: rainbows succumbed to whirling disease in Europe (Hedrick et al., 2003). When this parasite was introduced into North America, brown trout (themselves also introduced) were already well-adapted to it but indigenous rainbows and other salmonids were not and have suffered outbreaks of whirling disease as a consequence.
A third broad context in which immunology becomes relevant to extinction, and one that is much in evidence today, is the role played by diminution of diversity in key immune loci such as the MHC. This can result in vulnerability of endangered host species to general parasite attack and thus extinction (Radwan et al., 2010). Loci other than the MHC may also play major roles in dictating susceptibility to particular groups of parasites (Behnke et al., 2003), and polymorphism in non-MHC genes are relevant in resistance to both tuberculosis (Ottenhoff et al., 2005) and malaria (Hill, 2001). MHC genes are estimated to account for only about half of the genetic variability for resistance traits (Acevedo-Whitehouse and Cunningham, 2006).
Regardless of the immune locus involved, the role of random drift in diminishing allelic diversity in bottlenecked or fragmented host populations would seem to increase the risk of successful parasite attack and greatly increase extinction probabilities. There is a need to determine if other factors like genome-wide inbreeding depression are more important in causing extinction, to understand why some species depauperate in MHC diversity seemingly continue to thrive (Hedrick, 2003; Radwan et al., 2010), for increased transfer of information from the study of the main immunological models to endangered species, and to study other polymorphic genes involved in effective defense (Acevedo-Whitehouse and Cunningham, 2006).
Conclusion and Future Studies
A picture of the structure and function of immune systems across animal phyla is slowly emerging, but so many organisms remain unstudied we lack perspective on how representative our current picture is. Do all tunicates, for example, reveal evidence of pronounced genome reduction with respect to immune function, or is this a characteristic of just the few species studied to any extent? What kind of pressure from parasites or otherwise has driven the expansion of all the innate immunity genes evident in organisms like sea urchins, and how do these animals regulate and orchestrate effective responses given the multiplicity of defense molecules they possess? In their 100+ years life span, how frequently must marine bivalves mount immune responses and how do they avoid the problem of fast-evolving parasites from “locking on” and overrunning them? In the meantime, it is clear that the approaches taken to achieve immune defense are diverse, frequently innovative and often capable of generating diverse repertoires of defense molecules that blur the distinctions commonly made to distinguish adaptive from innate immunity. The diversity in immune systems among and within phyla is in and of itself a major macroevolutionary pattern that should become a more central part of how we characterize animal diversity, including in textbooks. We need to know what the full pattern actually is, and the pattern begs an explanation for the processes involved in generating it.
Study of model parasite–host systems shows that the strength of selection imposed on particular immune genes is strong, and can result in some of the fastest evolutionary rates known for metazoans. In other words, parasite–host immune interactions strongly influence anagenesis, the evolutionary changes occurring within parasite or host lineages. If we adopt a broader perspective with a longer time frame, it seems likely such intense interactions will be seen to have an impact on cladogenesis, the origin of lineages, as well. It seems the overall impact of infection and immunity should attract as much attention as predation, competition, or other biotic interactions in shaping the overall diversity of animal life. Needed are more empirical studies over longer time frames to provide more specific mechanistic insights as to how host immune responses drive diversification of parasites and how this can lead to speciation events, potentially in both parasite and host lineages. Further revelation of specific genes, often not considered part of the immune system per se, and how they facilitate defense against particular parasites and might favor evolutionary divergence among parasites are needed. Finally, placing immunology in a macroevolutionary perspective can hopefully provide insights for understanding today’s world in which a host of rapid changes greatly increase the odds for extinction of many animals.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by NIH grants AI024340, and by 1P20RR18754, the latter from the Institute Development Award (IDeA) Program of the National Center for Research Resources, NIH.
References
- Acevedo-Whitehouse K., Cunningham A. A. (2006). Is MHC enough for understanding wildlife immunogenetics? Trends in Ecol. Evol. 21, 433–438 10.1016/j.tree.2006.05.010 [DOI] [PubMed] [Google Scholar]
- Aeby G. S., Santavy D. L. (2006). Factors affecting susceptibility of the coral Montastraea faveolata to black-band disease. Mar. Ecol. Prog. Ser. 318, 103–110 10.3354/meps318103 [DOI] [Google Scholar]
- Agosta S. J. (2006). On ecological fitting, plant–insect associations, herbivore host shifts, and host plant selection. Oikos 114, 556–565 10.1111/j.2006.0030-1299.15025.x [DOI] [Google Scholar]
- Agosta S. J., Janz N., Brooks D. R. (2010). How specialists can be generalists: resolving the “parasite paradox” and implications for emerging infectious disease. Zoologia 27, 151–162 [Google Scholar]
- Anstee D. J. (2010). The relationship between blood groups and disease. Blood 115, 4635–4643 10.1182/blood-2010-01-261859 [DOI] [PubMed] [Google Scholar]
- Atkinson C. T., Samuel M. D. (2010). Avian malaria Plasmodium relictum in native Hawaiian forest birds: epizootiology and demographic impacts on ’apapane Himatione sanguinea. J. Avian Biol. 41, 357–366 10.1111/j.1600-048X.2009.04915.x [DOI] [Google Scholar]
- Augustin R., Sebastian S., Bosch T. C. G. (2010). How Hydra senses and destrys microbes. Semin. Immunol. 22, 54–58 10.1016/j.smim.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Ausubel F. M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979 10.1038/ni1253 [DOI] [PubMed] [Google Scholar]
- Azumi K., De Santis R., De Tomaso A., Rigoutsos I., Yoshizaki F., Pinto M. R., Marino R., Shida K., Ikeda M., Ikeda M., Arai M., Inoue Y., Shimizu T., Satoh N., Rokhsar D. S., Du Pasquier L., Kasahara M., Satake M., Nonaka M. (2003). Genomic analysis of immunity in a urochordate and the emergence of the vertebrate immune system: “waiting for Godot”. Immunogenetics 55, 570–581 10.1007/s00251-003-0606-5 [DOI] [PubMed] [Google Scholar]
- Barreiro L. B., Quintana-Murci L. (2010). From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 11, 17–30 10.1038/nrg2698 [DOI] [PubMed] [Google Scholar]
