Abstract
Surface proteins of Gram-positive bacteria play important roles during the pathogenesis of human infections and require sortase for anchoring to the cell-wall envelope. Sortase cleaves surface proteins at the LPXTG motif and catalyzes the formation of an amide bond between the carboxyl group of threonine (T) and the amino group of cell-wall crossbridges. The NMR structure of sortase reveals a unique β-barrel structure, in which the active-site sulfhydryl of cysteine-184 is poised for ionization by histidine-120, presumably enabling the resultant thiolate to attack the LPXTG peptide. Calcium binding near the active site stimulates catalysis, possibly by altering the conformation of a surface loop that recognizes newly translocated polypeptides. The structure suggests a mechanistic relationship to the papain/cathepsin proteases and should facilitate the design of new antiinfective agents.
Surface proteins of Gram-positive pathogens promote bacterial adhesion to specific organ tissues, resistance to phagocytic killing, or invasion of host cells (1). These mechanisms allow invading bacteria to establish an infection and to escape the host's immune surveillance. Many surface proteins of Gram-positive bacteria are anchored to the cell wall by a mechanism requiring a C-terminal sorting signal with a conserved LPXTG motif, a hydrophobic domain, and a tail of mostly positively charged residues (2, 3). During cell-wall anchoring, surface proteins of Staphylococcus aureus are cleaved between the threonine and the glycine of the LPXTG motif (4). The carboxyl group of threonine is amide-linked to the amino group of the pentaglycine crossbridges, thereby anchoring the C-terminal end of surface proteins to the staphylococcal cell-wall peptidoglycan (5, 6). It seems that lipid II [undecaprenyl pyrophosphate-MurNAc(-l-Ala-d-iGln-l-Lys(NH2-Gly5)-d-Ala-d-Ala)-β1–4-GlcNAc)], a membrane-anchored precursor of cell-wall synthesis (7, 8), serves as a substrate for surface protein anchoring (9). Surface proteins tethered to lipid II may subsequently be incorporated into the peptidoglycan by means of the transglycosylation and transpeptidation reactions of bacterial cell-wall synthesis (10).
S. aureus srtA mutants are defective in the anchoring of surface proteins and accumulate precursor molecules with uncleaved C-terminal sorting signals (11). srtA encodes sortase, a 206-residue polypeptide with an N-terminal membrane anchor (11). Knockout mutations in srtA abolish the ability of mutant staphylococci to anchor several different surface proteins to the cell wall (12). As compared with wild-type staphylococci, srtA mutants display defects in virulence, suggesting that cell-wall anchoring and the display of surface proteins are essential for the pathogenesis of S. aureus infections (12). Purified sortase cleaves LPXTG peptides at the peptide bond between the threonine and glycine (13). In the presence of NH2-Gly3, a substitute for the peptidoglycan substrate lipid II, sortase catalyzes an amide-bond exchange reaction to generate LPXT-Gly3 (14). Together, these data suggest that sortase catalyzes the anchoring of surface proteins to the cell-wall peptidoglycan of S. aureus (15).
Penicillin-binding proteins (PBPs), another group of bacterial transpeptidases, cleave cell-wall peptides at the amide bond of d-Ala-d-Ala (16, 17). The carboxyl group of the cleaved peptide is ester-linked to the hydroxyl group of the active-site serine residue (18, 19). The carboxylpeptidase and β-lactamase members of the PBP family resolve the acyl-enzyme by the nucleophilic attack of water, followed by the release of cleaved substrate (20). PBP transpeptidases bind cell-wall crossbridges, NH2-Gly5 in S. aureus (21, 22), and the nucleophilic attack of their amino group at the acyl-enzyme ester regenerates the active site, thereby linking the carboxyl group of d-Ala within wall peptides to the amino group of the crossbridge (10, 23). Penicillin and other β-lactam antibiotics act as a molecular mimic of d-Ala-d-Ala (24). The bulky structure of cleaved penicillinoic acid occupies the active site and prevents water and other nucleophiles from regenerating PBP acyl-enzymes (20). The three-dimensional structure of several PBPs and β-lactamases (with and without inhibitors) have been solved, revealing a similar overall fold (25–27).
It seems that sortase uses the sulfhydryl of cysteine-184 (C184) as an active-site nucleophile to cleave the peptide bond between threonine and glycine in LPXTG peptides. (i) Mutational exchange of C184 with alanine abolishes sortase activity in vivo and in vitro (14). (ii) Treatment of wild-type sortase with hydroxylamine resolves the acyl-enzyme intermediate and releases surface protein with a C-terminal threonine hydroxamate (13). (iii) Methyl methanethiosulfonates, reagents that form a disulfide with the sulfhydryl, prevent LPXTG cleavage by sortase (9), but activity can be restored by addition of DTT, a reductant that regenerates the active-site sulfhydryl (13). At present, nothing is known about the mechanism of catalysis beyond this cursory description, because the primary sequence of sortase is unrelated to any protein of known structure. To understand the sortase-catalyzed transpeptidation reaction in molecular detail, we have delineated the polypeptide domain within sortase that mediates catalysis and determined its three-dimensional solution structure with NMR spectroscopy.
