Abstract
A major limitation to the translation of tolerogenic therapies to clinical transplantation is a lack of biomarkers that can be used as surrogate measures for predicting the successful induction of immune tolerance which would allow for the safe withdrawal of immunosuppression. We have used three different mouse models of donor specific tolerance to skin grafts together with quantitative RT-PCR to search for potential biomarkers of tolerance using criteria based on the presence or activity of regulatory T cells and antigen presenting cells (APCs) within grafts or lymphoid organs. We find that significant differences in gene expression between tolerated and rejecting grafts are observed primarily within the grafted skin and not systemically or in the draining lymph node. The pattern of gene expression within long-term surviving tolerated grafts appear very similar to syngeneic grafts, with both having low levels of T cell and APC infiltration and a bias toward relative over-expression of “regulatory-associated” genes, while allografts destined for rejection show an overall increase in both “regulatory” and “effector” cell associated transcripts. We also, however, find an increase in a large number of regulatory genes, of both innate and T cell origin, even after grafting syngeneic skin. Taken together, these findings suggest that there may be no tissue biomarkers uniquely able to predict donor antigen specific tolerance per se, but that patterns of gene expression within tolerated grafts may be similar to those found in self tissues recovering from an inflammatory insult.
Keywords: transplantation tolerance, foxp3, regulatory T cell, RT-PCR, mouse model
Introduction
Transplantation is now recognized as the best option for treating a variety of diseases that lead to organ failure. The success of allogeneic transplantation has depended on the long-term use of non-specific immunosuppressive agents, which expose the recipient to a number of deleterious side effects, including infection and cancer. In order to facilitate the application of potentially tolerogenic therapies into clinical practice it would be helpful to identify and measure appropriate biomarkers predicting tolerance and graft acceptance. This would allow the identification of those individuals where tolerance has been established so enabling the safe reduction or withdrawal of immunosuppressive agents.
Transplantation tolerance can now be readily induced in a number of different rodent models, and it is becoming clear that such tolerance depends on the generation and activity of regulatory T cells (Treg; Qin et al., 1993; Cobbold et al., 2006). A number of different Treg populations have been described, and although we do not fully understand how they act, there seem to be a core set of genes whose expression is associated with regulation (Cobbold et al., 2003). The expression of many of these core genes seem to be linked to foxp3 expression (Sadlon et al., 2010), the “master” transcription factor for both natural and induced Treg, and therefore assays for these gene transcripts or products would seem to be primary candidates as biomarkers of tolerance. A secondary source of tolerance biomarkers might be those associated with the downstream action of Treg during maintenance of the tolerance state. The mechanisms by which Treg function in vivo are still unclear (Sakaguchi et al., 2009), but there is an emerging consensus that they act, at least in part, by modulating antigen presenting cells (APCs) from a pro-inflammatory to an anti-inflammatory or pro-tolerogenic state (Chen, 2006; Cobbold et al., 2010). Relevant APCs in this context may include not only the dendritic cells, but also other MHC-II+ cells in the graft such as macrophages and endothelial cells. Changes in the expression of a number of gene products have been associated with pro-tolerogenic antigen presentation, such as a relative increase in negative costimulation (e.g., PDL1; Guleria et al., 2005), and increased enzymatic degradation of essential amino acids (e.g., by IDO and arginase; Cobbold et al., 2009).
Although Treg seem to be essential to induce and maintain the tolerant state in vivo, we cannot assume that tolerance and activation/rejection behave as a binary switch between two alternative states. Tolerance is not just the lack of lymphocyte activation or of inflammation, as these can be achieved by immunosuppressive agents that fail to tolerize. Consequently, biomarkers reflecting a lack of inflammation may not be relevant to tolerance in the presence of immunosuppressive agents. Treg also seem to operate during any normal immune response, including the termination phase, and their presence alone is not sufficient to predict the development of tolerance. Indeed, many studies have shown that biomarkers for Treg (such as foxp3) often correlate with increasing inflammation and graft rejection rather than tolerance (Bunnag et al., 2008; Dijke et al., 2008; Huang et al., 2010), suggesting that Treg may play a physiological role in containing inflammation during an ongoing immune response. This suggests that the use of any Treg-based biomarker for tolerance may well depend on qualitative and contextual assessments, such as identifying an increased ratio of regulatory to inflammatory components in the context of a overall reduction in T cell and APC infiltration.
Preferred biomarkers would those that can be measured in readily accessible sources, such as blood or urine. There is accumulating evidence, however, that tolerance frequently depends on the activity of regulatory T cells locally within the target organ (Graca et al., 2002), and may not necessarily be associated with a systemic hyporesponsiveness to donor antigen (Cobbold et al., 2004). Although it may be possible to detect systemic secreted or excreted products associated with regulation (e.g., cytokines or amino acid metabolites), assays that depend on detecting Treg associated gene transcripts or cell associated products might require the use of tissue biopsies.
In this paper we investigated whether previously identified Treg and modulated APC associated gene transcripts could be used as biomarkers predicting tolerance in three different well characterized mouse skin grafting models. We found significant differences in gene expression within grafts such that we could distinguish allogeneic skin that was destined for tolerance rather than rejection, but we observed no such differences when we examined the spleen or draining lymph nodes of these recipients. Grafts destined for tolerance appeared very similar in their pattern of gene expression when compared to syngeneic grafts with both showing a reduced level of T cell and APC infiltration compared to rejecting grafts and a relative increase of Treg and modulated APC associated gene transcripts when compared to ungrafted, normal tail skin. These data suggest that the mechanisms that maintain allograft tolerance may be very similar to the local regulatory mechanisms acting to maintain self tolerance in the face of an inflammatory stimulus. This also means that there may be no unique gene signature able to form the basis of a reliable positive biomarker for transplantation tolerance.