- Bartholomay L. C., Waterhouse R. M., Mayhew G. F., Campbell C. L., Michel K., Zou Z., Ramirez J. L., Das S., Alvarez K., Arensburger P., Bryant B., Chapman S. B., Dong Y. M., Erickson S. M., Karunaratne S. H. P. P., Kokoza V., Kodira C. D., Pignatelli P., Shin S. W., Vanlandingham D. L., Atkinson P. W., Birren B., Christophides G. K., Clem R. J., Hemingway J., Higgs S., Megy K., Ranson H., Zdobnov E. M., Raikhel A. S., Christensen B. M., Dimopoulos G., Muskavitch M. A. T. (2010). Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science 330, 88–90 10.1126/science.1193162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne C. J. (2009). Successful parasitism of vector snail Biomphalaria glabrata by the human blood fluke (trematode) Schistosoma mansoni: a 2009 assessment. Mol. Biochem. Parasitol. 165, 8–18 10.1016/j.molbiopara.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J. M., Iraqi F., Menge D., Baker R. L., Gibson J., Wakelin D. (2003). Chasing the genes that control resistance to gastrointestinal nematodes. J. Helminthol. 77, 99–109 10.1079/JOH2003174 [DOI] [PubMed] [Google Scholar]
- Blais J., Rico C., van Oosterhout C., Cable J., Turner G. F., Bernatchez L. (2007). MHC adaptive divergence between closely related and sympatric African cichlids. PLoS ONE 2, e734. 10.1371/journal.pone.0000734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boëte C., Paul R. E., Koella J. C. (2004). Direct and indirect immunosuppression by a malaria parasite in its mosquito vector. Proc. R. Soc. Lond. B Biol. Sci. 271, 1611–1615 10.1098/rspb.2004.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen G. S. (1976). Experimental infection of North American mammals with epidemic Venezuelan Encephalitis Virus. Am. J. Trop. Med. Hyg. 25, 891–899 [DOI] [PubMed] [Google Scholar]
- Bozeman F. M., Sonenshine D. E., Williams M. S., Chadwick D. P., Lauer D. M., Elisberg B. L. (1981). Experimental infection of ectoparasitic arthropods with Rickettsia prowazekii (GVF-16 Strain) and transmission to flying squirrels. Am. J. Trop. Med. Hyg. 30, 253–263 [DOI] [PubMed] [Google Scholar]
- Brant S. V., Loker E. S. (2005). Can specialized pathogens colonize distantly related hosts? Schistosome evolution as a case study. PLoS Pathog. 1, 167–169 10.1371/journal.ppat.0010038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brites D., McTaggert S., Morris K., Anderson J., Thomas K., Colson I., Fabbro T., Little T. J., Ebert D., duPasquier L. (2008). The Dscam homologue of the crustacean Daphnia is diversified by alternative splicing like insects. Mol. Biol. Evol. 25, 1429–1439 10.1093/molbev/msn087 [DOI] [PubMed] [Google Scholar]
- Bruno J. F., Selig E. R., Casey K. S., Page C. A., Willis B. L., Harvell C. D., Sweatman H., Melendy A. M. (2007). Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol. 5, 1220–1227 10.1371/journal.pbio.0050124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan-Walker K., Leung T. L. F., Poulin R. (2007). Local adaptation of immunity against a trematode parasite in marine amphipod populations. Mar. Biol. 152, 687–695 10.1007/s00227-007-0725-x [DOI] [Google Scholar]
- Buchmann K., Lindenstrøm T. (2002). Interactions between monogenean parasites and their fish hosts. Int. J. Parasitol. 32, 309–319 10.1016/S0020-7519(01)00332-0 [DOI] [PubMed] [Google Scholar]
- Bush S. E., Clayton D. H. (2006). The role of body size in host specificity: reciprocal transfer experiments with feather lice. Evolution 60, 2158–2167 10.1111/j.0014-3820.2006.tb01853.x [DOI] [PubMed] [Google Scholar]
- Byers J. E., Blakeslee A. M. H., Linder E., Cooper A. B., Maguire T. J. (2008). Controls of spatial variation in the prevalence of trematode parasites infecting a marine snail. Ecology 89, 439–451 10.1890/06-1036.1 [DOI] [PubMed] [Google Scholar]
- Carius H. J., Little T. J., Ebert D. (2001). Genetic variation in a host–parasite association: potential for coevolution and frequency-dependent selection. Evolution 55, 1136–1145 10.1111/j.0014-3820.2001.tb00633.x [DOI] [PubMed] [Google Scholar]
- Cannon J. P., Haire R. N., Schnitker N., Mueller M. G., Litman G. W. (2004). Individual protochordates have unique immune-type receptor repertoires. Curr. Biol. 14, R465–R466 10.1016/j.cub.2004.06.009 [DOI] [PubMed] [Google Scholar]
- Cannon J. P., Litman G. W. (2009). Plasticity of the immunoglobulin domain in the evolution of immunity. Integr. Comp. Biol. 49, 187–196 10.1093/icb/icp018 [DOI] [PubMed] [Google Scholar]
- Combes C. (2001). Parasitism: The Ecology and Evolution of Intimate Interactions. Chicago, IL: The University of Chicago Press, 728 p [Google Scholar]
- Combes C., Théron A. (2000). Metazoan parasites and resource heterogeneity: constraints and benefits. Int. J. Parasitol. 30, 299–304 10.1016/S0020-7519(00)00012-6 [DOI] [PubMed] [Google Scholar]
- Cornet S., Franceschi N., Bollache L., Rigaud T., Sorci G. (2009). Variation and covariation in infectivity, virulence and immunodepression in the host–parasite association Gammarus pulex–Pomphorhynchus laevis. Proc. R. Soc. B Biol. Sci. 276, 4229–4236 10.1098/rspb.2009.1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M. M., Dios S., Alonso-Gutierrez J., Romero A., Novoa B., Figueras A. (2009). Evidence of high individual diversity on myticin C in mussel (Mytilus galloprovincialis). Dev. Comp. Immunol. 33, 162–170 10.1016/j.dci.2008.08.005 [DOI] [PubMed] [Google Scholar]
- Daszak P. (2010). Bats, in black and white. Science 329, 634–635 10.1126/science.1194089 [DOI] [PubMed] [Google Scholar]
- Daszak P., Cunningham A. A., Hyatt A. D. (2000). Wildlife ecology – emerging infectious diseases of wildlife – threats to biodiversity and human health. Science 287, 443–449 10.1126/science.287.5452.443 [DOI] [PubMed] [Google Scholar]
- Davidson C. R., Best N. M., Francis J. W., Cooper E. L., Wood T. C. (2008). Toll-like receptor genes (TLRs) from Capitella capitata and Helobdella robusta (Annelida). Dev. Comp. Immunol. 32, 608–612 10.1016/j.dci.2007.11.004 [DOI] [PubMed] [Google Scholar]
- de Bellocq J. G., Charbonnel N., Morand S. (2008). Coevolutionary relationship between helminth diversity and MHC class II polymorphism in rodents. J. Evol. Biol. 21, 1144–1150 10.1111/j.1420-9101.2008.01538.x [DOI] [PubMed] [Google Scholar]
- Detwiler J. T., Criscione C. D. (2010). An infectious topic in reticulate evolution: introgression and hybridization in animal parasites. Genes 1, 102–123 10.3390/genes1010102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vienne D. M., Hood M. E., Giraud T. (2009). Phylogenetic determinants of potential host shifts in fungal pathogens. J. Evol. Biol. 22, 2532–2541 10.1111/j.1420-9101.2009.01702.x [DOI] [PubMed] [Google Scholar]
- Dionne M., Miller K. M., Dodson J. J., Caron F., Bernatchez L. (2007). Clinal variation in MHC diversity with temperature: evidence for the role of host–pathogen interaction on local adaptation in Atlantic salmon. Evolution 61, 2154–2164 10.1111/j.1558-5646.2007.00178.x [DOI] [PubMed] [Google Scholar]
