Abstract
T cells recognize antigen fragments, presented to them by MHC molecules. It lies in the interest of the immune system to display a maximal diversity of these peptides and utilize all catabolic processes to generate them. Macroautophagy, a pathway that delivers cytoplasmic constituents for lysosomal degradation is no exception. In recent years, it has become apparent that macroautophagy assists in intra- and extracellular antigen processing for MHC class II presentation to CD4+ helper T cells. Surprisingly, however, macroautophagy also assists in antigen packaging for better cross-presentation on MHC molecules of bystander cells, which could be consistent with its role in unconventional protein secretion. These three pathways of antigen processing for MHC presentation via macroautophagy will be discussed in this review and cell biological aspects will be high-lighted that might explain, how the molecular machinery of macroautophagy might assist these diverse antigen processing pathways.
Keywords: multivesicular bodies, Atg, CD4+ T cells, MHC class II loading compartment, exosomes
Introduction
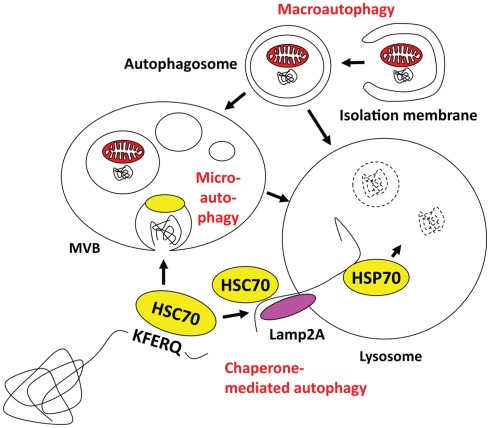
Autophagy describes a group of pathways that deliver cytoplasmic constituents for lysosomal degradation. Three main pathways exist with chaperone-mediated, micro- and macroautophagy (Münz, 2009; Levine et al., 2011; Romao and Münz, 2011; Figure 1). Current evidence suggests that mainly macroautophagy can mediate antigen processing for MHC presentation, while it still remains unclear if and to which extent the other two pathways might also contribute (Zhou et al., 2005). Both chaperone-mediated and microautophagy use cytosolic HSC70 chaperones for substrate recognition via a pentameric amino acid sequence (Massey et al., 2006; Sahu et al., 2011). KFERQ as the prototypic sequence in ribonuclease A and similar motifs that mediate HSC70 binding have been estimated to occur in 20–35% of all cytosolic proteins (Chiang and Dice, 1988). In the case of chaperone-mediated autophagy HSC70 links these substrates in a complex of co-chaperones to Lamp2a in the lysosomal membrane, across which this cargo then gets imported with the help of an intralysosomal HSP70 member (Agarraberes et al., 1997). For microautophagy, HSC70 attaches with its cargo to endosomal membranes and then buds into these endosomes for multivesicular body (MVB) formation (Sahu et al., 2011). MVB fusion with lysosomes leads then to degradation of microautophagy cargo. Although chaperone-mediated and microautophagy target HSC70 substrates to different vesicles, both are restricted to soluble protein delivery for lysosomal degradation. In contrast, macroautophagy is able to degrade larger cytoplasmic structures, like protein aggregates and cell organelles (Yang and Klionsky, 2010a). Depending on the substrate of macroautophagy, xenophagy of intracellular pathogens, mitophagy for mitochondria degradation, ribophagy for ribosome turnover, and other pathways can be distinguished. In all cases, however, a cup-shaped isolation membrane forms and engulfs cytoplasmic constituents to form double membrane surrounded vesicles, known as autophagosomes. Depending on the location of the substrate, autophagosomes can form from membranes of ER, Golgi, plasma membrane, and mitochondria (Tooze and Yoshimori, 2010). For autophagosome formation and their fusion with lysosomes more than 30 essential gene products of autophagy related genes (atgs) have now been identified in yeast, and some of these are transcriptionally regulated in parallel to lysosomal constituents (Settembre et al., 2011) or anti-oxidants (Komatsu et al., 2010; Fujita et al., 2011). I would like to high-light only five complexes containing Atgs that are involved in autophagosome initiation, elongation, and fusion with lysosomes (for a recent schematic overview see Levine et al., 2011). The first is involved in macroautophagy induced by starvation. It contains ULK1, 2, or 3, which are the mammalian homologs of the Atg1 serine/threonine kinase, as well as Atg13, FIP200, and Atg101 (Yang and Klionsky, 2010b). Upon starvation the mammalian target of rapamycin (mTOR) kinase is inhibited, which leads to dephosphorylation and translocation of the ULK complex to membranes to initiate autophagosome formation. Although the direct phosphorylation targets of ULK are still poorly characterized, the ULK complex recruits a second complex, containing Atg6/Beclin-1 to the site of autophagosome generation. This complex containing also the PI3 kinase VPS34, VPS15, and Atg14 marks the site of autophagosome generation with PI3P deposition. Atg6/Beclin-1 is normally sequestered from this complex by the anti-apoptotic Bcl-2 and -xL proteins, but gets released by for example the AMBRA1 protein to then stimulate macroautophagy. These PI3P patches recruit the FYVE domain-containing protein DFCP1, which together with the WIPI1-4 proteins in a third complex initiates cradle formation for isolation membrane generation at least at the ER (Hayashi-Nishino et al., 2009; Yla-Anttila et al., 2009). In these structures, two additional complexes for the ubiquitin-like proteins Atg8 and Atg12 assist isolation