Materials and Methods
Mice, skin grafting, and tolerance induction
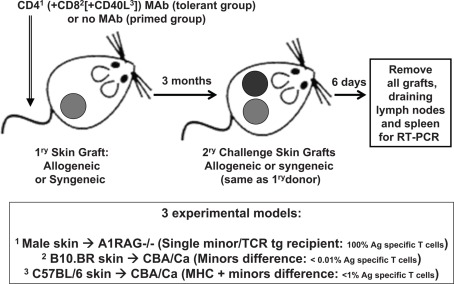
CBA/Ca, CBA.RAG1−/−, A1.RAG1−/−, B10.BR (all H-2k), C57BL/6 (H-2b) mice, were bred and maintained under SPF conditions in the animal facility of the Sir William Dunn School of Pathology, Oxford, UK. All procedures were conducted in accordance with the Home Office Animals (Scientific Procedures) Act of 1986. Tail skin was grafted on the lateral thoracic flank of recipient mice as previously described (Qin et al., 1990). Recipients in the tolerant groups were given 3 × 1 mg each, on days −1, +1, and +3 relative to their first grafts, of non-depleting monoclonal antibodies (Figure 1). A1.RAG1−/− recipients were given CD4 (clone YTS 177.9.6) antibody alone (Cobbold et al., 2004), B10.BR grafted CBA/Ca recipients were given the CD4 plus CD8 (clone YTS 105.18.10) antibodies (Qin et al., 1990), and CBA/Ca recipients of C57BL/6 skin were given a cocktail of CD4 plus CD8 plus CD154 (CD40L; clone MR1) antibodies (Daley et al., 2008). Control recipients received either allogeneic or syngeneic tail skin grafts without antibody treatment. After 100 days all recipients were given secondary challenge skin grafts from the same strain as the original donor graft on the opposite flank (without any additional antibody treatment). Recipients were sacrificed on day 6 after second grafting and their spleens, draining lymph nodes and all surviving grafts were taken for analysis by qRT-PCR.
Figure 1.
Outline of the experimental skin graft models used.
Quantitative real-time RT-PCR
Total RNA was prepared from tissues or whole skin grafts using SV Total RNA Lysis Buffer (Promega) and DNase-I treatment. cDNA was generated using the StrataScript First Strand Synthesis System (Stratagene), using random hexamer primers. Real-time RT-PCR and analysis were performed using the ABI/PRISM 7700 sequence detector system (Applied Biosystems) and inventoried “assay on demand” Taqman® gene expression assays (Table 1) in a low density array (TLDA) format as recommended (Applied Biosystems). Relative quantities (RQ) were calculated by the ΔΔCt method. Samples that gave no detectable signal were assigned Ct = 40. Samples were separately normalized either to a house keeping gene (hprt1) or to genes specifically expressed in T cells (cd3g) or APCs (cd74). All statistical analyses were performed on log transformed (RQ) data, although data is presented as a normal ratio between two samples of interest. Ratios were considered statistically significant when log[ratio + 2 × Σ(standard deviations)] were greater than 0 (P < 0.05).
Table 1.
Selection of genes for assay by Taqman qRT-PCR.
| TaqMan Assay | Gene | TaqMan Assay | Gene | TaqMan Assay | Gene | TaqMan Assay | Gene |
|---|---|---|---|---|---|---|---|
| Hs99999901_s1 | 18S10 | Mm01292449_m1 | Ccr103 | Mm00607939_s1 | Actb10 | Mm00812512_m1 | Prf1 |
| Mm99999915_g1 | Gapdh10 | Mm00438270_m1 | Ccr23 | Mm00446968_m1 | Hprt110 | Mm00442834_m1 | Gzmb7 |
| Mm00434371_m1 | Itga21 | Mm01216172_m1 | Ccr33 | Mm00439103_m1 | Gpr83 | Mm00651853_m1 | Stfa37 |
| Mm00442890_m1 | Itga31 | Mm00438271_m1 | Ccr43 | Mm00727638_s1 | Hig2 | Mm00440646_m1 | Furin7 |
| Mm00439770_m1 | Itga41 | Mm01216171_m1 | Ccr53 | Mm00441911_m1 | Cd40lg | Mm00439191_m1 | Gzma7 |
| Mm00439797_m1 | Itga51 | Mm99999114_s1 | Ccr63 | Mm00497237_m1 | Icosl | Mm00516884_m1 | Hp7 |
| Mm00434375_m1 | Itga61 | Mm01301785_m1 | Ccr73 | Mm00516023_m1 | Icam1 | Mm00656886_g1 | Mcpt15,7 |
| Mm00801807_m1 | Itgal1 | Mm99999115_s1 | Ccr83 | Mm00456990_m1 | Tln1 | Mm00487638_m1 | Cma1 (Mcpt5)5,7 |
| Mm00442916_m1 | Itgb71 | Mm02528165_s1 | Ccr93 | Mm00802831_m1 | Igf1r | Mm00469310_m1 | Ela27 |
| Mm00441291_m1 | Sell1 | Mm99999054_s1 | Cxcr33 | Mm00803629_m1 | Phb;Fyb | Mm00435860_m1 | Serpine17 |