Experimental Procedures
Purification of SrtAΔN59.
Two primers (5′-AAACATATGCAAGCTAAACCTCAAA-TTCC-3′) and (5′-AAGGATCCTTATTTGACTTCTGTAGCTACAA-3′) were used to amplify by PCR an srtA sequence from the chromosome of S. aureus OS2. The DNA fragment was digested with NdeI and BamHI, inserted into pET9a (Novagen), cut with NdeI and BamHI to generate pHTT25, transformed into Escherichia coli BL21(DE3), and selected on Luria agar with kanamycin (50 μg/ml). SrtAΔN59 for NMR studies was produced from 8 liters of minimal media culture grown at 37°C and incubated with 1 mM IPTG (isopropyl β-d-thiogalactoside) when the optical density at 600 nm reached 0.7. The cells were then harvested, suspended in 100 mM Tris⋅HCl (pH 7.2)/5 mM EDTA/2.5 mM DTT/5 mM benzamidine and lysed in a pressure cell. The lysate was centrifuged and the supernatant was applied to a DEAE-Sepharose Fast Flow XK-50 column (Amersham Pharmacia), and eluted with a salt concentration gradient of 0 to 1 M NaCl in 100 mM Tris⋅HCl, pH 7.2/5 mM EDTA/2.5 mM DTT. Pooled fractions were dialyzed against low-salt buffer, applied to an SP-Sepharose Fast Flow XK-50 column (Amersham Pharmacia), and eluted with a gradient of 0 to 1 M NaCl in 100 mM Tris⋅HCl, pH 7.2/5 mM EDTA/2.5 mM DTT. SrtAΔN59-containing fractions were then purified further by using a Sephacryl S-100 column (Amersham Pharmacia) equilibrated with 50 mM Tris⋅HCl, pH 6.2/100 mM NaCl/0.01% NaN3/20 mM CaCl2/3 mM DTT. For structural studies, isotopic labeling with 15N or 15N and 13C was accomplished by growing cells in minimal medium containing 15NH4Cl or [13C6]glucose as the sole nitrogen and carbon sources. Unlabeled purified protein was subjected to electrospray ionization-MS; an average mass of 16,595.12 Da was observed. This measurement is consistent with the predicted average mass of 16,595.98 for SrtAΔN59.
LPXTG Peptide Cleavage.
Dabcyl-LPETG-Edans {d-LPETG-e; Dabcyl is 4-{[4-(dimethylamino)phenyl]azo}benzoic acid and Edans is 5-[(2-aminoethyl)amino]naphthalene-1-sulfonic acid} was dissolved in 20% DMSO and added at a final concentration of 1–6 μM. Peptide cleavage was monitored as an increase of fluorescence with a FluoroMax-2 spectrometer (Instruments SA, Edison, NJ). The peptide was incubated in the presence or absence of 2 mM NH2-Gly3 and 5 μM SrtAΔN or SrtAΔN59 in buffer R (150 mM NaCl/5 mM CaCl2/50 mM Tris⋅HCl, pH 7.5) in a volume of 520 μl. The increase in fluorescence intensity was recorded as a function of time using excitation at 350 nm and recording the emission maximum at 495 nm.
Characterization of Cleaved Products.
A reaction mixture consisting of 10 μM fluorescent peptides and 15 μM recombinant enzymes in 520 μl of buffer R was incubated in either the presence or the absence of 2 mM NH2-Gly3 at 37°C for 16 h. The reaction was quenched by filtration through Centricon-10 filters (Millipore). The filtrate was subjected to RP-HPLC purification on C18 columns (2 × 250 mm, C18 Hypersil, Keystone Scientific, Bellefonte, PA). Separation was carried out with a gradient from 1% to 41% CH3CN (0.1% trifluoroacetic acid) in 41 min and from 41% to 100% for 10 min at a flow rate of 0.3 ml/min. Elution of peptides was monitored at 206 and 475 nm and fractions were collected every minute, dried under vacuum, and stored at 4°C for electrospray ionization-MS analysis, as described (6, 28).
NMR Spectroscopy and Structure Determination.