membrane elongation, substrate recruitment, and completion of autophagosomes. Atg12 is conjugated to Atg5 by the E1- and E2 like enzymes Atg7 and 10. In complex with Atg16L1 it associates with the outer autophagosome membrane to form the E3-like enzyme for lipid ligation of Atg8, which in higher eukaryotes has diversified into the family of LC3, GABARAP, and GATE16 proteins. Atg8 is proteolytically processed by Atg4 and then coupled in an E1- (Atg7), E2- (Atg3), and E3- (Atg12/5/16L1) assisted mechanism to phosphatidylethanolamine (PE) in the isolation membrane. Atg8 is thought to mediate membrane fusion for autophagosome elongation and substrate recruitment into the autophagosome. Once the autophagosome is completed Atg12/5/16L1 and Atg8 recycle from the outer autophagosomal membrane and only Atg8 stays with the inner autophagosomal membrane. Therefore, Atg8 can be used as a marker to follow autophagosomes. Fusion of these autophagosomes with lysosomes is then guided by additional Atg6/Beclin-1 complexes, containing besides Atg6, VPS34, and VPS15, UVRAG to promote fusion with lysosomes, and Rubicon to block autophagosome degradation (Liang et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). Thus, an elaborate machinery is involved in autophagosome generation and degradation to deliver cytoplasmic constituents for lyososomal hydrolysis.
Figure 1.
Macro-, micro-, and chaperone-mediated autophagy. Macroautophagy can engulf cytoplasmic material to form autophagosomes, which then can fuse with multivesicular bodies (MVBs) or directly with lysosomes. Cytosolic KFERQ-like signal peptide containing proteins may get transported in an HSC70 dependent fashion into MVBs by microautophagy, or via docking to Lamp2a and intravesicular HSP70 assisted import into lysosomes. The MVB content might also get degraded after fusion with lysosomes.
Substrate Recruitment into Autophagosomes
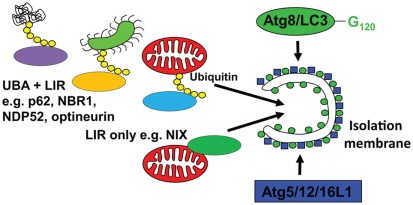
In order to understand, how macroautophagy can contribute to antigen processing, I have to discuss next which substrates are recruited to isolation membranes. Although it was originally assumed that at least during starvation conditions, autophagosomes would engulf non-selectively cytoplasm, it is now known that even under these conditions, a clear hierarchy for macroautophagic degradation to generate energy for the starving cell exists (Kristensen et al., 2008). Namely, proteasomes and protein aggregates are targeted early during starvation, ribosomes get degraded in a second wave, and only after prolonged starvation mitochondria get engulfed by autophagosomes. But what are the mechanisms mediating this selectivity? In higher eukaryotes, there seem to exist two pathways of substrate recruitment to autophagosomes, both anchoring cytoplasmic constituents to Atg8/LC3 (Figure 2). The respective anchor proteins contain a LC3/GABARAP interacting motif (LIR) of four amino acids (WXXL or WXXI; Kraft et al., 2010). One of these substrate recruiting pathways and its LIR proteins is evolutionary conserved down to yeast. For example, Atg32 in yeast and NIX in higher eukaryotes seem to associate with mitochondria as integral outer membrane proteins and target them for macroautophagy (Schweers et al., 2007; Sandoval et al., 2008; Kanki et al., 2009; Okamoto et al., 2009). NIX contains a LIR domain and recruits GABARAP-L1 only when this LIR domain is intact (Novak et al., 2010). Thus, organelle specific proteins can contain a LIR and mediate recruitment to autophagosomes. In addition, higher eukaryotes can utilize post-translational protein modifications to select macroautophagy substrates. Along these lines, ubiquitylation has been characterized in some detail so far. Aggregates of poly-ubiquitylated substrates, which are also degraded by proteasomes, are recruited to autophagosomes (Kirkin et al., 2009b). If macroautophagy is compromised these aggregates can accumulate in the cytosol and, if this happens in neurons, it can lead to neurodegenerative disease (Hara et al., 2006; Komatsu et al., 2006). The ubiquitin-ligase CHIP is involved in the ubiquitylation of these substrates and the chaperone BAG3, the ubiquitin-binding protein sequestosome 1/p62 and its interacting protein Alfy are involved in aggregate formation (Simonsen et al., 2004; Komatsu et al., 2007; Gamerdinger et al., 2009; Arndt et al., 2010). Furthermore, p62 contains a LIR domain and links protein aggregates to isolation membranes via LC3 (Bjorkoy et al., 2005; Pankiv et al., 2007). In addition to p62, at least three other proteins have been identified that anchor poly-ubiquitylated substrates to LC3 by containing both ubiquitin-binding domains (UBA or UBZ) and LIRs. These are NBR1, NDP52, and optineurin (Kirkin et al., 2009a; Thurston et al., 2009; Wild et al., 2011), and might recruit different ubiquitylated substrates to autophagosomes. Moreover, the versatility of substrate recruitment to macroautophagy could be further enhanced by combining these different anchor proteins with the extensive families of ubiquitin ligases. Along these lines the ubiquitin-ligase Parkin has been shown to be recruited to damaged mitochondria, for ubiquitylation