- Dishaw L. J., Mueller M. G., Gwatney N., Cannon J. P., Haire R. N., Litman R. T., Amemiya C. T., Ota T., Rowen L., Glusman G., Litman G. W. (2008). Genomic complexity of the variable region-containing chitin-binding proteins in amphioxus. BMC Genet. 9, 78. 10.1186/1471-2156-9-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishaw L. J., Ota T., Mueller M. G., Cannon J. P., Haire R. N., Gwatney N. R., Litman R. T., Litman G. W. (2010). The basis for haplotype complexity in VCBPs, an immune-type receptor in amphioxus. Immunogenetics 62, 623–631 10.1007/s00251-010-0464-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M., Fu Y., Yu C., Su J., Huang S., Wu X., Wei J., Yuan S., Shen Y., Xu A. (2005). Identification and characterisation of a homolog of an activation gene for the recombination activating gene 1 (RAG 1) in amphioxus. Fish Shellfish Immunol. 19, 165–174 10.1016/j.fsi.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Dong Y., Dimopoulos G. (2009). Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J. Biol. Chem. 284, 9835–9844 10.1074/jbc.M109.027896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Taylor H. E., Dimopoulos G. (2006). AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 4, 1137–1146 10.1371/journal.pbio.0040229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M. A., Brassil C. E., Hall S. R., Tessier A. J., Caceres C. E., Conner J. K. (2008). Parasite-mediated disruptive selection in a natural Daphnia population. BMC Evol. Biol. 8, 1–9 10.1186/1471-2148-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke B. (2004). Failed attempts at experimental transplantation and transmission of nocturnally-periodic simian Loa from monkey to man. Filaria J. 3, 5. 10.1186/1475-2883-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R. R., Harris N. C., Colwell R. K., Koh L. P., Sodhi N. S. (2009). The sixth mass coextinction: are most endangered species parasites and mutualists? Proc. R. Soc. B Biol. Sci. 276, 3037–3045 10.1098/rspb.2009.0413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G. (2010). A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol. 3, 450–460 10.1038/mi.2010.20 [DOI] [PubMed] [Google Scholar]
- Eizaguirre C., Lenz T. L., Sommerfeld R. D., Harrod C., Kalbe M., Milinski M. (2011). Parasite diversity, patterns of MHC II variation and olfactory based mate choice in diverging three-spined stickleback ecotypes. Evol. Ecol. 25, 605–622 10.1007/s10682-010-9424-z [DOI] [Google Scholar]
- Eizaguirre C., Yeates S. E., Lenz T. L., Kalbe M., Milinski M. (2009). MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol. Ecol. 18, 3316–3329 10.1111/j.1365-294X.2009.04243.x [DOI] [PubMed] [Google Scholar]
- Erwin D. H., Laflamme M., Tweedt S. M., Sperling E. A., Pisani D., Peterson K. J. (2011). The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science 334, 1091–1097 10.1126/science.1206375 [DOI] [PubMed] [Google Scholar]
- Evans J. D., Aronstein K., Chen Y. P., Hetru C., Imler J. L., Jiang H., Kanost M., Thompson G. J., Zou Z., Hultmark D. (2006). Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 15, 645–656 10.1111/j.1365-2583.2006.00682.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher C. L., Thornhill R. (2008). A parasite-driven wedge: infectious diseases may explain language and other biodiversity. Oikos 117, 1289–1297 10.1111/j.0030-1299.2008.16684.x [DOI] [Google Scholar]
- Flajnik M. F., Kasahara M. (2010). Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 11, 47–59 10.1038/ni0110-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming L., Rawlings D., Chenia H. (2007). Phenotypic and molecular characterisation of fish-borne Flavobacterium johnsoniae-like isolates from aquaculture systems in South Africa. Res. Microbiol. 158, 18–30 10.1016/j.resmic.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Frank S. A. (2002). Immunology and Evolution of Infectious Disease. Princeton, NJ: Princeton University Press, 348 p. [PubMed] [Google Scholar]
- Fritz R. S. (1999). Resistance of hybrid plants to herbivores: genes, environment, or both? Ecology 80, 382–391 10.1890/0012-9658(1999)080[0382:ROHPTH]2.0.CO;2 [DOI] [Google Scholar]
- Fugmann S. D. (2010). The origins of the Rag genes – from transposition to V(D)J recombination. Semin. Immunol. 22, 10–16 10.1016/j.smim.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fytrou A., Schofield P. G., Kraaijeveld A. R., Hubbard S. F. (2006). Wolbachia infection suppresses both host defence and parasitoid counter-defence. Proc. R. Soc. B Biol. Sci. 273, 791–796 10.1098/rspb.2005.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A. (2006). Antiviral response in pandemic influenza viruses. Emerging Infect. Dis. 12, 44–47 10.3201/eid1201.051186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnick S. W., Elgar M. A., Beveridge I., Coulson G. (2010). Foraging efficiency and parasite risk in eastern grey kangaroos (Macropus giganteus). Behav. Ecol. 21, 129–137 10.1093/beheco/arp162 [DOI] [Google Scholar]
- Genovese G., Friedman D. J., Ross M. D., Lecordier L., Uzureau P., Freedman B. I., Bowden D. W., Langefeld C. D., Oleksyk T. K., Knob A. L. U., Bernhardy A. J., Hicks P. J., Nelson G. W., Vanhollebeke B., Winkler C. A., Kopp J. B., Pays E., Pollak M. R. (2010). Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329, 841–845 10.1126/science.1193032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J., Buckley K. M., Nair S. V., Raftos D. A., Miller C., Majeske A. J., Hibino T., Rast J. P., Roth M., Smith L. C. (2010). Sp185/333: a novel family of genes and proteins involved in the purple sea urchin immune response. Dev. Comp. Immunol. 34, 235–245 10.1016/j.dci.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Giraud T., Gladieux P., Gavrilets S. (2010). Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol. Evol. 25, 387–395 10.1016/j.tree.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohar M., Gilois N., Graveline R., Garreau C., Sanchis V., Lereclus D. (2005). A comparative study of Bacillus cereus, Bacillus thuringiensis and Bacillus anthracis extracellular proteomes. Proteomics 5, 3696–3711 10.1002/pmic.200401225 [DOI] [PubMed] [Google Scholar]
- Goss E. M., Carbone I., Grunwald N. J. (2009). Ancient isolation and independent evolution of the three clonal lineages of the exotic sudden oak death pathogen Phytophthora ramorum. Mol. Ecol. 18, 1161–1174 10.1111/j.1365-294X.2009.04089.x [DOI] [PubMed] [Google Scholar]
- Graham A. L., Shuker D. M., Pollitt L. C., Auld S. K. J. R., Wilson A. J., Little T. J. (2011). Fitness consequences of immune responses: strengthening the empirical framework for ecoimmunology. Funct. Ecol. 25, 5–17 10.1111/j.1365-2435.2010.01777.x [DOI] [Google Scholar]
- Gray M. J., Miller D. L., Hoverman J. T. (2009). Ecology and pathology of amphibian ranaviruses. Dis. Aquat. Org. 87, 243–266 10.3354/dao02138 [DOI] [PubMed] [Google Scholar]
- Gross R., Vavre F., Heddi A., Hurst G. D., Zchori-Fein E., Bourtzis K. (2009). Immunity and symbiosis. Mol. Microbiol. 73, 751–759 10.1111/j.1365-2958.2009.06820.x [DOI] [PubMed] [Google Scholar]
- Guan Y., Zheng B. J., He Y. Q., Liu X. L., Zhuang Z. X., Cheung C. L., Luo S. W., Li P. H., Zhang L. J., Guan Y. J., Butt K. M., Wong K. L., Chan K. W., Lim W., Shortridge K. F., Yuen K. Y., Peiris J. S., Poon L. L. (2003). Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302, 276–278 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- Guzo D., Stoltz D. B. (1985). Obligatory multiparasitism in the tussock moth, Orgyia leucostigma. Parasitology 90, 1–10 10.1017/S0031182000052227 [DOI] [Google Scholar]
- Haldane J. B. S. (1949). Disease and evolution. Ric. Sci. 19(Suppl.), 68–76 [Google Scholar]
- Hamilton W. D., Zuk M. (1982). Heritable true fitness and bright birds – a role for parasites. Science 218, 384–387 10.1126/science.7123238 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt K., Kurtz J. (2005). Surface carbohydrate composition of a tapeworm in its consecutive intermediate hosts: individual variation and fitness consequences. Int. J. Parasitol. 35, 1499–1507 10.1016/j.ijpara.2005.08.011 [DOI] [PubMed] [Google Scholar]