All NMR experiments were performed at 306 K on Bruker DRX 500- and 600-MHz spectrometers. Spectra of the calcium-bound form of SrtAΔN59 were obtained by using either 15N or 15N/13C-labeled protein (2.5 mM SrtAΔN59 protein/50 mM Tris⋅HCl, pH 6.7/100 mM NaCl/0.01% NaN3/20 mM CaCl2/3 mM DTT/7% 2H2O). Near-complete assignment of the 1H, 13C, and 15N resonances was achieved by the application of multidimensional heteronuclear NMR experiments (29). All spectra were processed by using nmrpipe (30) and analyzed by using the program nmrview (31). Distance restraints were determined through analysis of three- and four-dimensional 15N- and 13C-edited nuclear Overhauser effect (NOE) spectroscopy (NOESY) experiments (mixing times, 80–100 ms). NOE restraints were grouped into four distance ranges: 1.8–2.7 Å (1.8–2.9 Å for distances involving 15N-bound protons), 1.8–3.3 Å (1.8–3.5 Å for distances involving 15N-bound protons), 1.8–5.0 Å, and 1.8–6.0 Å. The upper distance limits of restraints involving methyl protons were increased by adding 0.5 Å. Distances involving methyl protons, aromatic ring protons, and nonstereospecifically assigned methylene protons were represented as a (Σr6)−1/6 sum. 3JHNα couplings were measured quantitatively by using an HNHA experiment (32), and φ and ψ torsional angles were obtained with the program talos (33). Stereospecific assignments were determined by analyzing NOE and quantitative scalar coupling data. A total of 2,105 experimental restraints were used to calculate the solution structure (1,852 distance restraints, 21 χ1 angles, 1 χ2 angle, 67 ψ, 106 φ, and 58 3JHNα restraints) (Table 1). Structures were calculated by using distance geometry and simulated annealing protocols (34) with the program x-plor V. 3.843 (35). At the final stages of refinement, 41 hydrogen bonds were introduced between slowly exchanging amide protons and acceptor groups identified unambiguously from the ensemble of conformers. Near-complete backbone resonance assignments of calcium-free SrtAΔN59 were obtained by using HNCO, HNCA (36), HNCACB (37), CBCACONH (38) and CC(CO)NH (39) NMR experiments recorded on 15N/13C SrtAΔN59 after complete calcium removal.
Table 1.
Structural statistics
| NMR spectroscopy data | 〈SA〉 | SABEST |
|---|---|---|
| rms deviations from NOE interproton distance restraints, Å* | ||
| All (1,852) | 0.036 ± 0.002 | 0.039 |
| Protein interresidue sequential (|i − j| = 1) (539) | 0.037 ± 0.004 | 0.045 |
| Protein interresidue short range (1 < |i − j ≤ 5) (287) | 0.047 ± 0.006 | 0.043 |
| Protein interresidue long range (|i − j| > 5) (627) | 0.037 ± 0.002 | 0.040 |
| Protein intraresidue (399) | 0.028 ± 0.009 | 0.021 |
| rms deviations from hydrogen bonding restraints, ņ (82) | 0.028 ± 0.005 | 0.039 |
| rms deviations of the dihedral angle restraints, deg‡ (195) | 0.255 ± 0.085 | 0.312 |
| rms deviations of the 3JHN coupling constants, Hz (58) | 0.524 ± 0.038 | 0.573 |
| Deviations from idealized covalent geometry | ||
| Bonds, Å | 0.004 ± 0.0001 | 0.003 |
| Angles, deg | 0.450 ± 0.012 | 0.452 |
| Impropers, deg | 0.401 ± 0.023 | 0.389 |
| Measures of structure quality | ||
| procheck-NMR (ref. 58) | ||
| Most ravored regions, % | 66.4 | |
| Additionally allowed regions, % | 31.2 | |
| Generously allowed regions, % | 2.3 | |
| Disallowed regions, % | 0.0 | |
| Coordinate precision, ŧ | ||
| Protein backbone | 0.55 ± 0.11 | |
| All protein heavy atoms | 1.12 ± 0.16 |
The notation of the NMR structures is as follows: 〈SA〉 are the final 25 simulated annealing structures; SABEST is the lowest energy structure within the ensemble. The number of terms for each restraint is given in parentheses.
None of the structures exhibited distance violations greater than 0.5 Å, dihedral angle violations greater than 5°, or 3JHNα coupling constant violations greater than 2 Hz.
Two distance restraints were employed for each hydrogen bond. Hydrogen bond restraints were added at the very end of the refinement process based an analysis of the structures and amide solvent exchange data.
195 protein dihedral angle restraints (21 χ1 angles, 1 χ2 angle, 67 ψ, 106 φ).
The coordinate precision is defined as the average atomic rms deviation of the 25 individual SA structures and their mean coordinates. The values refer to residues K62 to V161 and K175 to V205.
Results and Discussion
Conservation of the Catalytically Active Domain of Sortase.
We have used the amino acid sequence of S. aureus sortase (SrtA) for blast searches and identified sortase homologs in the unfinished genome sequences of Gram-positive bacteria (the sequences are shown in Fig. 6, which is published as supplemental data on the PNAS web site, www.pnas.org). Sortase enzymes can be divided into two classes. Class A enzymes, including S. aureus SrtA, harbor an N-terminal segment of hydrophobic amino acids that functions as both a signal peptide for secretion and a stop-transfer signal for membrane anchoring (12). Class B enzymes contain an N-terminal signal peptide as well as a C-terminal segment of hydrophobic amino acids that could serve as a membrane anchor. Thus, whereas class A sortases adopt a type II membrane topology (N terminus inside, C terminus outside the cytoplasm), class B enzymes are thought to represent type I membrane proteins (N terminus outside, C terminus inside). Signal peptide, membrane anchor, and a short linker domain of sortase enzymes (S. aureus SrtA residues 26–59) display no amino acid conservation. In contrast, the core (SrtA residues 60–206) is present in all sortase homologs examined, suggesting that this domain may comprise the catalytically active domain (supplemental Fig. 6).