and macroautophagic degradation (Narendra et al., 2008, 2010; Dagda et al., 2009). In addition, the multitude of proteins that bridge ubiquitin with LC3 allows their cooperation in substrate recruitment to autophagosomes. Along these lines, NDP52 recruits in addition to LC3 the Tank-binding kinase 1 (TBK1), which in turn phosphorylates optineurin to bind with higher affinity to LC3 (Thurston et al., 2009; Wild et al., 2011). This cooperation for efficient macroautophagy occurs at distinct regions of pathogenic Salmonella bacteria, which have escaped to the cytosol. Furthermore, p62 binds also to cytosolic Salmonella, but at separate regions of the bacteria (Wild et al., 2011). Thus three anchor proteins seem to be involved in xenophagy of this pathogen. Interestingly, no homologs of p62, NBR1, NDP52, and optineurin have been identified in yeast (Kraft et al., 2010). Therefore, ubiquitin-independent, but not ubiquitin dependent substrate recruitment to autophagosomes seems to be conserved between higher and lower eukaryotes. Accordingly, ubiquitylation rather inhibits than accelerates macroautophagy of ribosomes in yeast (Kraft et al., 2008). These recent studies provide some information about the mechanistic underpinning of substrate selection into autophagosomes, and suggest that in contrast to previous assumptions macroautophagy is rarely non-selective.
Figure 2.
Substrate recruitment into autophagosomes. Substrates are imported into emerging autophagosomes (isolation membrane) via anchoring to Atg8/LC3, which is covalently attached to the autophagosomal membrane by the E3-like ligase Atg5/12/16L1. This anchoring occurs through a LC3 interacting region (LIR), either via proteins that directly bind to the cargo (LIR only, e.g., NIX for mitophagy) or via proteins that interact with polyubiquitin tags on substrates, like Salmonella bacteria (green), mitochondria (red), or protein aggregates (UBA + LIR, e.g., p62, NBR1, NDP52, and optineurin for xeno- and mitophagy).
Intracellular Antigen Processing via Macroautophagy
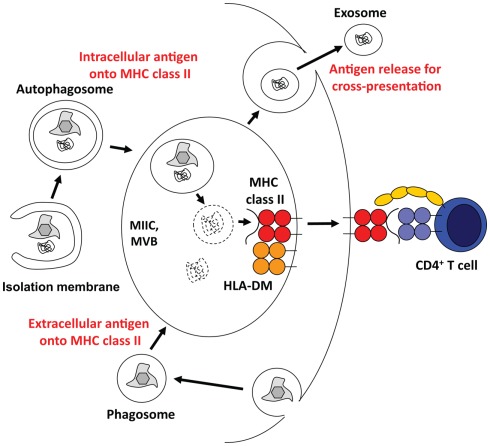
The immune system of higher eukaryotes uses the same degradation machineries that are involved in organelle and protein turnover for antigen processing. Accordingly, proteasomal products are presented on MHC class I molecules to CD8+ T cells, and lysosomal degradation provides protein fragments for MHC class II presentation to CD4+ T cells (Sijts and Kloetzel, 2011; Van Den Hoorn et al., 2011). Therefore, it is probably not very surprising that autophagic pathways that lead to lysosomal degradation of cytoplasmic constituents contribute to antigen processing for MHC class II presentation (Figure 3). Indeed, 20–30% of natural MHC class II ligands are derived from cytosolic and nuclear antigens (Marrack et al., 1993; Rammensee et al., 1999; Muntasell et al., 2002). Starvation induced stimulation of macroautophagy significantly enhances MHC class II presentation of cytosolic and nuclear antigens at the expense of membrane proteins (Dengjel et al., 2005). Among the cytosolic proteins, whose peptides are presented on MHC class II molecules, are also LC3 and Gabarap, suggesting autophagosome fusion with MHC class II loading compartments (Dengjel et al., 2005; Suri et al., 2008). Indeed, 70–80% of autophagosomes fuse with MIICs (Schmid et al., 2007). Furthermore, fusion of a model antigen to LC3 enhances its presentation on MHC class II molecules up to 20-fold, while its MHC class I presentation remains largely unaffected (Schmid et al., 2007). Among other tissues this frequent delivery of autophagosome cargo to MIICs occurs also in thymic epithelial cells (Kasai et al., 2009). The functional relevance of this cytoplasmic constituent delivery to MIICs in the thymus is underscored by the finding that both positive and negative selection of CD4+, but not CD8+ T cells are compromised in macroautophagy-deficient thymii (Nedjic et al., 2008). Some, but not all CD4+ T cell receptors are not positively selected by Atg5 deficient thymic epithelial cells, suggesting that some MHC class II ligands required for positive selection get generated in a macroautophagy dependent pathway. Furthermore, MHC class II ligands for negative selection, eliminating autoimmune T cells, are also dependent on macroautophagy. As a result, the T cell repertoire selected through macroautophagy negative thymii causes signs of autoimmunity, including severe colitis (Nedjic et al., 2008). Along these lines, Atg16L1 mutations, which presumably compromise macroautophagy, are enriched in Crohn’s disease patients, who suffer from extreme colitis (Hampe et al., 2007; Rioux et al., 2007). Interestingly, dendritic cells (DCs) from these patients are deficient in MHC class II up-regulation upon maturation (Cooney et al., 2010). Modulation of this intracellular antigen processing via macroautophagy inhibition with a cell permeable spliceosomal U1-70K ribonucleoprotein peptide seems also to ameliorate systemic autoimmunity, including lupus-like symptoms in mice (Page et al., 2011). All these data suggest that self-protein derived ligand processing and presentation on MHC class II requires macroautophagy.