- Hanington P. C., Lun C. M., Adema C. M., Loker E. S. (2010a). Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei. Int. J. Parasitol. 40, 819–831 10.1016/j.ijpara.2009.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanington P. C., Forys M. A., Drago J. W., Zhang S. M., Adema C. M., Loker E. S. (2010b). Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc. Natl. Acad. Sci. U.S.A. 107, 21087–21092 10.1073/pnas.1011242107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauton C., Smith V. J. (2007). Adaptive immunity in invertebrates: a straw house without a mechanistic foundation. Bioessays 29, 1138–1146 10.1002/bies.20650 [DOI] [PubMed] [Google Scholar]
- Hedrick P. W. (2003). “The major histocompatibility complex (MHC) in declining populations: an example of adaptive variation,” in Reproduction Science and Integrated Conservation, eds Holt W. V., Pickard A. R., Rodger J. C., Wildt D. E. (Cambridge, UK: Cambridge University Press; ), 97–113 [Google Scholar]
- Hedrick R. P., McDowell T. S., Marty G. D., Fosgate G. T., Mukkatira K., Myklebust K., El-Matbouli M. (2003). Susceptibility of two strains of rainbow trout (one with suspected resistance to whirling disease) to Myxobolus cerebralis infection. Dis. Aquat. Org. 55, 37–44 10.3354/dao055037 [DOI] [PubMed] [Google Scholar]
- Hemmrich G., Miller D. J., Bosch T. C. G. (2007). The evolution of immunity: a low life perspective. Trends Immunol. 28, 449–454 10.1016/j.it.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Hibino T., Loza-Coll M., Messier C., Majeske A. J., Cohen A. H., Terwilliger D. P., Buckley K. M., Brocton V., Nair S. V., Berney K., Fugmann S. D., Anderson M. K., Pancer Z., Cameron R. A., Smith L. C., Rast J. P. (2006). The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 300, 349–365 10.1016/j.ydbio.2006.08.065 [DOI] [PubMed] [Google Scholar]
- Hill A. V. (2001). Immunogenetics and genomics. Lancet 357, 2037–2041 10.1016/S0140-6736(00)04588-8 [DOI] [PubMed] [Google Scholar]
- Hoberg E. P., Brooks D. R. (2008). A macroevolutionary mosaic: episodic host-switching, geographical colonization and diversification in complex host–parasite systems. J. Biogeogr. 35, 1533–1550 10.1111/j.1365-2699.2008.01951.x [DOI] [Google Scholar]
- Holmes C. B., Losina E., Walensky R. P., Yazdanpanah Y., Freedberg K. A. (2003). Review of human immunodeficiency virus type 1-related opportunistic infections in sub-Saharan Africa. Clin. Infect. Dis. 36, 652–662 10.1086/367655 [DOI] [PubMed] [Google Scholar]
- Horak P., Kolarova L. (2005). Molluscan and vertebrate immune responses to bird schistosomes. Parasite Immunol. 27, 247–255 10.1111/j.1365-3024.2005.00776.x [DOI] [PubMed] [Google Scholar]
- Horvath P., Barrangou R. (2010). CRISPR/Cas, the immune system of bacteria and Archaea. Science 327, 167–170 10.1126/science.1179555 [DOI] [PubMed] [Google Scholar]
- Huang S., Yuan S., Guo L., Yu Y., Li J., Wu T., Liu T., Yang M., Wu K., Liu H., Ge J., Huang H., Dong M., Yu C., Chen S., Xu A. (2008). Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 18, 1112–1126 10.1101/gr.069674.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L. (1998). Protein phylogenies provide evidence of a radical discontinuity between arthropod and vertebrate immune systems. Immunogenetics 47, 283–296 10.1007/s002510050360 [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Piontkivska H. (2008). Functional diversification of the toll-like receptor gene family. Immunogenetics 60, 249–256 10.1007/s00251-008-0283-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E., Urbach J. M., Ausubel F. M. (2010). Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 10, 47–58 10.1038/nri2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. A., Tinsley R. C. (2003). Parasite infectivity to hybridising host species: a link between hybrid resistance and allopolyploid speciation? Int. J. Parasitol. 33, 137–144 10.1016/S0020-7519(02)00255-2 [DOI] [PubMed] [Google Scholar]
- Janz N., Nylin S., Wahlberg N. (2006). Diversity begets diversity: host expansions and the diversification of plant-feeding insects. BMC Evol. Biol. 6, 1–10 10.1186/1471-2148-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. P., Malenke J. R., Clayton D. H. (2009). Competition promotes the evolution of host generalists in obligate parasites. Proc. R. Soc. B Biol. Sci. 276, 3921–3926 10.1098/rspb.2008.0793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbe M., Kurtz J. (2006). Local differences in immunocompetence reflect resistance of sticklebacks against the eye fluke Diplostomum pseudospathaceum. Parasitology 132, 105–116 10.1017/S0031182005008681 [DOI] [PubMed] [Google Scholar]
- Kanost M. R., Jiang H. B., Yu X. Q. (2004). Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 198, 97–105 10.1111/j.0105-2896.2004.0121.x [DOI] [PubMed] [Google Scholar]
- Karraker N. E., Ruthig G. R. (2009). Effect of road deicing salt on the susceptibility of amphibian embryos to infection by water molds. Environ. Res. 109, 40–45 10.1016/j.envres.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Keesing F., Brunner J., Duerr S., Killilea M., LoGiudice K., Schmidt K., Vuong H., Ostfeld R. S. (2009). Hosts as ecological traps for the vector of Lyme disease. Proc. R. Soc. B Biol. Sci. 276, 3911–3919 10.1098/rspb.2009.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh L. P., Dunn R. R., Sodhi N. S., Colwell R. K., Proctor H. C., Smith V. S. (2004). Species coextinctions and the biodiversity crisis. Science 305, 1632–1634 10.1126/science.1101101 [DOI] [PubMed] [Google Scholar]
- Komar N., Langevin S., Hinten S., Nemeth N., Edwards E., Hettler D., Davis B., Bowen R., Bunning M. (2003). Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerging Infect. Dis. 9, 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs J., Masur H. (2008). HIV related opportunistic infections: still relevant after 25 years of AIDS progress. Enferm. Infecc. Microbiol. Clin. 26, 323–324 10.1157/13123835 [DOI] [PubMed] [Google Scholar]
- Kraaijeveld A. R., Godfray H. C. J. (1999). Geographic patterns in the evolution of resistance and virulence in Drosophila and its parasitoids. Am. Nat. 153, S61–S74 10.1086/303212 [DOI] [PubMed] [Google Scholar]
- Krasnov B. R., Mouillot D., Khokhlova I. S., Shenbrot G. I., Poulin R. (2005). Covariance in species diversity and facilitation among non-interactive parasite taxa: all against the host. Parasitology 131, 557–568 10.1017/S0031182005007912 [DOI] [PubMed] [Google Scholar]
- Kuris A. M., Goddard J. H. R., Torchin M. E., Murphy N., Gurney R., Lafferty K. D. (2007). An experimental evaluation of host specificity: the role of encounter and compatibility filters for a rhizocephalan parasite of crabs. Int. J. Parasitol. 37, 539–545 10.1016/j.ijpara.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Labropoulou V., Douris V., Stefanou D., Magrioti C., Swevers L., Iatrou K. (2008). Endoparasitoid wasp bracovirus-mediated inhibition of hemolin function and lepidopteran host immunosuppression. Cell. Microbiol. 10, 2118–2128 10.1111/j.1462-5822.2008.01195.x [DOI] [PubMed] [Google Scholar]