Sortase Activity Is Stimulated by Calcium Ions.
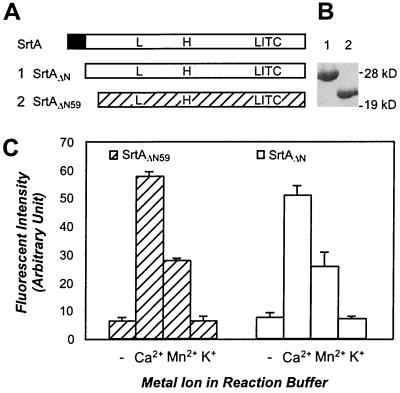
To test whether the conserved domain of sortase is catalytically active, two enzymes were purified. SrtAΔN is a soluble enzyme lacking the N-terminal signal peptide of S. aureus SrtA (residues 2–29) (13), whereas SrtAΔN59 is truncated further and contains only the presumed catalytic core of sortase (residues 60–206; Fig. 1). Enzymes were incubated with d-LPETG-e, a substrate peptide bearing the LPXTG motif tethered to a fluorophore (denoted by e) and fluorescence quencher (denoted by d) (13). Substrate cleavage was monitored as an increase in fluorescence at 495 nm, the emission wavelength of the Edans fluorophore (e) (40). SrtAΔN and SrtAΔN59 displayed similar enzyme activities, indicating that removal of nonconserved residues had no effect on cleavage of the polypeptide substrate (Fig. 1C). Both SrtAΔN and SrtAΔN59 activities were stimulated 8-fold by the addition of calcium ions to the reaction buffer (Fig. 1C). The concentration of calcium (Ca2+) required for the increase in sortase activity is about 2 mM. Mn2+ and Mg2+ can substitute in part for Ca2+, whereas the addition of several other divalent (Fe2+, Zn2+, Cd2+, and Co2+) or monovalent (K+) cations failed to stimulate peptide cleavage. The role of calcium ions in activating sortase in vivo remains to be elucidated. S. aureus encounters extracellular calcium concentrations of 1.5 mM or higher upon entry into human tissues. It is conceivable that sortase activity and the rate of surface protein anchoring to the bacterial envelope are increased during infection.
Figure 1.
Truncated sortase enzymes, SrtAΔN and SrtAΔN59, cleave LPXTG peptides and are activated by calcium ions. (A) Primary structure of wild-type sortase (SrtA), SrtAΔN (1) and SrtAΔN59 (2) and the position of the conserved leucine (L97), histidine (H120), and cysteine (C184) residues. The N-terminal membrane anchor of S. aureus SrtA is indicated as a black box. (B) Coomassie blue-stained SDS/PAGE displays the migration of purified SrtAΔN (lane 1) and SrtAΔN59 (lane 2). (C) SrtAΔN59 and SrtAΔN were incubated with d-LPETG-e peptide, and cleavage was measured as an increase in fluorescence. Addition of 2 mM Ca2+ or Mn2+ ions to the reaction buffer stimulated enzyme activity 8-fold, whereas K+ had no effect on cleavage. (− marks control.)
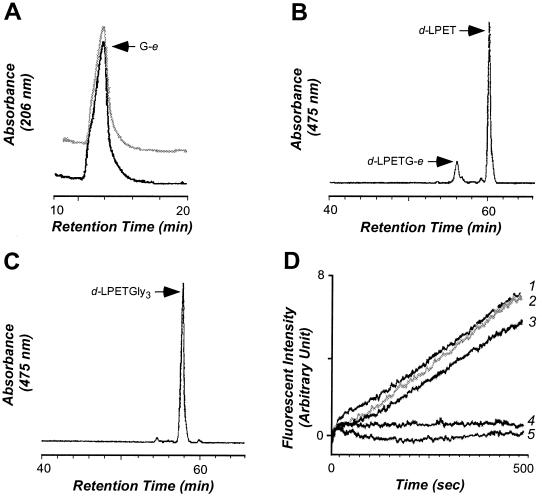
In the absence of a peptidoglycan substrate, i.e., the pentaglycine crossbridge of S. aureus muropeptides, SrtAΔN hydrolyzes the peptide bond between threonine and glycine of the LPXTG motif (14). When NH2-Gly3 is added as a substitute for peptidoglycan, sortase catalyzes a transpeptidation reaction and forms an amide bond between the carboxyl group of threonine and the amino group of NH2-Gly3 (14). To test whether SrtAΔN59 is capable of catalyzing both of these reactions, we separated enzyme substrates and products by RP-HPLC and identified peptides by electrospray ionization-MS (Fig. 2). When incubated in the presence or absence of NH2-Gly3, SrtAΔN59 cleaved d-LPETG-e and generated the G-e product, indicating that the peptide bond between threonine and glycine had been faithfully cut (Fig. 2A). In the absence of peptidoglycan substrate, SrtAΔN59 hydrolyzed d-LPETG-e at the correct peptide bond, as revealed by the appearance of the d-LPET product (Fig. 2B). In the presence of NH2-Gly3, SrtAΔN59 generated only d-LPET-Gly3 but not d-LPET (Fig. 2C). Thus, the removal of nonconserved N-terminal sequences does not alter the catalytic properties of sortase.