| Mm01204601_m1 | Selplg1 | Mm01292123_m1 | Cxcr43 | Mm00448831_s1 | Sod3 | Mm00438094_g1 | Cd148 |
| Mm00497118_m1 | Aass2 | Mm00432086_m1 | Cxcr53 | Mm00516004_m1 | Hmox1 | Mm00442346_m1 | Tlr28 |
| Mm00475988_m1 | Arg12 | Mm00472858_m1 | Cxcr63 | Mm00847448_s1 | Rap1a | Mm00546288_s1 | Tlr58 |
| Mm00477592_m1 | Arg22 | Mm00442206_s1 | Xcr13 | Mm00487448_s1 | Fut4 | Mm00446193_m1 | Tlr98 |
| Mm00500289_m1 | Bcat12 | Mm00432102_m1 | Bmp74 | Mm01330673_g1 | Fut7 | Mm01291777_m1 | Ahr9 |
| Mm00802192_m1 | Bcat22 | Mm00801778_m1 | Ifng4 | Mm00449152_m1 | Tyrobp | Mm00438095_m1 | Cd3g9,10 |
| Mm00473573_m1 | Cdo12 | Mm00439616_m1 | Il104 | Mm00438867_m1 | Fcer1a5 | Mm00658576_m1 | Cd749,10 |
| Mm00516688_m1 | Ddc2 | Mm00434169_m1 | Il12a4 | Mm00445212_m1 | Kit5 | Mm00649916_m1 | Ms4a4b9 |
| Mm00456709_m1 | Hal2 | Mm00439619_m1 | Il17a4 | Mm00451600_g1 | Pth5 | Mm00459296_m1 | Ms4a6c9 |
| Mm00456104_m1 | Hdc2 | Mm00434228_m1 | Il1b4 | Mm00432631_m1 | Cort | Mm00508099_m1 | Tmem176b9 |
| Mm00515786_m1 | Il4i12 | Mm00446185_m1 | Il1rn4 | Mm00480990_m1 | Rnf128 (Grail) | Mm00463324_g1 | Pilra9 |
| Mm00492586_m1 | Indo2 | Mm99999222_m1 | Il24 | Mm00802100_m1 | Alox5ap | Mm00652421_m1 | Pilrb19 |
| Mm00524206_m1 | Indol12 | Mm00444241_m1 | Il224 | Mm00469161_m1 | Hebp1 | Mm00655955_gH | Pira69 |
| Mm00500918_m1 | Pah2 | Mm00518984_m1 | Il23a4 | Mm01298628_m1 | Skap1 | Mm00776306_mH | Klra69 |
| Mm00451856_g1 | Tdh2 | Mm00445259_m1 | Il44 | Mm00493634_m1 | Tgfbi | Mm00452054_m1 | Cd2749 |
| Mm00451266_m1 | Tdo22 | Mm00439646_m1 | Il54 | Mm00457979_m1 | Zbp1 | Mm00435532_m1 | Pdcd19 |
| Mm00546816_m1 | Tha12 | Mm00446190_m1 | Il64 | Mm00656724_m1 | Egr16 | Mm00711660_m1 | Cd809 |
| Mm00493794_m1 | Tph12 | Mm00434305_m1 | Il94 | Mm00456650_m1 | Egr26 | Mm00444543_m1 | Cd869 |
| Mm00440485_m1 | Nos22 | Mm00441724_m1 | Tgfb14 | Mm00475164_m1 | Foxp36 | Mm00486849_m1 | Ctla49 |
| Mm00522563_m1 | Mat1a2 | Mm00436952_m1 | Tgfb24 | Mm00484683_m1 | Gata36 | Mm00514644_m1 | S1pr19 |
| Mm00506137_m1 | Mat2b2 | Mm00434189_m1 | Il12rb14 | Mm00515191_m1 | Irf16 | Mm00488795_m1 | Clec4a2 |
| Mm00444228_m1 | Ccl203 | Mm00434200_m1 | Il12rb24 | Mm00516431_m1 | Irf46 | Mm00490931_m1 | Clec4n |
| Mm00436446_g1 | Ccl63 | Mm00434223_m1 | Il17ra4 | Mm03682796_m1 | Rorc6 | Mm00496572_m1 | Gp49a |
| Mm00436450_m1 | Cxcl23 | Mm00439622_m1 | Il1r24 | Mm00443103_m1 | Rora6 | Mm00656925_m1 | S100a9 |
| Mm00469294_m1 | Ebi33 | Mm00519942_m1 | Il23r4 | Mm00450960_m1 | Tbx216 | Mm00802901_m1 | Lgals3 |
| Mm01216147_m1 | Ccr13 | Mm01212875_m1 | Penk | Mm00491292_g1 | Zbtb326 (ROG) | Mm00436767_m1 | Spp1 |
1Adhesion molecule, 2Amino acid metabolism, 3Chemokine/chemokine receptor, 4Cytokine/cytokine receptor, 5Mast cells, 6Transcription factor, 7Protease/proteolysis, 8Receptor/immunity/defense, 9Signal transduction/immunity defence, 10Normalizing controls.
Selection of markers for T cells and APCs as indicators of immune responses
The Taqman qRT-PCR gene transcript assays that were chosen for analysis are shown in (Table 1). They were selected on the basis of the following criteria: “signature genes” which were mainly those considered a “master” transcription factors or cytokines for functionally relevant T cell subsets (Th1, Th2, Tr1, Th17, and Treg); chemokine receptors and adhesion molecules likely to be important in any differential trafficking or accumulation of T cell or APC subsets in grafts or lymph nodes; enzymes implicated in the consumption of essential amino acids by APCs under regulatory conditions, mast cell genes (as mast cells have been previously implicated in tolerance; Lu et al., 2006) and additional genes we had previously shown to be associated with either regulatory T cells or modulated dendritic cells by serial analysis of gene expression (Cobbold et al., 2003).