- Lafferty K. D., Kuris A. M. (2009a). Parasitic castration: the evolution and ecology of body snatchers. Trends Parasitol. 25, 564–572 10.1016/j.pt.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Lafferty K. D., Kuris A. M. (2009b). Parasites reduce food web robustness because they are sensitive to secondary extinction as illustrated by an invasive estuarine snail. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 1659–1663 10.1098/rstb.2008.0220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry C., Garant D., Duchesne P., Bernatchez L. (2001). ‘Good genes as heterozygosity’: the major histocompatibility complex and mate choice in Atlantic salmon (Salmo salar). Proc. R. Soc. Lond. B Biol. Sci. 268, 1279–1285 10.1098/rspb.2001.1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B. P., Little T. J. (2009). Immunity in a variable world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 15–26 10.1098/rstb.2008.0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. A. (2006). Linking immune defenses and life history at the levels of the individual and the species. Integr. Comp. Biol. 46, 1000–1015 10.1093/icb/icl047 [DOI] [PubMed] [Google Scholar]
- Lee K. A., Klasing K. C. (2004). A role for immunology in invasion biology. Trends Ecol. Evol. 19, 523–529 10.1016/j.tree.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 10.1146/annurev.immunol.25.022106.141615 [DOI] [PubMed] [Google Scholar]
- Levinton J. S. (2001). Genetics, Paleontology and Macroevolution. Cambridge, MA: Cambridge University Press, 634 p [Google Scholar]
- Lie K. J. (1982). Survival of Schistosoma mansoni and other trematode larvae in the snail Biomphalaria glabrata – a discussion of the interference theory. Trop. Geogr. Med. 34, 111–122 [PubMed] [Google Scholar]
- Light J. E., Hafner M. S. (2008). Codivergence in heteromyid rodents (Rodentia: Heteromyidae) and their sucking lice of the genus Fahrenholzia (Phthiraptera: Anoplura). Syst. Biol. 57, 449–465 10.1080/10635150802169610 [DOI] [PubMed] [Google Scholar]
- Lindstrom K. M., Foufopoulos J., Parn H., Wikelski M. (2004). Immunological investments reflect parasite abundance in island populations of Darwin’s finches. Proc. R. Soc. Lond. B Biol. Sci. 271, 1513–1519 10.1098/rspb.2004.2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman G. W., Rast J. P., Fugmann S. D. (2010). The origins of vertebrate adaptive immunity. Nat. Rev. Immunol. 10, 543–553 10.1038/nri2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loker E. S. (1994). On being a parasite in an invertebrate host – a short survival course. J. Parasitol. 80, 728–747 10.2307/3283252 [DOI] [PubMed] [Google Scholar]
- Loker E. S. (2010). “Gastropod immunobiology,” in Invertebrate Immunity, ed. Söderhall K. (Austin/New York: Landes Bioscience/Springer; ), 17–43 [Google Scholar]
- Loker E. S., Moyo H. G., Gardner S. L. (1981). Trematode-gastropod associations in 9 non-lacustrine habitats in the Mwanza region of Tanzania. Parasitology 83, 381–399 10.1017/S0031182000085383 [DOI] [Google Scholar]
- MacColl A. D. C. (2009). Parasites may contribute to ‘magic trait’ evolution in the adaptive radiation of three-spined sticklebacks, Gasterosteus aculeatus (Gasterosteiformes: Gasterosteidae). Biol. J. Linn. Soc. 96, 425–433 10.1111/j.1095-8312.2008.01123.x [DOI] [Google Scholar]
- Mangeat B., Turelli P., Liao S. Y., Trono D. (2004). A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to vif action. J. Biol. Chem. 279, 14481–14483 10.1074/jbc.C400060200 [DOI] [PubMed] [Google Scholar]
- Maan E., Seehausen O. (2010). Mechanisms of species divergence through visual adaptation and sexual selection: perspectives from a cichlid model system. Curr. Zool. 56, 285–299 [Google Scholar]
- Martinsen E. S., Perkins S. L., Schall J. J. (2008). A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol. Phylogenet. Evol. 47, 261–273 10.1016/j.ympev.2007.11.012 [DOI] [PubMed] [Google Scholar]
- Matsunaga T., Rahman A. (1998). What brought the adaptive immune system to vertebrates? The jaw hypothesis and the seahorse. Immunol. Rev. 166, 177–186 10.1111/j.1600-065X.1998.tb01262.x [DOI] [PubMed] [Google Scholar]
- Matthews B., Harmon L. J., M’Gonigle L., Marchinko K. B., Schaschl H. (2010). Sympatric and allopatric divergence of MHC Genes in Threespine Stickleback. PLoS ONE 5, e10948. 10.1371/journal.pone.0010948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. (1963). Animal Species and Evolution. Cambridge, MA: Belknap Press of Harvard University Press [Google Scholar]
- McClintock M. K., Jacob S., Zelano B., Ober C. (2002). Preferences for human odors are associated with paternally inherited HLA alleles. Horm. Behav. 41, 451. [DOI] [PubMed] [Google Scholar]
- McKeever D. J. (2009). Bovine immunity – a driver for diversity in Theileria parasites? Trends Parasitol. 25, 269–276 10.1016/j.pt.2009.03.005 [DOI] [PubMed] [Google Scholar]
- McKenzie V. J., Townsend A. R. (2007). Parasitic and infectious disease responses to changing global nutrient cycles. EcoHealth 4, 384–396 10.1007/s10393-007-0131-3 [DOI] [Google Scholar]
- McVicar A. H., Fletcher T. C. (1970). Serum factors in Raja radiata toxic to Acanthobothrium quadripartitum (Cesoda–Tetraphyllidea), a parasite specific to R. naevus. Parasitology 61, 55–63 [Google Scholar]
- Messier-Solek C., Buckley K. M., Rast J. P. (2010). Highly diversified innate receptor systems and new forms of animal immunity. Semin. Immunol. 22, 39–47 10.1016/j.smim.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Milinski M. (2006). The major histocompatibility complex, sexual selection, and mate choice. Annu. Rev. Ecol. Evol. Syst. 37, 159–186 10.1146/annurev.ecolsys.37.091305.110242 [DOI] [Google Scholar]
- Milinski M., Griffiths S., Wegner K. M., Reusch T. B. H., Haas-Assenbaum A., Boehm T. (2005). Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc. Natl. Acad. Sci. U.S.A. 102, 4414–4418 10.1073/pnas.0408264102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. J., Ehrman L., Schneider D. (2010). Infectious speciation revisited: impact of symbiont depletion on female fitness and mating behavior of Drosophila paulistorum. PLoS Pathog. 6, e1001214. 10.1371/journal.ppat.1001214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. J., Hemmrich G., Ball E. E., Hayward D. C., Khalturin K., Funayama N., Agata K., Bosch T. C. G. (2007). The innate immune repertoire in Cnidaria – ancestral complexity and stochastic gene loss. Genome Biol. 8, R59.1–R59.13 10.1186/gb-2007-8-6-r105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooring M. S., Fitzpatrick T. A., Fraser I. C., Benjamin J. E., Reisig D. D., Nishihira T. T. (2003). Insect-defense behavior by desert bighorn sheep. Southwest. Nat. 48, 635–643 [DOI] [Google Scholar]
- Morales-Hojas R., Vieira C. P., Reis M., Vieira J. (2009). Comparative analysis of five immunity-related genes reveals different levels of adaptive evolution in the virilis and melanogaster groups of Drosophila. Heredity 102, 573–578 10.1038/hdy.2009.11 [DOI] [PubMed] [Google Scholar]
- Morgan J. A., DeJong R. J., Kazibwe F., Mkoji G. M., Loker E. S. (2003). A newly-identified lineage of Schistosoma. Int. J. Parasitol. 33, 977–985 10.1016/S0020-7519(03)00132-2 [DOI] [PubMed] [Google Scholar]
- Neill D. R., Wong S. H., Bellosi A., Flynn R. J., Daly M., Langford T. K. A., Bucks C., Kane C. M., Fallon P. G., Pannell R., Jolin H. E., McKenzie A. N. J. (2010). that mediates type-2 immunity. Nature 464, 1367–1370 10.1038/nature08900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. E., Virok D. P., Wood H., Roshick C., Johnson R. M., Whitmire W. M., Crane D. D., Steele-Mortimer O., Kari L., McClarty G., Caldwell H. D. (2005). Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc. Natl. Acad. Sci. U.S.A. 102, 10658–10663 10.1073/pnas.0401664102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra M. L., Powell A. E., Rosengarten R. D., Moreno M., Grimwood J., Lakkis F. G., Dellaporta S. L., Buss L. W. (2009). A hypervariable invertebrate allodeterminant. Curr. Biol. 19, 583–589 10.1016/j.cub.2009.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm S. V., Nishiguchi M. K. (2008). The evolutionary ecology of a sepiolid squid-vibrio association: from cell to environment. Vie Milieu 58, 175–184 [PMC free article] [PubMed] [Google Scholar]