Figure 2.
Truncated sortase SrtAΔN59 catalyzes the transpeptidation reaction of surface protein anchoring. (A) SrtAΔN59 was incubated with the polypeptide substrate d-LPETG-e in the presence or absence of peptidoglycan substrate (NH2-Gly3), and reaction products were separated by RP-HPLC on C18 columns (see Experimental Procedures). Products that eluted at 14 min (1% acetonitrile) were analyzed by electrospray ionization-MS, revealing the sequence G-e. (B) The fluorescent product (475 nm) generated during the absence of NH2-Gly3 eluted at 61 min (76% acetonitrile) and is composed of d-LPET. (C) Fluorescent product generated in the presence of NH2-Gly3 eluted at 58 min (59% acetonitrile) with the structure d-LPET-Gly3. (D) SrtAΔN59 catalyzes the transpeptidation reaction in the presence of 5 mM NH2-Gly3 (trace 1) at a faster rate than hydrolysis of the substrate peptide d-LPETG-e in the absence of the peptidoglycan substrate (trace 3). Transpeptidation is inhibited by 5 mM MTSET, a methyl methanethiosulfonate reagent (5). As a control, we measured the rate of transpeptidation when SrtAΔN (trace 2) or no enzyme (trace 4) was added. Reaction mixtures contained 5 μM d-LPETG-e and 10 μM SrtAΔN or SrtAΔN59 in 520 μl of buffer R at 37°C.
The Structure of Sortase.
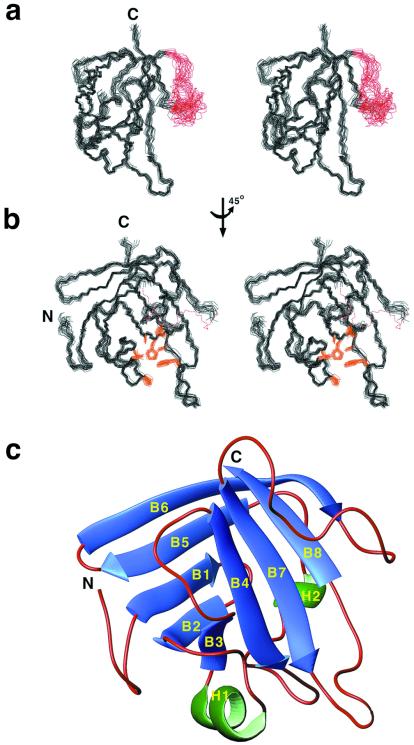
Having localized the catalytic domain of sortase to SrtAΔN59, we determined its three-dimensional structure with NMR spectroscopy (41, 42) to gain insights into the mechanism of transpeptidation. A total of 25 conformers representing the structure of SrtAΔN59 were calculated from 2,105 experimental restraints (complete statistics are presented in Table 1). Sortase possesses a heretofore unseen eight-stranded β-barrel fold that includes two short helices and several loops (Fig. 3). The structure is precisely defined by the NMR data, with the exception of a loop that connects strands β6 and β7. Several residues within this loop exhibit resonance line broadening and are either mobile or directly involved in calcium binding (described below). Strands β7 and β8 of sortase form the floor of a hydrophobic depression, with walls constructed by amino acids located in loops connecting strands β3–β4, β2–β3, β6–β7, and β7–β8 (Fig. 3c). We propose that the LPXTG motif within sorting signals of secreted proteins is cleaved by the sulfhydryl group of C184 located within this pocket. Consistent with this hypothesis is the strict conservation of H120 and C184 within the putative active site, the detrimental effects of a C184A mutation (14), and the observation that methyl methanethiosulfonate (43), a compound preferentially reactive with thiolate ions (44), abolishes sortase activity (9).
Figure 3.
SrtAΔN59 adopts a unique three-dimensional β-barrel structure. (a) Wall-eyed stereoview of the ensemble of 25 SrtAΔN59 conformers. Residues K62 to V161 and K175 to V205 (amino acid numbering is for full-length sortase) are well ordered in solution (black). The structurally disordered loop (K162–G174) connecting strands β5 and β6 is colored red. (b) Amino acids within the active site are well defined by the NMR data. The panel shows the ensemble of conformers rotated by 45° degrees with the active-site side chains colored gold (A92, P94, L97, A118, H120, I182, C184, W194). The β5–β6 loop is represented by a red dashed line. (c) Ribbon drawing of the structure of SrtAΔN59. β-strands and helices are colored blue and green, respectively. Beginning at the N terminus, the β1 strand (G74–I78) is followed by a short hairpin and lies antiparallel to strand β2 (I83–Y88). The β2 and β3 strands (V101–A104) are positioned in parallel and are tethered by a 310 helix (P94–L97) which crosses over the surface of the enzyme to form the lateral wall of the active site. Strands β3 and β4 (Q113–G119) lie antiparallel with respect to one another and are followed by a long loop and a second α-helix, which assume a circuitous path to position strand β5 (S140–V146) for antiparallel alignment with strand β1. Strand β6 (E149–K155) is then connected by a short hairpin turn for antiparallel pairing, followed by a long loop structure to connect it to strand β7 (K177–T183), which is aligned parallel to β4. The active-site sulfhydryl, C184, is positioned at the end of β7, which also includes LITC184, the signature sequence of sortase enzymes. The structure is completed by a loop (D185–W194), which connects strands β7 and β8 (E195–F200) for antiparallel pairing. Analysis with the program dali reveals no structural homologs (no structures with Z scores >2.1).