Results
Mouse models of tolerance
We chose models where the only source of donor antigen was the skin graft itself, as we have already shown that these models depend on the continued presence of Treg within the graft for the maintenance of the tolerance state (Graca et al., 2002; Cobbold et al., 2006). Such tolerance is defined by the ability to accept fresh donor-type skin grafts even though normal, but systemic T cell reactivity to donor antigen generally appears to remain intact when assayed in vitro (Cobbold et al., 1996), so that conventional in vitro tests have not provided any useful biomarkers in such models. All three models used the same fully tolerance permissive CBA/Ca gene background recipients, but varied in the frequency of donor antigen specific T cells from 100% (A1.RAG transgenic recipients given syngeneic male skin and non-depleting CD4 antibody) to ∼1% (CBA/Ca recipients given MHC and minor mismatched C567BL/6 skin and both CD4, CD8, and CD40L antibodies) to <<0.1% (CBA/Ca recipients given multiple minor mismatched B10.BR skin and CD4 plus CD8 antibodies; Figure 1). In order to compare intact grafts (on day 6 after grafting) that we knew were destined to be accepted or rejected we focused on an analysis of secondary challenge grafts in recipients that had been previously tolerized by grafting and antibody co-administration or that had been primed by prior skin grafting alone. We also included a group of recipients given only syngeneic primary and secondary skin grafts so that we could potentially distinguish antigen specific and non-antigen specific components of any response. We also analyzed spleen and draining lymph nodes from all these mice at the same time.
Limitations of Foxp3 as a potential biomarker of tolerance
We first analyzed the differential expression of the “master” Treg gene Foxp3 (Hori et al., 2003). No significant differences in foxp3 between tolerant and rejecting recipients were observed in any of the three models in the spleen or draining lymph nodes. Total Foxp3 (when normalized to house keeping gene hprt1; Table 2) was unable to distinguish between any of the challenge grafts destined for tolerance compared to those primed for rejection (Table 2). When Foxp3 values were normalized to the level of T cell infiltration, as indicated by CD3γ expression, there was some indication that a higher proportion of T cells expressed the Treg associated gene in the two models with the higher frequency of donor specific T cells, but there was still a high degree of variation between different individuals in the TCR transgenic group at this time point, particularly in the CD3γ content, which therefore failed to reach statistical significance. If such individual variation, either between individuals or over time, of relevant transcripts were a general finding this would limit their predictive value as biomarkers. The only striking increase in Foxp3, whether normalized to hprt1 or CD3γ, was observed when the originally long-term surviving tolerated allogeneic skin was compared with a similarly long-term accepted syngeneic graft in the TCR transgenic model where all T cells were specific for donor antigen. Therefore, Foxp3 does not seem to reliably correlate with transplantation tolerance in these models.
Table 2.
Foxp3 expression in grafts is not a reliable indicator of tolerance.
| Comparison |
Foxp3/ Hprt1 ratio |
Foxp3/ Cd3g ratio |
|
|---|---|---|---|
| B10BR → CBA | Tol vs Rej challenge grafts | 0.41 | 1 |
| (n = 4/group) | Tol vs Syn challenge grafts | 0.76 | 1.1 |
| Tol vs Syn original grafts | 2.1 | 1 | |
| C57BL/6 → CBA | Tol vs Rej challenge grafts | 1.6 | 3.6* |
| (n = 4/group) | Tol vs Syn challenge grafts | 0.8 | 3.2 |
| Tol vs Syn original grafts | 1.1 | 1 | |
| Male → A1RAG | Tol vs Rej challenge grafts | 0.8 | 5.3 |
| (n = 6/group) | Tol vs Syn challenge grafts | 0.6 | 0.9 |
| Tol vs Syn original grafts | 38.2* | 25.5* | |
*P < 0.05, all other ratios were non-significant.
Other genes that potentially distinguish tolerant and rejecting skin
We performed similar comparisons between challenge grafts given to tolerant and primed recipients for a set of more than 150 genes representative of the following categories: amino acid metabolism, signature T cell subset transcription factors, signature cytokines, chemokine receptors, adhesion molecules, mast cell genes, and additional genes associated with regulatory T cell subsets or modulated APCs previously identified by serial analysis of gene expression (Table 1). Surprisingly, none of the genes tested were found to differ significantly between tolerant, rejecting or syngeneic grafted recipients when samples from the spleen or draining lymph node were compared, which strongly supports the hypothesis (Cobbold et al., 2006) that tolerance and immune regulation act primarily within the local grafted tissue in these models. We did find in all three models that rejecting grafts tended to exhibit higher levels, when compared to tolerated grafts, of both CD3γ and CD74 gene transcripts, when normalized to a house keeping gene such as hprt1 (Table 3), suggesting an increased infiltration of T cells and APC in grafts destined for rejection. In the MHC mismatched model, where CD3γ and CD74 were most significantly associated with rejection (Table 4), the additional genes upregulated in rejecting grafts included Th1 related genes such as Ifng and Gzmb, the Th2 related Il4 and Th17 inducing Il6. Some genes usually associated with regulation were also over-expressed in rejecting grafts, including Indo, Nos2, Arg1, Indol1, and foxp3 (as discussed earlier). Note that none of these differences were observed in the draining lymph nodes.
Table 3.
Infiltration of skin grafts by T cells and APCs.
| Comparison |
CD3g/ Hprt1 ratio |
CD74/ Hprt1 ratio |
|
|---|---|---|---|
| B10BR → CBA | Rej vs Tol challenge grafts | 6.1* | 3.8* |
| (n = 4/group) | Tol vs Syn challenge grafts | 1.4 | 1.2 |
| Tol vs Syn original grafts | 2.3 | 1 | |
| C57BL/6 → CBA | Rej vs Tol challenge grafts | 16.6* | 7.1* |
| (n = 4/group) | Tol vs Syn challenge grafts | 4.2 | 1.8 |
| Tol vs Syn original grafts | 1.1 | 0.9 | |
| Male → A1RAG | Rej vs Tol challenge grafts | 8.3 | 2.6 |
| (n = 6/group) | Tol vs Syn challenge grafts | 8.6 | 2.3 |
| Tol vs Syn original grafts | 1.5 | 0.2 | |
*P < 0.05, all other ratios were non-significant.