Figure 3.
Multivesicular bodies (MVBs) at the core of antigen processing with the help of the macroautophagic molecular machinery. Macroautophagy and/or its molecular machinery support antigen processing via three main pathways. They transport cytoplasmic material into MHC class II loading compartments (MIICs), assist phagosome fusion with MIICs, and might allow for the release of antigen for efficient cross-presentation.
In addition, also pathogen derived antigens get processed via macroautophagy for MHC class II presentation. The nuclear antigen 1 (EBNA1) of the Epstein Barr virus (EBV) was found to be presented by EBV transformed B cells on MHC class II molecules to EBNA1 specific CD4+ T cells, which can be found in most healthy carriers of this virus (Paludan et al., 2005; Leung et al., 2010). The nuclear localization of this protein limits access to macroautophagic processing for MHC class II presentation, but nevertheless sensitizes EBV transformed B cells to recognition by some EBNA1 specific CD4+ T cells. Similarly, some bacterial antigens that escape after endocytosis or are injected into the cytosol via secretion systems, can be processed for MHC class II presentation via macroautophagy. This applies to the mycobacterial Ag85B antigen, which is more efficiently presented by DCs after rapamycin induced macroautophagy up-regulation (Jagannath et al., 2009). These DCs are then more potent in inducing protective immune responses via vaccination. Furthermore, outer proteins of Yersinia, which are injected into the cytosol by the bacterial pathogen, are presented on MHC class II molecules and this presentation is sensitive to pharmacological inhibition of macroautophagy (Russmann et al., 2010). Additional evidence for intracellular antigen processing for MHC class II presentation by infected cells comes from herpes simplex virus (HSV) infection. HSV encodes with ICP34.5 a gene product that interferes with macroautophagy (Orvedahl et al., 2007). Interestingly, infection with HSV carrying mutant ICP34.5, which no longer inhibits macroautophagy, is more rapidly controlled with elevated CD4+ T cell expansion (Leib et al., 2009). Accordingly, HSV antigens are more efficiently presented on MHC class II molecules after infection with virus lacking the macroautophagy compromising domain in ICP34.5. Along the same lines, HSV infection is less well controlled in mice with macroautophagy deficiency in DCs due to an inefficient CD4+ T cell expansion (Lee et al., 2010). These studies suggest that intracellular antigen processing via macroautophagy can lead to MHC class II presentation to CD4+ T cells.
In addition, one report exists that suggests that macroautophagy can also assist intracellular antigen processing for MHC class I presentation (English et al., 2009).
Extracellular Antigen Processing via Macroautophagy
However, in recent years it has become apparent that also extracellular antigen processing is affected by macroautophagy. Indeed, the in vivo phenotype of diminished HSV specific CD4+ T cell priming after infection of mice with macroautophagy-deficient DCs was accompanied by decreased MHC class II presentation of extracellular antigen after endocytosis (Lee et al., 2010). In these Atg5 deficient DCs phagosomes did not efficiently fuse with lysosomes, and thus did not sufficiently recruit hydrolases for the degradation of the endocytosed cargo for processing toward MHC class II presentation (Figure 3). Similarly, siRNA mediated silencing of Atg8/LC3 compromises extracellular HIV p24 processing for MHC class II presentation to CD4+ T cells (Blanchet et al., 2010). These authors suggested that macroautophagy is required to efficiently target endocytosed antigen to so-called immunoamphisomes, in which MHC class II molecules could be preferentially loaded. This defect in extracellular antigen processing by macroautophagy-deficient DCs has been observed in a third system. Salmonella antigen presentation on MHC class II molecules and MHC class II up-regulation during maturation of DCs in general was macroautophagy dependent after nucleotide-binding oligomerization domain-containing 2 (NOD2) receptors were stimulated (Cooney et al., 2010). Interestingly, the Atg16L1 mutation associated with Crohn’s disease also conferred this diminished antigen processing for MHC class II presentation to DCs. These data suggest that macroautophagy or its molecular machinery contributes to efficient delivery of phagosomes to lysosomes for extracellular antigen processing. However, the mechanism of this phagocytosis assistance by Atgs remains obscure.