Active Site of Sortase.
The first chemical step in sortase-mediated protein attachment is the formation of a thioacyl intermediate with the concomitant cleavage of the TG peptide bond within the sorting sequence. Although structurally unrelated, our results indicate that this step is mechanistically related to the proteolytic reactions of the papain/cathepsin protein family. The active sites of these proteases contain three conserved residues, C25, H159, and N175 (45–47); before substrate binding, C25 is held in an active configuration through a thiolate–imidazolium ion interaction with H159 (48). Analogously, C184 of sortase may be activated by the imidazole ring of H120, which in all probability facilitates thiolate formation and subsequent nucleophilic attack on the carbonyl carbon at the scissile peptide bond. In the structure of sortase solved in the absence of substrate, the side chains of C184 and H120 do not interact, with C184 projecting away from H120 into the hydrophobic pocket (Fig. 4A). It would thus seem that during catalysis, substrate binding initiates a subtle conformational rearrangement in these side chains (for example, rotation of the χ1 and χ2 angles of C184 and H120, respectively), enabling sulfhydryl proton extraction and subsequent nucleophilic attack. Substrate-induced activation of sortase may be advantageous, preventing spurious proteolysis reactions without the need for more elaborate inactivation mechanisms, such as prosegment occlusion of the active site as observed in the zymogen structures of papain and cathepsin. In the cysteine proteases, a hydrogen bond between the side-chain oxygen of N175 and the Nɛ2 of H159 has been proposed to stabilize the active site (49). Analogously, N98 in sortase may stabilize the positioning of H120, because in sortase the chemical shift positions of the imidazole nitrogens of H120 indicate that it is fully protonated, and 48% of the conformers in the ensemble contain a hydrogen bond between the Nɛ2 proton of H120 to the side-chain oxygen of N98. This interaction would not seem to be essential, because N98 is poorly conserved in the sortase family, and N175 in the cysteine proteases can be replaced by glutamine or alanine without complete loss of enzymatic activity (49).
Figure 4.
The active site of SrtAΔN59 and its calcium-binding site. (A) Expanded view of the active site. As for the papain/cathepsin family, N98, H120, and C184 may form a catalytic triad that mediates the transpeptidation reaction and are positioned within a large hydrophobic pocket suitable for sorting signal binding. (B) Representative histogram of the Cα-carbon chemical shift differences between the calcium-free and -bound forms of sortase plotted as a function of residue number. These data and observed chemical shift changes in the amide nitrogen and proton atoms were used to determine the calcium-binding surface of sortase. The largest chemical shift changes occur in amino acids V168, L169, and N114; they are 2.1 (Cα), 7.3 (N), and 1.3 (HN) ppm, respectively. We estimate that calcium binds to sortase with a Kd of ≈10−4 to 10−3 M. The secondary structure of sortase is indicated. The absence of a bar indicates that the chemical shift of the α-carbon is unassigned in either the calcium-bound or calcium-free form. (C) Ribbon drawing of sortase showing the putative calcium-binding site and calcium-sensitive active-site loop. Three acidic side chains (E105, E108, D112) are poised to bind calcium as judged by localized, large-amplitude calcium-dependent changes in their chemical shifts and in surrounding amino acids. Several amino acids distal to the calcium-binding site exhibit broadening of their backbone atoms only in the presence of calcium (indicated by magenta spheres). These amino acids experience micro- to millisecond fluctuations in their magnetic environments, which presumably result either from calcium-dependent movements of the loop connecting strands β6 and β7 or from the binding of a second cation. The active-site side chains of H120, C184, and W194 are shown for reference. (D) Electrostatic surface of the SrtAΔN59 active site shown in a similar orientation as in C (acidic and basic surfaces are colored red and blue, respectively.) An active-site groove can readily accommodate a polypeptide substrate denoted by a white line. The hypothetical positioning of the sorting signal is derived by analogy with the papain/cathepsin protease family.