Table 4.
Genes associated with rejection in C57BL/6 → CBA (fully allogeneic) skin grafts.
| Gene | Rej:Tol 2nd graft ratio | Rej:Tol lymph node ratio | |
|---|---|---|---|
| Cd3g | 13.2* | 1.1 | |
| Cd74 | 7.06* | 1.05 | |
| Genes over-expressed in rejecting compared to tolerated grafts normalized to Hprt1 | Indo | 121.0* | 0.6 |
| Nos2 | 118.0* | 1.3 | |
| Ifng | 86.9* | 1.5 | |
| Gzmb | 84.0* | ND | |
| Arg1 | 18.5* | 2.6 | |
| Il4 | 31.7* | 0.5 | |
| Il6 | 27.6* | 1.5 | |
| Indol1 | 14.8* | 1.0 |
*P < 0.05 (n = 6/group), all other ratios were non-significant.
ND, not determined.
The corollary of these data is that tolerated skin grafts tended to have a reduced infiltration by T cells and APCs as indicated by CD3γ and CD74 expression, and none of the genes tested ( normalized to house keeping genes) were positively correlated with tolerance. We therefore estimated the relative contribution of different T cell and APC differentiation pathways by normalizing T cell expressed transcripts to CD3γ and APC related transcripts to CD74. In the TCR transgenic model 5 T cell associated transcripts were found to be significantly over-expressed within tolerant compared to rejecting grafts (Rorc, Gata3, Egr2, Rnf128, and Tgfb1; Table 5) when normalized to CD3γ while Bmp7 was the only over-expressed APC related gene (normalized to MHC-II invariant chain, CD74). When we examined the C57BL/6 → CBA/Ca model we also observed over-expression of Rorc, Gata3, and Tgfb1 in tolerated grafts (Table 6), while Gata3 and Bmp7 were differential in the B10.BR-CBA model (Table 7). Over-expression of the energy related genes Egr2 (Harris et al., 2004) and Rnf128 (Grail; Anandasabapathy et al., 2003) was only observed in the TCR transgenic model where there were no non-antigen specific T cells present to overwhelm the antigen specific signal. A variety of amino acid catabolizing enzymes were also relatively increased (normalized to CD74) in tolerated MHC and minors different skin grafts (Tables 6 and 7). The tolerance associated genes in common suggest a weak bias away from Th1 responses to Th17 or NK cells (Rorc), Th2 cells (Gata3), and Treg activity (Tgfb1), but there is little evidence of the dominant signature containing the variety of regulatory-associated genes that one might have expected. Perhaps even more surprisingly, the secondary challenge MHC mismatched skin grafts in tolerant recipients were almost indistinguishable by their patterns of gene expression to equivalent syngeneic grafts (Table 6).
Table 5.
Differential gene expression in male → female A1.RAG−/− skin grafts.
| Gene | Ratio | |
|---|---|---|
| T cell related genes over-expressed in tolerated compared to rejecting grafts (Cd3g normalized) | Rorc | 23.43* |
| Gata3 | 13.85* | |
| Egr2 | 9.76* | |
| Rnf128 (grail) | 8.40* | |
| Tgfb1 | 5.93* | |
| Foxp3 | 5.32 | |
| APC related genes over-expressed in tolerated compared to rejecting grafts (Cd74 normalized) | Bmp7 | 5.01* |
*P < 0.05 (n = 6/group), all others ratios non-significant.
Table 6.
Differential gene expression in C57BL/6 → CBA (fully allogeneic) skin grafts.
| Gene | Tol:Rej 2nd grafts ratio | Tol:Rej lymph nodes ratio | Tolerant:syngeneic 2nd grafts ratio | |
|---|---|---|---|---|
| T cell genes upregulated in tolerated compared to rejecting grafts normalized to Cd3g | Ccl5 | 45.9* | ND | ND |
| Gata3 | 41.0* | 1.3 | 0.66 | |
| Rora | 40.2* | ND | ND | |
| Ccl20 | 40.1* | ND | ND | |
| Rorc | 29.6* | 2.7 | 0.40 | |
| Itga6 | 22.5* | ND | ND | |
| Itga3 | 18.0* | ND | ND | |
| Il9 | 14.6* | 1.4 | 1.74 | |
| Tgfb1 | 9.4* | 1.0 | 0.91 | |
| Ccr9 | 7.0* | ND | ND | |
| Rap1a | 6.7* | ND | ND | |
| APC genes upregulated in tolerated compared to rejecting grafts normalized to Cd74 | Tdh | 51.2* | 0.9 | 1.63 |
| Ddc | 34.8* | 0.4 | 2.05 | |
| Tgfb2 | 21.9* | 1.7 | 0.96 | |
| Hal | 16.1* | 1.2 | 0.60 | |
| Cdo1 | 12.5* | 2.9 | 1.09 | |
| Bcat2 | 10.8* | 1.7 | 0.78 | |
| Bcat1 | 7.4* | 0.8 | 0.67 | |
*P < 0.05 (n = 4/group), all other ratios were non-significant.
ND, not determined.
Table 7.