The two main hypothesis that have been put forward to explain enhanced phagosome processing with the help of the macroautophagic machinery are amphisome formation prior to lysosome fusion and recruitment of parts of the macroautophagic machinery to the phagosomal membrane for enhanced fusion with lysosomes. Along the lines of the first hypothesis, NOD1 and 2 stimulation was suggested to recruit Atg16L1 directly to the site of bacterial entry for rapid amphisome formation and degradation of the invading pathogen (Travassos et al., 2010). Again mutant Atg16L1, as present in Crohn’s disease patients, is not capable to localize to sites of Shigella entry. In addition, reactive oxygen species (ROS) and diacylglycerol production at the phagosome membrane was proposed to promote engulfment of phagosomes by autophagosomes or amphisome formation after fusion of these two vesicular compartments (Huang et al., 2009; Shahnazari et al., 2010). ROS was directly produced at the phagosome by NADPH oxidases for this function. These signaling events at the phagosomes were required for efficient Salmonella degradation. As an alternative for phagosome fusion with autophagosomes for more efficient delivery of endocytosed cargo via amphisomes, direct coupling of the macroautophagic machinery, namely Atg8/LC3, to the phagosome membrane was proposed (Sanjuan et al., 2007). This Atg8/LC3 recruitment occurred only on phagosomes containing toll-like receptor (TLR) ligands, primarily TLR2 and 4 agonists. It enhanced phagosome fusion with lysosomes. This study tried to exclude amphisome formation by documenting that the respective phagosomes were surrounded by a single membrane, which did not contain any ER markers. However, autophagosome membranes can be recruited from Golgi, mitochondria and plasma membrane in addition to the ER (Tooze and Yoshimori, 2010) and amphisomes like MVBs are expected to be surrounded by only one membrane with additional intravesicular membranes. Thus, this study could not entirely exclude that the observed Atg8/LC3 recruitment is still due to autophagosome formation, but if Atg8/LC3 would be directly coupled to phagosomes it could assist enhanced membrane fusion with lysosomes (Nakatogawa et al., 2007). The signaling downstream of TLRs that would mediate this Atg8/LC3 recruitment and the function of enhanced phagosome fusion with lysosomes via this mechanism for antigen processing toward MHC presentation, however, remains unclear and requires further investigation. Thus, extracellular antigen processing for MHC class II, but not class I is enhanced by macroautophagy, probably via promoting more efficient fusion of phagosomes with lysosomes. However, the mechanism, by which the macroautophagic machinery achieves this, remains unclear.
Support of Cross-Presentation by Macroautophagy
In contrast to the role of the macroautophagy machinery for extracellular antigen presentation on MHC class II molecules in antigen presenting cells, autophagosomes seem to play a role for intracellular antigen release by the antigen donor cell that favors cross-presentation on MHC class I molecules. Both cross-presentation of viral and tumor antigens seems to benefit from intact macroautophagy in the infected or transformed cells, respectively (Li et al., 2008; Uhl et al., 2009). Interestingly, the viral antigens that were investigated were from influenza A virus, which stabilizes autophagosomes by preventing their fusion with lysosomes (Gannage et al., 2009). Along these lines, autophagosomes were found to be efficient antigen carriers, when isolated from tumor cells (Li et al., 2008). Therefore, autophagosome stabilization might enhance the ability of antigen donor cells to provide antigen for efficient cross-presentation. But how do autophagosomes and their content exit cells for cross-presentation? One possibility might be macroautophagy dependent unconventional secretion of proteins. In yeast, macroautophagy dependent secretion of the acyl coenzyme A-binding protein (ACBP) occurs signal sequence independent and requires macroautophagy (Duran et al., 2010; Manjithaya et al., 2010). Interestingly, this process requires components of vesicular fusion with the plasma membrane, but not fusion with the vacuole, yeast’s lysosome equivalent. It is tempting to speculate that antigen can be secreted by an unconventional pathway from MVBs that receive input from autophagosomes. In this case, inner autophagosomal membranes, which become intravesicular membranes of MVBs might exit cells as exosomes (Figure 3). Exosomes, only one of many antigen transfer vectors, have been reported to be highly immunogenic (Thery et al., 2009). Along these lines, drusen, extracellular amorphous deposits in Age-related macular degeneration, were found to be positive for both exosome and macroautophagy markers (Wang et al., 2009). The source of these drusen, the retinal pigment epithelium was found to display markers of elevated macroautophagy, diminished lysosomal activity and increased exocytosis, which might lead to enhanced secretion of inner autophagic membranes as exosomes. Thus autophagosomes might package antigens efficiently for cross-presentation on MHC class I molecules.