Sortase presumably examines newly translocated polypeptides, searching for a LPXTG motif. Once the recognition sequence is found, the polypeptide is transferred to the cell wall (3). The sagittal dimensions of the active site can readily accommodate the sorting signal, forming a groove constructed by several hydrophobic side chains (A92, L94, A118, I182, L169; Fig. 4A). By analogy to the papain/cathepsin proteins, we expect the N-terminal end of a bound peptide to lie near the beginning of strand β8, traversing over the active site, and exiting at the saddle formed by H120. After covalent linkage through a thioacyl bond to the threonine carbonyl group, the incoming amine of the cell-wall crossbridge may then be deprotonated by H120 for attack on the covalent intermediate. This hypothesis is consistent with the mechanism of the papain/cathepsin proteins, where the active-site histidine may function to protonate the leaving amine or act as a general base during the deacylation reaction (50).
Calcium Binds Near the Active Site of Sortase.
To gain insights into the molecular basis of enzymatic activation by divalent cations, we compared the NMR spectra of sortase in the presence and absence of calcium, reasoning that large amplitude changes would occur in amino acids proximal to an ion-binding site. The largest calcium-dependent chemical shift changes are localized to the backbone atoms of amino acids within the β3–β4 and β6–β7 loops (Fig. 4B). Inspection of the electrostatic surface potential of these amino acids indicates that they form a continuous acidic surface near the active site (Fig. 4 C and D). A structurally well ordered calcium-binding site seems to be formed by the side chains of E105, E108, and E112 within the β3–β4 loop and possibly by E171 from the β6–β7 loop (Fig. 4C). A second interaction surface is formed by several amino acids in the β6–β7 loop, which experience milli- to microsecond calcium-dependent modulations in their magnetic environments, as judged by their resonance line broadening only in the presence of calcium (represented as magenta spheres in Fig. 4C). These amino acids either form a second weaker ion-binding site or participate in slow-motion conformational dynamics as a result of ion binding. It should be emphasized that the NMR technique does not directly reveal the positions of calcium ions within the structure of sortase and that more quantitative methods are required to determine both the dissociation constants and stoichiometry of binding. However, our results clearly indicate that calcium binding significantly perturbs the wall of the groove leading to the active site, implying that cations activate sortase by facilitating substrate binding. The elucidation of additional structures with and without calcium, and the LPETG peptide and crossbridge substrates, will complement the results presented here and enable the development of compounds that inhibit cell-wall anchoring of surface proteins in Gram-positive bacteria. These compounds may act as antiinfectives that should be useful for the treatment or the prevention of human infections caused by Gram-positive bacteria. Such strategies are urgently needed because many infectious microbes have developed resistance to currently used antibiotics (51).
Supplementary Material
Acknowledgments
We thank Dr. R. Peterson and Mr. K. Januszyk for technical assistance. H.T.-T. is supported by the Postdoctoral Training Program in Microbial Pathogenesis, National Institute of Allergy and Infectious Diseases (Grant AI07323). This work was supported by the U.S. Public Health Service, National Institutes of Health, National Institute of Allergy and Infectious Diseases, Infectious Diseases Branch Grant AI38897 (to O.S.) and Department of Energy Grant DE-FC-03–87ER60615 (to R.T.C.).
Abbreviations
- PBP
penicillin-binding proteins
- SrtA
sortase
- d or Dabcyl
4-{[4-(dimethylamino)phenyl]azo}benzoic acid
- e or Edans
5-[(2-aminoethyl)amino]naphthalene-1-sulfonic acid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1IJA).
References
- 1.Cossart P, Lecuit M. EMBO J. 1998;17:3797–3806. doi: 10.1093/emboj/17.14.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneewind O, Model P, Fischetti V A. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 3.Schneewind O, Mihaylova-Petkov D, Model P. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarre W W, Schneewind O. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 5.Schneewind O, Fowler A, Faull K F. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 6.Ton-That H, Faull K F, Schneewind O. J Biol Chem. 1997;272:22285–22292. doi: 10.1074/jbc.272.35.22285. [DOI] [PubMed] [Google Scholar]
- 7.Higashi Y, Strominger J L, Sweeley C C. Proc Natl Acad Sci USA. 1967;57:1878–1884. doi: 10.1073/pnas.57.6.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higashi Y, Strominger J L, Sweeley C C. J Biol Chem. 1970;245:3697–3702. [PubMed] [Google Scholar]
- 9.Ton-That H, Schneewind O. J Biol Chem. 1999;274:24316–24320. doi: 10.1074/jbc.274.34.24316. [DOI] [PubMed] [Google Scholar]