Differential gene expression in B10BR → CBA (multiple minors) skin grafts.
| Gene | Ratio | |
|---|---|---|
| T cell genes over-expressed in tolerant compared to rejecting 2nd grafts (Cd3g normalized) | Gata3 | 11.1* |
| Tgfb1 | 4.3 | |
| APC genes over-expressed in tolerant compared to rejecting 2nd grafts (Cd74 normalized) | Aass | 42.7* |
| Bmp7 | 11.2* | |
| Hal | 6.4* | |
| Mcpt5 | 4.7* | |
| Tdh | 14.2 | |
*P < 0.05 (n = 4/group), all other ratios non-significant.
This latter observation suggested two possibilities – first, that tolerated skin grafts, once fully accepted and healed in, no longer require active regulation and are effectively ignored by the immune system, or second, that both allogeneic and syngeneic skin require similar active immune regulation to maintain their tolerant status. We have already shown, by transferring skin grafts to secondary RAG1−/− recipients and subsequent depletion of Treg that tolerated grafts contain primed effector cells that are actively held in check by Treg (Cobbold et al., 2006), so the first possibility seems unlikely. We found there were a large number of genes upregulated early during the process of skin grafting (Table 8), even in T cell deficient recipients, many of which may also have an immunoregulatory function [e.g., Bcat1, Hdc, Arg1, Ebi3, Gzma, Tdh, and Tmem176b (TORID)]. We then looked for additional genes expressed by long-term surviving syngeneic skin grafts on CBA/Ca recipients with an intact immune system compared with freshly harvested normal tail skin we found that the syngeneic grafts were highly enriched for Treg associated gene transcripts (Table 9), including Foxp3, Gata3, Il10, and Zbtb32 (ROG) and modulated APCs (IL4i1 and Nos2). This latter result may represent an amplification of the normal bias toward regulatory T cells that has been previously described in the skin and which has been suggested to maintain (self) tolerance in the face of an inflammatory stimulus (Dudda et al., 2008). In other words, it is possible that self tolerance to certain skin antigens is also dependent on active regulation.
Table 8.
Genes over-expressed in skin during the process of grafting and in the absence of adaptive immunity.
| Gene | Ratio | |
|---|---|---|
| Genes upregulated in skin during the process of grafting in the absence of adaptive immunity (Hprt1 normalized) |
Spp1 | 83.8* |
| Bcat1 | 52.4* | |
| Il1b | 39.3* | |
| Clec4a2 | 30.2* | |
| Ms4a6c | 27.6* | |
| Gp49a;Lilrb4 | 20.9* | |
| Tyrobp | 19.1* | |
| Pilra | 18.5* | |
| Clec4n | 17.3* | |
| Zbp1 | 13.5* | |
| Ccr1 | 12.6* | |
| Hdc | 11.6* | |
| Cd80 | 10.8* | |
| Arg1 | 10.4* | |
| Ccr7 | 8.7* | |
| Pira6 | 8.3* | |
| Cd86 | 8.3* | |
| Ebi3 | 7.9* | |
| Hp | 7.9* | |
| Gzma | 6.2* | |
| Mcpt5 | 6.1* | |
| Tlr2 | 5.2* | |
| Alox5ap | 4.9* | |
| Ms4a4b | 4.4* | |
| Tdh | 4.1* | |
| Tmem176b | 4.1* | |
| Irf1 | 3.8* | |
| Ccl6 | 3.7* | |
| Hmox1 | 3.5* | |
| Foxp3 | 0.5 | |
| Cd3g | 0.5 | |
| CD74 | 1.5 | |
*P < 0.05 (n = 4/group), other ratios non-significant.
Table 9.
Syngeneic skin grafts over-express genes associated with regulatory cells.
| Gene | Ratio | |
|---|---|---|
| Genes over-expressed in syngeneic original grafts vs normal skin (Hprt1 normalized)+ | Foxp3 | 40.1* |
| Il4i1 | 17.1* | |
| Zbtb32(ROG) | 13.0* | |
| Nos2 | 4.7* | |
| Il10 | 4.7* | |
| Gata3 | 3.3* |
*P < 0.05 (n = 4/group).
+This table excludes all genes listed in Table 8 as over-expressed in the absence of adaptive immunity.
Discussion
Although foxp3 represents the best marker currently available for identifying regulatory T cells, we found that it could not be used as a reliable biomarker to indicate transplantation tolerance even when we analyzed the grafted tissue itself. Indeed, absolute foxp3 levels were often higher in grafts undergoing rejection, as has been previously reported (Dijke et al., 2007; Yang et al., 2007; Bunnag et al., 2008). One reason for this might be that foxp3 can be transiently expressed in activated effector T cells (Wang et al., 2007). An alternative possibility is that foxp3+ regulatory T cells are attracted to all sites of inflammation where they act to limit immune pathology, but it is the balance of activation vs regulation that determines the eventual outcome of rejection or tolerance, respectively. One way to estimate this balance would be to normalize the foxp3 expression for the number of T cells present, but this approach was still unable to provide a clear correlation with graft outcome, suggesting that the numbers of effector and regulatory T cells within a graft may be closely coupled and finely balanced.
There are two findings from this study that suggest that it may not be possible to identify biomarkers that uniquely identify tolerance in recipients of allografts. First, the only significant differences in gene expression between tolerated, rejecting and syngenic responses were observed within the grafted organ itself, and not in draining lymph nodes or spleens. This may be due to alloantigen specific T cells preferentially accumulating at the main site of antigen, i.e., within the graft itself (Graca et al., 2002; Cobbold et al., 2004). In order to observe donor specific tolerance associated biomarkers systemically it may be necessary for the alloantigen to also be systemically distributed, as may be seen, for example, in some liver transplant recipients that develop macrochimerism (Starzl et al., 1992), by inducing tolerance by chronic administration of soluble peptide antigen (Apostolou and von Boehmer, 2004), or by providing a systemic alloantigen boost to enrich Treg in the lymphoid tissues (Bemelman et al., 1998). Second, the pattern of gene expression that distinguished tolerated from rejecting grafts, i.e., a reduced inflammatory infiltrate and a bias toward regulatory T cell and APC associated transcripts, was indistinguishable from that seen in syngeneic grafts (except in the TCR transgenic model where all T cells were donor antigen specific).