Conclusion
In this review, I have discussed three pathways, by which macroautophagy can assist in antigen processing for MHC presentation to T cells. Two affect intracellular antigens, and deliver those either directly for MHC class II presentation by the antigen expressing cells or packages them for cross-presentation by MHC molecules of bystander cells. The third affects processing of endocytosed extracellular antigens, promoting phagosome fusion with lysosomes for antigen processing toward MHC class II presentation by phagocytes. Interestingly, in the two antigen processing pathways for MHC class II presentation the lysosome targeting abilities of the macroautophagic machinery are utilized, while for cross-presentation prevention of lysosome fusion and redirection of autophagosome content to exocytosis might be required. At the core of all these diverse pathways, MVBs might play a key role, both as MIICs and sources of unconventional secretion. Thus, all these antigen processing pathways supported by macroautophagy might use the macroautophagic machinery to efficiently generate these vesicles and transport cargo into them. Therefore, a more detailed analysis of how the macroautophagic machinery contributes to MVB generation might proof useful for a better understanding of antigen processing for MHC presentation via macroautophagy.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work in my laboratory is in part supported by the National Cancer Institute (R01CA108609), Cancer Research Switzerland (KFS-02652-08-2010), the Association for International Cancer Research (11-0516), the Sassella Foundation (10/02), the Vontobel Foundation and the Swiss National Science Foundation (310030_126995).
References
- Agarraberes F. A., Terlecky S. R., Dice J. F. (1997). An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J. Cell Biol. 137, 825–834 10.1083/jcb.137.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V., Dick N., Tawo R., Dreiseidler M., Wenzel D., Hesse M., Furst D. O., Saftig P., Saint R., Fleischmann B. K., Hoch M., Hohfeld J. (2010). Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 20, 143–148 10.1016/j.cub.2009.12.015 [DOI] [PubMed] [Google Scholar]
- Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet F. P., Moris A., Nikolic D. S., Lehmann M., Cardinaud S., Stalder R., Garcia E., Dinkins C., Leuba F., Wu L., Schwartz O., Deretic V., Piguet V. (2010). Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 32, 654–669 10.1016/j.immuni.2010.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H. L., Dice J. F. (1988). Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J. Biol. Chem. 263, 6797–6805 [PubMed] [Google Scholar]
- Cooney R., Baker J., Brain O., Danis B., Pichulik T., Allan P., Ferguson D. J., Campbell B. J., Jewell D., Simmons A. (2010). NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16, 90–97 10.1038/nm.2069 [DOI] [PubMed] [Google Scholar]
- Dagda R. K., Cherra S. J., III, Kulich S. M., Tandon A., Park D., Chu C. T. (2009). Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 284, 13843–13855 10.1074/jbc.M808515200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J., Schoor O., Fischer R., Reich M., Kraus M., Muller M., Kreymborg K., Altenberend F., Brandenburg J., Kalbacher H., Brock R., Driessen C., Rammensee H. G., Stevanovic S. (2005). Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 7922–7927 10.1073/pnas.0501190102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran J. M., Anjard C., Stefan C., Loomis W. F., Malhotra V. (2010). Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188, 527–536 10.1083/jcb.200911154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English L., Chemali M., Duron J., Rondeau C., Laplante A., Gingras D., Alexander D., Leib D., Norbury C., Lippe R., Desjardins M. (2009). Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 10, 480–487 10.1038/ni.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Maeda D., Xiao Q., Srinivasula S. M. (2011). Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 1427–1432 10.1073/pnas.1014156108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M., Hajieva P., Kaya A. M., Wolfrum U., Hartl F. U., Behl C. (2009). Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 28, 889–901 10.1038/emboj.2009.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannage M., Dormann D., Albrecht R., Dengjel J., Torossi T., Ramer P. C., Lee M., Strowig T., Arrey F., Conenello G., Pypaert M., Andersen J., Garcia-Sastre A., Münz C. (2009). Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6, 367–380 10.1016/j.chom.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F. M., Briggs J., Gunther S., Prescott N. J., Onnie C. M., Hasler R., Sipos B., Folsch U. R., Lengauer T., Platzer M., Mathew C. G., Krawczak M., Schreiber S. (2007). A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 10.1038/ng1954 [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006). Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- Huang J., Canadien V., Lam G. Y., Steinberg B. E., Dinauer M. C., Magalhaes M. A., Glogauer M., Grinstein S., Brumell J. H. (2009). Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. U.S.A. 106, 6226–6231 10.1073/pnas.0811045106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath C., Lindsey D. R., Dhandayuthapani S., Xu Y., Hunter R. L., Jr., Eissa N. T. (2009). Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 15, 267–276 10.1038/nm.1928 [DOI] [PubMed] [Google Scholar]
- Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. (2009). Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai M., Tanida I., Ueno T., Kominami E., Seki S., Ikeda T., Mizuochi T. (2009). Autophagic compartments gain access to the MHC class II compartments in thymic epithelium. J. Immunol. 183, 7278–7285 10.4049/jimmunol.0804087 [DOI] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y. S., Bjorkoy G., Nunn J. L., Bruun J. A., Shvets E., Mcewan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Overvatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. (2009a). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Kirkin V., Mcewan D. G., Novak I., Dikic I. (2009b). A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269 10.1016/j.molcel.2009.04.026 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y. S., Ueno I., Sakamoto A., Tong K. I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. (2010). The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12, 213–223 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006). Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007). Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Kraft C., Deplazes A., Sohrmann M., Peter M. (2008). Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10, 602–610 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- Kraft C., Peter M., Hofmann K. (2010). Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 12, 836–841 10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- Kristensen A. R., Schandorff S., Hoyer-Hansen M., Nielsen M. O., Jaattela M., Dengjel J., Andersen J. S. (2008). Ordered organelle degradation during starvation-induced autophagy. Mol. Cell Proteomics 7, 2419–2428 10.1074/mcp.M800184-MCP200 [DOI] [PubMed] [Google Scholar]
- Lee H. K., Mattei L. M., Steinberg B. E., Alberts P., Lee Y. H., Chervonsky A., Mizushima N., Grinstein S., Iwasaki A. (2010). In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32, 227–239 10.1016/j.immuni.2009.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib D. A., Alexander D. E., Cox D., Yin J., Ferguson T. A. (2009). Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J. Virol. 83, 12164–12171 10.1128/JVI.01676-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C. S., Haigh T. A., Mackay L. K., Rickinson A. B., Taylor G. S. (2010). Nuclear location of an endogenously expressed antigen, EBNA1, restricts access to macroautophagy and the range of CD4 epitope display. Proc. Natl. Acad. Sci. U.S.A. 107, 2165–2170 10.1073/pnas.0909448107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H. W. (2011). Autophagy in immunity and inflammation. Nature 469, 323–335 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang L. X., Yang G., Hao F., Urba W. J., Hu H. M. (2008). Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 68, 6889–6895 10.1158/0008-5472.CAN-07-6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Lee J. S., Inn K. S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J. U. (2008). Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10, 776–787 10.1038/ncb1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R., Anjard C., Loomis W. F., Subramani S. (2010). Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 188, 537–546 10.1083/jcb.200911149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack P., Ignatowicz L., Kappler J. W., Boymel J., Freed J. H. (1993). Comparison of peptides bound to spleen and thymus class II. J. Exp. Med. 178, 2173–2183 10.1084/jem.178.6.2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A. C., Zhang C., Cuervo A. M. (2006). Chaperone-mediated autophagy in aging and disease. Curr. Top. Dev. Biol. 73, 205–235 10.1016/S0070-2153(05)73007-6 [DOI] [PubMed] [Google Scholar]
- Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., Akira S., Noda T., Yoshimori T. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 10.1038/ncb1846 [DOI] [PubMed] [Google Scholar]
- Muntasell A., Carrascal M., Serradell L., Veelen Pv. P., Verreck F., Koning F., Raposo G., Abian J., Jaraquemada D. (2002). HLA-DR4 molecules in neuroendocrine epithelial cells associate to a heterogeneous repertoire of cytoplasmic and surface self peptides. J. Immunol. 169, 5052–5060 [DOI] [PubMed] [Google Scholar]
- Münz C. (2009). Enhancing immunity through autophagy. Annu. Rev. Immunol. 27, 423–429 10.1146/annurev.immunol.021908.132537 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010). PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedjic J., Aichinger M., Emmerich J., Mizushima N., Klein L. (2008). Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 455, 396–400 10.1038/nature07208 [DOI] [PubMed] [Google Scholar]
- Novak I., Kirkin V., Mcewan D. G., Zhang J., Wild P., Rozenknop A., Rogov V., Lohr F., Popovic D., Occhipinti A., Reichert A. S., Terzic J., Dotsch V., Ney P. A., Dikic I. (2010). Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009). Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 10.1016/j.devcel.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Orvedahl A., Alexander D., Talloczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D., Levine B. (2007). HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1, 23–35 10.1016/j.chom.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Page N., Gros F., Schall N., Decossas M., Bagnard D., Briand J. P., Muller S. (2011). HSC70 blockade by the therapeutic peptide P140 affects autophagic processes and endogenous MHCII presentation in murine lupus. Ann. Rheum. Dis. 70, 837–843 10.1136/ard.2010.139832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Münz C. (2005). Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307, 593–596 10.1126/science.1104904 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Overvatn A., Bjorkoy G., Johansen T. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Rammensee H., Bachmann J., Emmerich N. P., Bachor O. A., Stevanovic S. (1999). SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 10.1007/s002510050595 [DOI] [PubMed] [Google Scholar]