- Nyman T. (2010). To speciate, or not to speciate? Resource heterogeneity, the subjectivity of similarity, and the macroevolutionary consequences of niche-width shifts in plant-feeding insects. Biol. Rev. 85, 393–411 10.1111/j.1469-185X.2009.00109.x [DOI] [PubMed] [Google Scholar]
- Olson P. D., Caira J. N., Jensen K., Overstreet R. M., Palm H. W., Beveridge I. (2010). Evolution of the trypanorhynch tapeworms: parasite phylogeny supports independent lineages of sharks and rays. Int. J. Parasitol. 40, 223–242 10.1016/j.ijpara.2009.07.012 [DOI] [PubMed] [Google Scholar]
- Ottenhoff T. H. M., Verreck F. A. W., Hoeve M. A., van de Vosse E. (2005). Control of human host immunity to mycobacteria. Tuberculosis 85, 53–64 10.1016/j.tube.2004.09.011 [DOI] [PubMed] [Google Scholar]
- Pais R., Lohs C., Wu Y., Wang J., Aksoy S. (2008). The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74, 5965–5974 10.1128/AEM.00741-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z., Amemiya C. T., Ehrhardt G. R. A., Ceitlin J., Gartland G. L., Cooper M. D. (2004). Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430, 174–180 10.1038/nature02740 [DOI] [PubMed] [Google Scholar]
- Parra Z. E., Baker M. L., Schwarz R. S., Deakin J. E., Lindblad-Toh K., Miller R. D. (2007). A unique T cell receptor discovered in marsupials. Proc. Natl. Acad. Sci. U.S.A. 104, 9776–9781 10.1073/pnas.0609106104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish C. R., Holmes E. C., Morens D. M., Park E. C., Burke D. S., Calisher C. H., Laughlin C. A., Saif L. J., Daszak P. (2008). Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72, 457–470 10.1128/MMBR.00004-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. (2009). The Biology of Genomes, 5–9 May 2009, Cold Spring Harbor, New York. The bug and the bacterium: interdependent genomes. Science 324, 1253. 10.1126/science.324.5924.162a [DOI] [PubMed] [Google Scholar]
- Philips H. L., Clarkson M. J. (1998). Experimental infection of pregnant ewes with Chlamydia pecorum. Infect. Immun. 66, 2818–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimm S. L., Raven P. (2000). Biodiversity extinction by numbers. Nature 403, 843–845 10.1038/35002708 [DOI] [PubMed] [Google Scholar]
- Pfeffer M., Dobler G. (2010). Emergence of zoonotic arboviruses by animal trade and migration. Parasit. Vectors 3, 1–15 10.1186/1756-3305-3-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin R., Keeney D. B. (2008). Host specificity under molecular and experimental scrutiny. Trends Parasitol. 24, 24–28 10.1016/j.pt.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Poulin R., Thomas F. (1999). Phenotypic variability induced by parasites: extent and evolutionary implications. Parasitol. Today 15, 28–32 10.1016/S0169-4758(99)01554-9 [DOI] [PubMed] [Google Scholar]
- Price P. W. (1980). Evolutionary Biology of Parasites. Princeton, NJ: Princeton University Press, 237 p [Google Scholar]
- Prugnolle F., Manica A., Charpentier M., Guegan J. F., Guernier V., Balloux F. (2005). Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 15, 1022–1027 10.1016/j.cub.2005.02.038 [DOI] [PubMed] [Google Scholar]
- Qureshi S. T., Medzhitov R. (2003). Toll-like receptors and their role in experimental models of microbial infection. Genes Immun. 4, 87–94 10.1038/sj.gene.6363937 [DOI] [PubMed] [Google Scholar]
- Radwan J., Biedrzycka A., Babik W. (2010). Does reduced MHC diversity decrease viability of vertebrate populations? Biol. Conserv. 143, 537–544 10.1016/j.biocon.2010.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast J., Litman G. (2010). Evolution of innate and adaptive immune recognition structures. Semin. Immunol. 22, 1–2 10.1016/j.smim.2010.01.001 [DOI] [PubMed] [Google Scholar]
- Refrégier G., Le Gac M., Jabbour F., Widmer A., Shykoff J. A., Yockteng R., Hood M. E., Giraud T. (2008). Cophylogeny of the anther smut fungi and their caryophyllaceous hosts: prevalence of host shifts and importance of delimiting parasite species for inferring cospeciation. BMC Evol. Biol. 8, 100. 10.1186/1471-2148-8-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rest J. S., Mindell D. P. (2003). SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 3, 219–225 10.1016/j.meegid.2003.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud T., Moret Y. (2003). Differential phenoloxidase activity between native and invasive gammarids infected by local acanthocephalans: differential immunosuppression? Parasitology 127, 571–577 10.1017/S0031182003004050 [DOI] [PubMed] [Google Scholar]
- Rivkin H., Kroemer J. A., Bronshtein A., Belausov E., Webb B. A., Chejanovsky N. (2006). Response of immunocompetent and immunosuppressed Spodoptera littoralis larvae to baculovirus infection. J. Gen. Virol. 87, 2217–2225 10.1099/vir.0.81918-0 [DOI] [PubMed] [Google Scholar]
- Roeder T., Stanisak M., Gelhaus C., Bruchhaus I., Grotzinger J., Leippe M. (2010). Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Dev. Comp. Immunol. 34, 203–209 10.1016/j.dci.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Rogozin I. B., Iyer L. M., Liang L. Z., Glazko G. V., Liston V. G., Pavlov Y. I., Aravind L., Pancer Z. (2007). Evolution and diversification of lamprey antigen receptors: evidence for involvement of AID-APOBEC family cytosine deaminase. Nature Immunol. 8, 647–656 10.1038/nrg2186 [DOI] [PubMed] [Google Scholar]
- Rohr J. R., Schotthoefer A. M., Raffel T. R., Carrick H. J., Halstead N., Hoverman J. T., Johnson C. M., Johnson L. B., Lieske C., Piwoni M. D., Schoff P. K., Beasley V. R. (2008). Agrochemicals increase trematode infections in a declining amphibian species. Nature 455, 1235–1239 10.1038/nature07281 [DOI] [PubMed] [Google Scholar]
- Rolff J., Siva-Jothy M. T. (2002). Copulation corrupts immunity: a mechanism for a cost of mating in insects. Proc. Natl. Acad. Sci. U.S.A. 99, 9916–9918 10.1073/pnas.152271999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooijakkers S. H., Ruyken M., Roos A., Daha M. R., Presanis J. S., Sim R. B., van Wamel W. J., van Kessel K. P., van Strijp J. A. (2005). Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6, 920–927 10.1038/ni1235 [DOI] [PubMed] [Google Scholar]
- Rosengard A. M., Liu Y., Nie Z., Jimenez R. (2002). Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc. Natl. Acad. Sci. U.S.A. 99, 8808–8813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton T. B., Lazzaro B. P., Schlenke T. A., Evans J. D., Hultmark D., Clark A. G. (2007). Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 39, 1461–1468 10.1038/ng.2007.60 [DOI] [PubMed] [Google Scholar]
- Saenz S. A., Siracusa M. C., Perrigoue J. G., Spencer S. P., Urban J. F., Tocker J. E., Budelsky A. L., Kleinschek M. A., Kastelein R. A., Kambayashi T., Bhandoola A., Artis D. (2010). IL25 elicits a multipotent progenitor cell population that promotes T(H)2 cytokine responses. Nature 464, 1362–1366 10.1038/nature08901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu S., Das B. K., Mishra B. K., Pradhan J., Samal S. K., Sarangi N. (2008). Effect of dietary Curcuma longa on enzymatic and immunological profiles of rohu, Labeo rohita (Ham.), infected with Aeromonas hydrophila. Aquac. Res. 39, 1720–1730 10.1111/j.1365-2109.2008.02048.x [DOI] [Google Scholar]