Although a lack of inflammation in tolerated grafts is an important observation, it may not represent a useful predictive biomarker in the context of clinical transplantation at the point where the recipients are still on high doses of immunosuppressive and anti-inflammatory drugs. The similarity between tolerated allografts and syngeneic grafts also suggests that transplantation tolerance is probably maintained by the same mechanisms that are actively maintaining self tolerance in normal peripheral tissues. While allo-tolerance is being maintained, at least in part, by an antigen specific regulatory T cell population (as observed in the monospecific TCR transgenic model), in a recipient with an intact polyclonal repertoire the frequency of allospecific regulatory cells, even within grafts, will likely be too low to provide a distinct and detectable gene signature above the background of regulation mediated by innate and self reactive components. This background response to syngeneic grafts may be very similar to the normal process of limiting an inflammatory or healing response throughout the body, so any systemically detectable tolerance associated gene products may be uninformative with respect to the state of the allograft.
A detection of systemic regulatory gene products may, however, provide some indication of whether a particular immunosuppressive regimen is permissive rather than inhibitory of immune regulatory mechanisms in general. Published biomarkers [from “Reprogramming the Immune System for Establishment of Tolerance” (RISET) and “International Tolerance Network” (ITN) Turka et al., 2010] may therefore be more indicative of the systemic responses of individual patients to immunosuppressive agents and the type of graft they received than tolerance to the graft alloantigens per se.
We used skin grafts throughout these experiments as these represent our most robust and well characterized model systems of donor antigen specific tolerance that is completely dependent on the presence of Tregs within the tolerated tissue (Graca et al., 2002; Cobbold et al., 2006), but it is possible that the vascularized organ grafts most often used clinically may differ from skin grafts, particularly in their initial healing and angiogenic responses. Skin grafts, however, are already re-vascularized by day 4, and we found that tolerated allografts were similar in their regulatory gene signature to syngeneic grafts at both 6 and +100 days post grafting, when any initial vascularization, wound healing and ischemic reperfusion responses would have resolved. We would therefore have no reason to expect that our findings from skin grafting would differ, in general terms, to the transplantation of vascularized organs.
Acknowledgments
We would like to thank Sue Humm for expert technical assistance and the PSB staff for excellent animal care. This work was funded by a research fellowship from EP Abrahams Trust (Stephen P. Cobbold), a programme grant from MRC UK “Therapeutic Immunoregulation,” and an EU framework 6 network grant (“Reprogramming the Immune System for Establishment of Tolerance”: RISET).
References
- Anandasabapathy N., Ford G. S., Bloom D., Holness C., Paragas V., Seroogy C., Skrenta H., Hollenhorst M., Fathman C. G., Soares L. (2003). GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4( T cells. Immunity 18, 535–547 10.1016/S1074-7613(03)00084-0 [DOI] [PubMed] [Google Scholar]
- Apostolou I., von Boehmer H. (2004). In vivo instruction of suppressor commitment in naive T cells. J. Exp. Med. 199, 1401–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemelman F., Honey K., Adams E., Cobbold S., Waldmann H. (1998). Bone marrow transplantation induces either clonal deletion or infectious tolerance depending on the dose. J. Immunol. 160, 2645–2648 [PubMed] [Google Scholar]
- Bunnag S., Allanach K., Jhangri G. S., Sis B., Einecke G., Mengel M., Mueller T. F., Halloran P. F. (2008). FOXP3 expression in human kidney transplant biopsies is associated with rejection and time post transplant but not with favorable outcomes. Am. J. Transplant. 8, 1423–1433 10.1111/j.1600-6143.2008.02268.x [DOI] [PubMed] [Google Scholar]
- Chen W. (2006). Dendritic cells and (CD4+) CD25+ T regulatory cells: crosstalk between two professionals in immunity versus tolerance. Front. Biosci. 11, 1360–1370 [DOI] [PubMed] [Google Scholar]
- Cobbold S. P., Adams E., Farquhar C. A., Nolan K. F., Howie D., Lui K. O., Fairchild P. J., Mellor A. L., Ron D., Waldmann H. (2009). Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 12055–12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold S. P., Adams E., Graca L., Daley S., Yates S., Paterson A., Robertson N. J., Nolan K. F., Fairchild P. J., Waldmann H. (2006). Immune privilege induced by regulatory T cells in transplantation tolerance. Immunol. Rev. 213, 239–255 [DOI] [PubMed] [Google Scholar]
- Cobbold S. P., Adams E., Marshall S. E., Davies J. D., Waldmann H. (1996). Mechanisms of peripheral tolerance and suppression induced by monoclonal antibodies to CD4 and CD8. Immunol. Rev. 149, 5–33 [DOI] [PubMed] [Google Scholar]
- Cobbold S. P., Adams E., Nolan K. F., Regateiro F. S., Waldmann H. (2010). Connecting the mechanisms of T-cell regulation: dendritic cells as the missing link. Immunol. Rev. 236, 203–218 [DOI] [PubMed] [Google Scholar]