- Rioux J. D., Xavier R. J., Taylor K. D., Silverberg M. S., Goyette P., Huett A., Green T., Kuballa P., Barmada M. M., Datta L. W., Shugart Y. Y., Griffiths A. M., Targan S. R., Ippoliti A. F., Bernard E. J., Mei L., Nicolae D. L., Regueiro M., Schumm L. P., Steinhart A. H., Rotter J. I., Duerr R. H., Cho J. H., Daly M. J., Brant S. R. (2007). Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 10.1038/ng2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao S., Münz C. (2011). Autophagy of pathogens alarms the immune system and participates in its effector functions. Swiss. Med. Wkly. 141, w13198. [DOI] [PubMed] [Google Scholar]
- Russmann H., Panthel K., Kohn B., Jellbauer S., Winter S. E., Garbom S., Wolf-Watz H., Hoffmann S., Grauling-Halama S., Geginat G. (2010). Alternative endogenous protein processing via an autophagy-dependent pathway compensates for Yersinia-mediated inhibition of endosomal major histocompatibility complex class II antigen presentation. Infect. Immun. 78, 5138–5150 10.1128/IAI.00155-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R., Kaushik S., Clement C. C., Cannizzo E. S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A. M., Santambrogio L. (2011). Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 20, 131–139 10.1016/j.devcel.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H., Thiagarajan P., Dasgupta S. K., Schumacher A., Prchal J. T., Chen M., Wang J. (2008). Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235 10.1038/nature07006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan M. A., Dillon C. P., Tait S. W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J. L., Withoff S., Green D. R. (2007). Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., Münz C. (2007). MHC class II antigen loading compartments continuously receive input from autophagosomes. Immunity 26, 79–92 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers R. L., Zhang J., Randall M. S., Loyd M. R., Li W., Dorsey F. C., Kundu M., Opferman J. T., Cleveland J. L., Miller J. L., Ney P. A. (2007). NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A. 104, 19500–19505 10.1073/pnas.0708818104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A. (2011). TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnazari S., Yen W. L., Birmingham C. L., Shiu J., Namolovan A., Zheng Y. T., Nakayama K., Klionsky D. J., Brumell J. H. (2010). A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 8, 137–146 10.1016/j.chom.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijts E. J., Kloetzel P. M. (2011). The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell. Mol. Life Sci. 68, 1491–1502 10.1007/s00018-011-0657-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Birkeland H. C., Gillooly D. J., Mizushima N., Kuma A., Yoshimori T., Slagsvold T., Brech A., Stenmark H. (2004). Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell. Sci. 117, 4239–4251 10.1242/jcs.01287 [DOI] [PubMed] [Google Scholar]
- Suri A., Walters J. J., Rohrs H. W., Gross M. L., Unanue E. R. (2008). First signature of islet β-cell-derived naturally processed peptides selected by diabetogenic class II MHC molecules. J. Immunol. 180, 3849–3856 [DOI] [PubMed] [Google Scholar]
- Thery C., Ostrowski M., Segura E. (2009). Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 10.1038/nri2567 [DOI] [PubMed] [Google Scholar]
- Thurston T. L., Ryzhakov G., Bloor S., Von Muhlinen N., Randow F. (2009). The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221 10.1038/ni.1800 [DOI] [PubMed] [Google Scholar]
- Tooze S. A., Yoshimori T. (2010). The origin of the autophagosomal membrane. Nat. Cell Biol. 12, 831–835 10.1038/ncb0910-831 [DOI] [PubMed] [Google Scholar]
- Travassos L. H., Carneiro L. A., Ramjeet M., Hussey S., Kim Y. G., Magalhaes J. G., Yuan L., Soares F., Chea E., Le Bourhis L., Boneca I. G., Allaoui A., Jones N. L., Nunez G., Girardin S. E., Philpott D. J. (2010). Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11, 55–62 10.1038/ni.1823 [DOI] [PubMed] [Google Scholar]
- Uhl M., Kepp O., Jusforgues-Saklani H., Vicencio J. M., Kroemer G., Albert M. L. (2009). Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 16, 991–1005 10.1038/cdd.2009.8 [DOI] [PubMed] [Google Scholar]
- Van Den Hoorn T., Paul P., Jongsma M. L., Neefjes J. (2011). Routes to manipulate MHC class II antigen presentation. Curr. Opin. Immunol. 23, 88–95 10.1016/j.coi.2010.11.002 [DOI] [PubMed] [Google Scholar]
- Wang A. L., Lukas T. J., Yuan M., Du N., Tso M. O., Neufeld A. H. (2009). Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS ONE 4, e4160. 10.1371/journal.pone.0004160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P., Farhan H., Mcewan D. G., Wagner S., Rogov V. V., Brady N. R., Richter B., Korac J., Waidmann O., Choudhary C., Dotsch V., Bumann D., Dikic I. (2011). Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 10.1126/science.1205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D. J. (2010a). Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822 10.1038/ncb2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D. J. (2010b). Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22, 124–131 10.1016/j.ceb.2010.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla-Anttila P., Vihinen H., Jokitalo E., Eskelinen E. L. (2009). 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5, 1180–1185 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. (2009). Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468–476 10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Li P., Lott J. M., Hislop A., Canaday D. H., Brutkiewicz R. R., Blum J. S. (2005). Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity 22, 571–581 10.1016/j.immuni.2005.03.009 [DOI] [PubMed] [Google Scholar]