- 10.Strominger J L, Izaki K, Matsuhashi M, Tipper D J. Fed Proc. 1967;26:9–18. [PubMed] [Google Scholar]
- 11.Mazmanian S K, Liu G, Ton-That H, Schneewind O. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 12.Mazmanian S K, Liu G, Jensen E R, Lenoy E, Schneewind O. Proc Natl Acad Sci USA. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. . (First published April 18, 2000; 10.1073/pnas.080520697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ton-That H, Liu G, Mazmanian S K, Faull K F, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ton-That H, Mazmanian H, Faull K F, Schneewind O. J Biol Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- 15.Mazmanian, S. K., Ton-That, H. & Schneewind, O. (2001) Mol. Microbiol., in press. [DOI] [PubMed]
- 16.Munoz E, Ghuysen J-M, Lehy-Bouille M, Petit J-F, Heymann H, Bricas E, Lefrancier P. Biochemistry. 1966;5:3748–3764. [Google Scholar]
- 17.Izaki K, Matsuhashi M, Strominger J L. Proc Natl Acad Sci USA. 1966;55:656–663. doi: 10.1073/pnas.55.3.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozarich J W, Strominger J L. J Biol Chem. 1978;253:1272–1278. [PubMed] [Google Scholar]
- 19.Rasmussen J R, Strominger J L. Proc Natl Acad Sci USA. 1978;75:84–88. doi: 10.1073/pnas.75.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghuysen J-M. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 21.Matsuhashi M, Dietrich C P, Strominger J L. Proc Natl Acad Sci USA. 1965;54:587–594. doi: 10.1073/pnas.54.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petit J-F, Munoz E, Ghuysen J M. Biochemistry. 1966;5:2764–2776. doi: 10.1021/bi00872a037. [DOI] [PubMed] [Google Scholar]
- 23.Waxman D J, Strominger J L. Annu Rev Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 24.Tipper D J, Strominger J L. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strynadka N C J, Adachi H, Jensen S E, Johns K, Sielecki A, Betzel C, Sutoh K, James M N G. Nature (London) 1992;359:700–704. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 26.Herzberg O. J Mol Biol. 1991;217:701–719. doi: 10.1016/0022-2836(91)90527-d. [DOI] [PubMed] [Google Scholar]
- 27.Gordon E, Mouz N, Duee E, Dideberg O. J Mol Biol. 2000;299:477–485. doi: 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 28.Navarre W W, Ton-That H, Faull K F, Schneewind O. J Biol Chem. 1999;274:15847–15856. doi: 10.1074/jbc.274.22.15847. [DOI] [PubMed] [Google Scholar]
- 29.Ilangovan, U., Iwahara, J., Ton-That, H., Schneewind, O. & Clubb, R. T. (2001) J. Biomol. NMR, in press. [DOI] [PubMed]
- 30.Delaglio F. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Johnson B A, Blevins R A. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 32.Vuister G W, Bax A. J Am Chem Soc. 1993;101:7772–7777. [Google Scholar]
- 33.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 34.Nilges M, Clore G M, Gronenborn A M. FEBS Lett. 1988;229:129–136. doi: 10.1016/0014-5793(88)80559-3. [DOI] [PubMed] [Google Scholar]
- 35.Brünger T A. x-plor: A System for X-ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1993. [Google Scholar]
- 36.Grzesiek S, Bax A. J Magn Reson. 1992;96:432–440. [Google Scholar]
- 37.Wittekind M, Mueller L. J Magn Reson. 1993;101:201–205. [Google Scholar]
- 38.Grzesiek S, Bax A. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 39.Grzesiek S, Anglister J, Bax A. J Magn Reson. 1993;101:114–119. [Google Scholar]
- 40.Matayoshi E D, Wang G T, Krafft G A, Erickson J. Science. 1989;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 41.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 42.Clore G M, Gronenborn A M. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 43.Smith D J, Maggio E T, Kenyon G L. Biochemistry. 1975;14:764–771. doi: 10.1021/bi00675a019. [DOI] [PubMed] [Google Scholar]
- 44.Roberts D D, Lewis S D, Ballou D P, Olson S T, Shafer J A. Biochemistry. 1986;25:5595–5601. doi: 10.1021/bi00367a038. [DOI] [PubMed] [Google Scholar]
- 45.Drenth J, Jansonius J N, KoeKoek R, Swen H M, Wolthers B G. Nature (London) 1968;218:929–932. doi: 10.1038/218929a0. [DOI] [PubMed] [Google Scholar]
- 46.Storer A C, Menard R. Methods Enzymol. 1994;244:487–500. doi: 10.1016/0076-6879(94)44035-2. [DOI] [PubMed] [Google Scholar]
- 47.Somoza J R, Zhan H, Bowman K K, Yu L, Mortara K D, Palmer J T, Clark J M, McGrath M E. Biochemistry. 2000;39:12543–12551. doi: 10.1021/bi000951p. [DOI] [PubMed] [Google Scholar]
- 48.Lewis S D, Johnson F A, Shafer J A. Biochemistry. 1981;20:48–51. doi: 10.1021/bi00504a009. [DOI] [PubMed] [Google Scholar]
- 49.Vernet T, Tessier D C, Chatellier J, Plouffe C, Lee T S, Thomas D Y, Storer A C, Menard R. J Biol Chem. 1995;270:16645–16652. doi: 10.1074/jbc.270.28.16645. [DOI] [PubMed] [Google Scholar]
- 50.Bromme D, Bonneau P R, Purisima E, Lachance P, Hajnik S, Thomas D Y, Storer A C. Biochemistry. 1996;35:3970–3979. doi: 10.1021/bi9523015. [DOI] [PubMed] [Google Scholar]
- 51.Sieradzki K, Roberts R B, Haber S W, Tomasz A. N Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski R A, Rullmann J A, MacArthur M W, Kaptein R, Thorton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.