- Salzet M., Tasiemski A., Cooper E. (2006). Innate immunity in lophotrochozoa: the annelids. Curr. Pharm. Des. 12, 3043–3050 10.2174/138161206777947551 [DOI] [PubMed] [Google Scholar]
- Sapp K. K., Loker E. S. (2000). Mechanisms underlying digenean-snail specificity: role of miracidial attachment and host plasma factors. J. Parasitol. 86, 1012–1019 10.1645/0022-3395(2000)086[1020:ACSOMU]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Scharsack J. P., Koch K., Hammerschmidt K. (2007). Who is in control of the stickleback immune system: interactions between Schistocephalus solidus and its specific vertebrate host. Proc. R. Soc. B Biol. Sci. 274, 3151–3158 10.1098/rspb.2007.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P. (2008). Parasite immune evasion: a momentous molecular war. Trends Ecol. Evol. 23, 318–326 10.1016/j.tree.2008.02.011 [DOI] [PubMed] [Google Scholar]
- Schmidt O., Theopold U., Strand M. (2001). Innate immunity and its evasion and suppression by hymenopteran endoparasitoids. Bioessays 23, 344–351 10.1002/bies.1049 [DOI] [PubMed] [Google Scholar]
- Schmucker D., Chen B. (2009). Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 23, 147–156 10.1101/gad.1752909 [DOI] [PubMed] [Google Scholar]
- Schroder H. C., Natalio F., Wiens M., Tahir M. N., Shukoor M. I., Tremel W., Belikov S. I., Krasko A., Muller W. E. (2008). The 2′-5′-oligoadenylate synthetase in the lowest metazoa: isolation, cloning, expression and functional activity in the sponge Lubomirskia baicalensis. Mol. Immunol. 45, 945–953 10.1016/j.molimm.2007.07.036 [DOI] [PubMed] [Google Scholar]
- Schulenburg H., Ewbank J. J. (2004). Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol. Biol. 4, 49. 10.1186/1471-2148-4-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte R. D., Makus C., Hasert B., Michiels N. K., Schulenburg H. (2010). Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc. Natl. Acad. Sci. U.S.A. 107, 7359–7364 10.1073/pnas.1003113107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet B. T., Johnston J. B., Brunetti C. R., Barrett J. W., Everett H., Cameron C., Sypula J., Nazarian S. H., Lucas A., McFadden G. (2003). Poxviruses and immune evasion. Annu. Rev. Immunol. 21, 377–423 10.1146/annurev.immunol.21.120601.141049 [DOI] [PubMed] [Google Scholar]
- Seppala O., Karvonen A., Valtonen E. T. (2007). Phenotypic variation in infectivity of Diplostomum spathaceum cercariae within a population. J. Parasitol. 93, 1244–1246 10.1645/GE-1187R.1 [DOI] [PubMed] [Google Scholar]
- Shaw S. R. (1988). Euphorine phylogeny – the evolution of diversity in host utilization by parasitoid wasps (Hymenoptera, Braconidae). Ecol. Entomol. 13, 323–335 10.1111/j.1365-2311.1988.tb00363.x [DOI] [Google Scholar]
- Shelby K. S., Cui L., Webb B. A. (1998). Polydnavirus-mediated inhibition of lysozyme gene expression and the antibacterial response. Insect Mol. Biol. 7, 265–272 10.1111/j.1365-2583.1998.00071.x [DOI] [PubMed] [Google Scholar]
- Smith C. R., Smith C. D., Robertson H. M., Helmkampf M., Zimin A., Yandell M., Holt C., Hu H. E., Abouheif E., Benton R., Cash E., Croset V., Currie C. R., Elhak E., Elsik C. G., Fave M. N., Fernandes V., Gibson J. D., Garaur D., Gronenberg W., Grubbs K. J., Hagen D. E., Viniegra A. S. I., Johnson B. R., Johnson R. M., Khila A., Kim J. W., Mathis K. A., Munoz-Torres M. C., Murphy M. C., Mustard J. A., Nakamura R., Niehuis O., Nigam S., Overson R. P., Placek J. E., Rajakumar R., Reese J. T., Suen G., Tao S., Torres C. W., Tsutsui N. D., Viljakainen L., Wolschin F., Gadau J. (2011). Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc. Natl. Acad. Sci. U.S.A. 108, 5667–5672 10.1073/pnas.1018857108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. (2005). Major histocompatibility complex and mate choice in a monogamous rodent. Behav. Ecol. Sociobiol. (Print) 58, 181–189 10.1007/s00265-005-0909-7 [DOI] [Google Scholar]
- Southgate V. R., Brown D. S., Warlow A., Knowles R. J., Jones A. (1989). The influence of Calicophoron microbothrium on the susceptibility of Bulinus tropicus to Schistosoma bovis. Parasitol. Res. 75, 381–391 10.1007/BF00931134 [DOI] [PubMed] [Google Scholar]
- Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E. A., Mitros T., Richards G. S., Conaco C., Dacre M., Hellsten U., Larroux C., Putnam N. H., Stanke M., Adamska M., Darling A., Degnan S. M., Oakley T. H., Plachetzki D. C., Zhai Y. F., Adamski M., Calcino A., Cummins S. F., Goodstein D. M., Harris C., Jackson D. J., Leys S. P., Shu S. Q., Woodcroft B. J., Vervoort M., Kosik K. S., Manning G., Degnan B. M., Rokhsar D. S. (2010). The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 10.1038/nature09201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S. M. (1998). Macroevolution: Pattern and Process. Baltimore, MD: The Johns Hopkins University Press, 370 p [Google Scholar]
- Streicker D. G., Turmelle A. S., Vonhof M. J., Kuzmin I. V., McCracken G. F., Rupprecht C. E. (2010). Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 329, 676–679 10.1126/science.1188836 [DOI] [PubMed] [Google Scholar]
- Stukenbrock E. H., McDonald B. A. (2008). The origins of plant pathogens in agro-ecosystems. Annu. Rev. Phytopathol. 46, 75–100 10.1146/annurev.phyto.010708.154114 [DOI] [PubMed] [Google Scholar]
- Sullivan J. T., Richards C. S. (1981). Schistosoma mansoni, NIH-SM-PR-2 strain, in susceptible and nonsusceptible stocks of Biomphalaria glabrata: comparative histology. J. Parasitol. 67, 702–708 10.2307/3280445 [DOI] [PubMed] [Google Scholar]
- Summers K., McKeon S., Sellars J., Keusenkothen M., Morris J., Gloeckner D., Pressley C., Price B., Snow H. (2003). Parasitic exploitation as an engine of diversity. Biol. Rev. Camb. Philos. Soc. 78, 639–675 10.1017/S146479310300616X [DOI] [PubMed] [Google Scholar]
- Sung J. M., Lloyd D. H., Lindsay J. A. (2008). Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology 154, 1949–1959 10.1099/mic.0.2007/015289-0 [DOI] [PubMed] [Google Scholar]
- Taraschewski H. (2006). Hosts and parasites as aliens. J. Helminthol. 80, 99–128 10.1079/JOH2006364 [DOI] [PubMed] [Google Scholar]
- Tenor J. L., Aballay A. (2008). A conserved Toll-like receptor for Caenorhabditis elegans innate immunity. EMBO Rep. 9, 103–109 10.1038/sj.embor.7401104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F., Guldner E., Renaud F. (2000). Differential parasite (Trematoda) encapsulation in Gammarus aequicauda (Amphipoda). J. Parasitol. 86, 650–654 10.2307/3284895 [DOI] [PubMed] [Google Scholar]
- Thomas J. A., Telfer M. G., Roy D. B., Preston C. D., Greenwood J. J. D., Asher J., Fox R., Clarke R. T., Lawton J. H. (2004). Comparative losses of British butterflies, birds and plants and the global extinction crisis. Science 303, 1879–1881 10.1126/science.1095046 [DOI] [PubMed] [Google Scholar]
- Thompson J. N. (2005). The Geographic Mosaic of Coevolution. Chicago, IL: The University of Chicago Press [Google Scholar]
- Thompson J. N. (2009). The coevolving web of life. Am. Nat. 173, 125–140 10.1086/595752 [DOI] [PubMed] [Google Scholar]
- Tompkins D. M., Clayton D. H. (1999). Host resources govern the specificity of swiftlet lice: size matters. J. Anim. Ecol. 68, 489–500 10.1046/j.1365-2656.1999.00297.x [DOI] [Google Scholar]
- Torchin M. E., Lafferty K. D., Kuris A. M. (2002). Parasites and marine invasions. Parasitology 124, S137–S151 10.1017/S0031182002001506 [DOI] [PubMed] [Google Scholar]