- Cobbold S. P., Castejon R., Adams E., Zelenika D., Graca L., Humm S., Waldmann H. (2004). Induction of foxP3( regulatory T cells in the periphery of T cell receptor transgenic mice tolerized to transplants. J. Immunol. 172, 6003–6010 [DOI] [PubMed] [Google Scholar]
- Cobbold S. P., Nolan K. F., Graca L., Castejon R., Le Moine A., Frewin M., Humm S., Adams E., Thompson S., Zelenika D., Paterson A., Yates S., Fairchild P. J., Waldmann H. (2003). Regulatory T cells and dendritic cells in transplantation tolerance: molecular markers and mechanisms. Immunol. Rev. 196, 109–124 [DOI] [PubMed] [Google Scholar]
- Daley S. R., Cobbold S. P., Waldmann H. (2008). Fc-disabled anti-mouse CD40L antibodies retain efficacy in promoting transplantation tolerance. Am. J. Transplant. 8, 2265–2271 10.1111/j.1600-6143.2008.02382.x [DOI] [PubMed] [Google Scholar]
- Dijke I. E., Caliskan K., Korevaar S. S., Maat A. P., Zondervan P. E., Balk A. H., Weimar W., Baan C. C. (2008). FOXP3 mRNA expression analysis in the peripheral blood and allograft of heart transplant patients. Transpl. Immunol. 18, 250–254 [DOI] [PubMed] [Google Scholar]
- Dijke I. E., Velthuis J. H., Caliskan K., Korevaar S. S., Maat A. P., Zondervan P. E., Balk A. H., Weimar W., Baan C. C. (2007). Intragraft FOXP3 mRNA expression reflects antidonor immune reactivity in cardiac allograft patients. Transplantation 83, 1477–1484 10.1097/01.tp.0000264997.53153.8b [DOI] [PubMed] [Google Scholar]
- Dudda J. C., Perdue N., Bachtanian E., Campbell D. J. (2008). Foxp3+ regulatory T cells maintain immune homeostasis in the skin. J. Exp. Med. 205, 1559–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graca L., Cobbold S. P., Waldmann H. (2002). Identification of regulatory T cells in tolerated allografts. J. Exp. Med. 195, 1641–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guleria I., Khosroshahi A., Ansari M. J., Habicht A., Azuma M., Yagita H., Noelle R. J., Coyle A., Mellor A. L., Khoury S. J., Sayegh M. H. (2005). A critical role for the programmed death ligand 1 in fetomaternal tolerance. J. Exp. Med. 202, 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. E., Bishop K. D., Phillips N. E., Mordes J. P., Greiner D. L., Rossini A. A., Czech M. P. (2004). Early growth response gene-2, a zinc-finger transcription factor, is required for full induction of clonal anergy in CD4+ T cells. J. Immunol. 173, 7331–7338 [DOI] [PubMed] [Google Scholar]
- Hori S., Nomura T., Sakaguchi S. (2003). Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 10.1126/science.1079490 [DOI] [PubMed] [Google Scholar]
- Huang Y., Shan J., Zhang C., Zhang J., Feng L., Li S., Li Y. (2010). Peripheral blood T regulatory cell counts may not predict transplant rejection. BMC Immunol. 11:40. 10.1186/147-2172-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L. F., Lind E. F., Gondek D. C., Bennett K. A., Gleeson M. W., Pino-Lagos K., Scott Z. A., Coyle A. J., Reed J. L., Van Snick J., Strom T. B., Zheng X. X., Noelle R. J. (2006). Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature 442, 997–1002 10.1038/nature05010 [DOI] [PubMed] [Google Scholar]
- Qin S., Cobbold S. P., Pope H., Elliott J., Kioussis D., Davies J., Waldmann H. (1993). “Infectious” transplantation tolerance. Science 259, 974–977 10.1126/science.8094901 [DOI] [PubMed] [Google Scholar]
- Qin S. X., Wise M., Cobbold S. P., Leong L., Kong Y. C., Parnes J. R., Waldmann H. (1990). Induction of tolerance in peripheral T cells with monoclonal antibodies. Eur. J. Immunol. 20, 2737–2745 [DOI] [PubMed] [Google Scholar]
- Sadlon T. J., Wilkinson B. G., Pederson S., Brown C. Y., Bresatz S., Gargett T., Melville E. L., Peng K., D’Andrea R. J., Glonek G. G., Goodall G. J., Zola H., Shannon M. F., Barry S. C. (2010). Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J. Immunol. 185, 1071–1081 10.4049/jimmunol.1000082 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S., Wing K., Onishi Y., Prieto-Martin P., Yamaguchi T. (2009). Regulatory T cells: how do they suppress immune responses? Int. Immunol. 21, 1105–1111 [DOI] [PubMed] [Google Scholar]
- Starzl T. E., Demetris A. J., Trucco M., Ramos H., Zeevi A., Rudert W. A., Kocova M., Ricordi C., Ildstad S., Murase N. (1992). Systemic chimerism in human female recipients of male livers. Lancet 340, 876–877 10.1016/0140-6736(92)93286-V [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turka L. A., Wood K., Bluestone J. A. (2010). Bringing transplantation tolerance into the clinic: lessons from the ITN and RISET for the establishment of tolerance consortia. Curr. Opin. Organ Transplant. 15, 441–448 10.1097/MOT.0b013e32833bd371 [DOI] [PubMed] [Google Scholar]
- Wang J., Ioan-Facsinay A., van der Voort E. I., Huizinga T. W., Toes R. E. (2007). Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur. J. Immunol. 37, 129–138 [DOI] [PubMed] [Google Scholar]
- Yang H., Ding R., Sharma V. K., Hilaire F. S., Lagman M., Li B., Thomas D. A., Luo X., Song P., Stauffer C., August P., Suthanthiran M. (2007). Hyperexpression of Foxp3 and IDO during acute rejection of islet allografts. Transplantation 83, 1643–1647 10.1097/01.tp.0000263991.74052.46 [DOI] [PubMed] [Google Scholar]