- Trachtenberg E., Korber B., Sollars C., Kepler T. B., Hraber P. T., Hayes E., Funkhouser R., Fugate M., Theiler J., Hsu Y. S., Kunstman K., Wu S., Phair J., Erlich H., Wolinsky S. (2003). Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9, 928–935 10.1038/nm893 [DOI] [PubMed] [Google Scholar]
- Trowsdale J., Parham P. (2004). Defense strategies and immunity-related genes. Eur. J. Immunol. 34, 7–17 10.1002/eji.200324693 [DOI] [PubMed] [Google Scholar]
- Tunaz H., Stanley D. (2009). An immunological axis of biocontrol: infections in field-trapped insects. Naturwissenschaften 96, 1115–1119 10.1007/s00114-009-0572-3 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I. (2007). The human microbiome project. Nature 449, 804–810 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeWoude S., Troyer J., Poss M. (2010). Restrictions to cross-species transmission of lentiviral infection gleaned from studies of FIV. Vet. Immunol. Immunopathol. 134, 25–32 10.1016/j.vetimm.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn G. S., Edelaar P., Weissing F. J. (2009). On the origin of species by natural and sexual selection. Science 326, 1704–1707 10.1126/science.1181661 [DOI] [PubMed] [Google Scholar]
- Vidal-Martinez V. M., Osoriosarabia D., Overstreet R. M. (1994). Experimental infection of Contracaecum multipapillatum (Nematoda, Anisakinae) from Mexico in the domestic cat. J. Parasitol. 80, 576–579 10.2307/3283194 [DOI] [PubMed] [Google Scholar]
- Viljakainen L., Evans J. D., Hasselmann M., Rueppell O., Tingek S., Pamilo P. (2009). Rapid evolution of immune proteins in social insects. Mol. Biol. Evol. 26, 1791–1801 10.1093/molbev/msp086 [DOI] [PubMed] [Google Scholar]
- Vorburger C., Sandrock C., Gouskov A., Castaneda L. E., Ferrari J. (2009). Genotypic variation and the role of defensive endosymbionts in an all parthenogenetic host–parasitoid interaction. Evolution 63, 1439–1450 10.1111/j.1558-5646.2009.00660.x [DOI] [PubMed] [Google Scholar]
- Walker J. C. (1979). Austrobilharzia terrigalensis – schistosome dominant in interspecific interactions in the molluscan host. Int. J. Parasitol. 9, 137–140 10.1016/0020-7519(79)90104-8 [DOI] [Google Scholar]
- Wang F., Ma Y. Y., Barrett J. W., Gao X. J., Loh J., Barton E., Virgin H. W., McFadden G. (2004). Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 5, 1266–1274 10.1038/ni1132 [DOI] [PubMed] [Google Scholar]
- Webb B., Fisher T., Nusawardani T. (2009). The natural genetic engineering of polydnaviruses. Ann. N. Y. Acad. Sci. 1178, 146–156 10.1111/j.1749-6632.2009.05023.x [DOI] [PubMed] [Google Scholar]
- Wedekind C., Gessner M. O., Vazquez F., Maerki M., Steiner D. (2010). Elevated resource availability sufficient to turn opportunistic into virulent fish pathogens. Ecology 91, 1251–1256 10.1890/09-1067.1 [DOI] [PubMed] [Google Scholar]
- Wegner K. M. (2008). Historical and contemporary selection of teleost MHC genes: did we leave the past behind? J. Fish Biol. 73, 2110–2132 10.1111/j.1095-8649.2008.02051.x [DOI] [Google Scholar]
- Wegner K. M., Kalbe M., Kurtz J., Reusch T. B. H., Milinski M. (2003). Parasite selection for immunogenetic optimality. Science 301, 1343. 10.1126/science.1088293 [DOI] [PubMed] [Google Scholar]
- Weissenbock H., Hubalek Z., Bakonyi T., Nowotny N. (2010). Zoonotic mosquito-borne flaviviruses: worldwide presence of agents with proven pathogenicity and potential candidates of future emerging diseases. Vet. Microbiol. 140, 271–280 10.1016/j.vetmic.2009.08.025 [DOI] [PubMed] [Google Scholar]
- Welchman D. P., Aksoy S., Jiggins F., Lemaitre B. (2009). Insect immunity: from pattern recognition to symbiont-mediated host defense. Cell Host Microbe 6, 107–114 10.1016/j.chom.2009.07.008 [DOI] [PubMed] [Google Scholar]
- Wheatley B. P. (1980). Malaria as a possible selective factor in the speciation of macaques. J. Mammal. 61, 307–311 10.2307/1380052 [DOI] [PubMed] [Google Scholar]
- Wheeler R. J. (2010). The trypanolytic factor-mechanism, impacts and applications. Trends Parasitol. 26, 457–464 10.1016/j.pt.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Wiegel F. W., Perelson A. S. (2004). Some scaling principles for the immune system. Immunol. Cell Biol. 82, 127–131 10.1046/j.0818-9641.2004.01229.x [DOI] [PubMed] [Google Scholar]
- Wiens M., Korzhev M., Perovic-Ottstadt S., Luthringer B., Brandt D., Klein S., Muller W. E. G. (2007). Toll-like receptors are part of the innate immune defense system of sponges (Demospongiae: Porifera). Mol. Biol. Evol. 24, 792–804 10.1093/molbev/msl208 [DOI] [PubMed] [Google Scholar]
- Winkler I. S., Mitter C., Scheffer S. J. (2009). Repeated climate-linked host shifts have promoted diversification in a temperate clade of leaf-mining flies. Proc. Natl. Acad. Sci. U.S.A. 106, 18103–18108 10.1073/pnas.0904852106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinska J., Lively C. M., Spaak P. (2008). Parasites in hybridizing communities: the Red Queen again? Trends Parasitol. 24, 121–126 10.1016/j.pt.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Wolinska J., Spaak P. (2009). The cost of being common: evidence from natural Daphnia populations. Evolution 63, 1893–1901 10.1111/j.1558-5646.2009.00663.x [DOI] [PubMed] [Google Scholar]
- Wood M. J., Cosgrove C. L., Wilkin T. A., Knowles S. C., Day K. P., Sheldon B. C. (2007). Within-population variation in prevalence and lineage distribution of avian malaria in blue tits, Cyanistes caeruleus. Mol. Ecol. 16, 3263–3273 10.1111/j.1365-294X.2007.03362.x [DOI] [PubMed] [Google Scholar]
- Wood-Charlson E. M., Weis V. M. (2009). The diversity of C-type lectins in the genome of a basal metazoan, Nematostella vectensis. Dev. Comp. Immunol. 33, 881–889 10.1016/j.dci.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Woolhouse M. E. J., Haydon D. T., Antia R. (2005). Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20, 238–244 10.1016/j.tree.2005.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt K. B., Campos P. F., Gilbert M. T., Kolokotronis S. O., Hynes W. H., DeSalle R., Ball S. J., Daszak P., MacPhee R. D., Greenwood A. D. (2008). Historical mammal extinction on Christmas Island (Indian Ocean) correlates with introduced infectious disease. PLoS ONE 3, e3602. 10.1371/journal.pone.0003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Cox-Foster D. L. (2005). Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. U.S.A. 102, 7470–7475 10.1073/pnas.0406904102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino T. P., Dinguirard N., Kunert J., Hokk C. H. (2008). Molecular and functional characterization of a tandem-repeat galectin from the freshwater snail Biomphalaria glabrata, intermediate host of the human blood fluke Schistosoma mansoni. Gene 411, 46–58 10.1016/j.gene.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zmasek C. M., Godzik A. (2010). Domain architecture evolution of pattern-recognition receptors. Immunogenetics 62, 263–272 10.1007/s00251-010-0428-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. M., Adema C. M., Kepler T. B., Loker E. S. (2004). Diversification of Ig superfamily genes in an invertebrate. Science 305, 251–254 10.1126/science.1097540 [DOI] [PubMed] [Google Scholar]
- Zietara M. S., Lumme J. (2002). Speciation by host switch and adaptive radiation in a fish parasite genus Gyrodactylus (Monogenea, Gyrodactylidae). Evolution 56, 2445–2458 10.1111/j.0014-3820.2002.tb00170.x [DOI] [PubMed] [Google Scholar]