Abstract
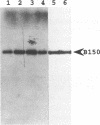
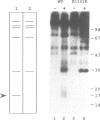
In a previous study, yeast RNA polymerase II(B) was affinity labeled with two nucleotide derivatives (III and VIII) (1). In both cases, the labeled site was localized to the C-terminal part of the B150 subunit. The potential target lysyl residues of derivative III were mapped to the conserved domain H, between Asn946 and Met999. In the present work, we have mutagenized to arginine the five lysines present in domain H. Three lysines can be replaced, individually or simultaneously, without affecting cell growth, and each mutated enzyme can still be affinity labeled. Hence one or both of the other two lysyl residues, Lys979 and Lys987, is the target of the affinity reagent. These two lysines were each found to be essential for cell viability. Derivative VIII labeled another domain in addition to domain H. Supported by analogous results obtained for E. coli RNA polymerase using derivative VIII (2), we hypothesized that the second domain labeled by this derivative in the B150 subunit was domain I. Mutagenesis of the unique lysine present in domain I demonstrated that Lys 1102 was the target of derivative VIII. These results indicate that in both prokaryotic and eukaryotic RNA polymerases, domains H and I are in close proximity and participate to the active site.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartholomew B., Dahmus M. E., Meares C. F. RNA contacts subunits IIo and IIc in HeLa RNA polymerase II transcription complexes. J Biol Chem. 1986 Oct 25;261(30):14226–14231. [PubMed] [Google Scholar]
- Coulter D. E., Greenleaf A. L. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem. 1985 Oct 25;260(24):13190–13198. [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Hartmann G. R., Maximova T. G., Mustaev A. A., Schäffner A. R., Sieber H., Zaychikov E. F. Highly selective affinity labelling of RNA polymerase B (II) from wheat germ. FEBS Lett. 1986 May 12;200(2):287–290. doi: 10.1016/0014-5793(86)81154-1. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Lukhtanov E. A., Mustaev A. A., Zaychikov E. F., Abdukayumov M. N., Rabinov I. V., Richter V. I., Skoblov Y. S., Chistyakov P. G. Studies of the functional topography of Escherichia coli RNA polymerase. A method for localization of the sites of affinity labelling. Eur J Biochem. 1989 Apr 1;180(3):577–585. doi: 10.1111/j.1432-1033.1989.tb14684.x. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlev M., Lee J., Zalenskaya K., Nikiforov V., Goldfarb A. Blocking of the initiation-to-elongation transition by a transdominant RNA polymerase mutation. Science. 1990 May 25;248(4958):1006–1009. doi: 10.1126/science.1693014. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Riva M., Carles C., Sentenac A., Grachev M. A., Mustaev A. A., Zaychikov E. F. Mapping the active site of yeast RNA polymerase B (II). J Biol Chem. 1990 Sep 25;265(27):16498–16503. [PubMed] [Google Scholar]
- Riva M., Schäffner A. R., Sentenac A., Hartmann G. R., Mustaev A. A., Zaychikov E. F., Grachev M. A. Active site labeling of the RNA polymerases A, B, and C from yeast. J Biol Chem. 1987 Oct 25;262(30):14377–14380. [PubMed] [Google Scholar]
- Ruet A., Sentenac A., Fromageot P., Winsor B., Lacroute F. A mutation of the B220 subunit gene affects the structural and functional properties of yeast RNA polymerase B in vitro. J Biol Chem. 1980 Jul 10;255(13):6450–6455. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saris C. J., van Eenbergen J., Jenks B. G., Bloemers H. P. Hydroxylamine cleavage of proteins in polyacrylamide gels. Anal Biochem. 1983 Jul 1;132(1):54–67. doi: 10.1016/0003-2697(83)90425-6. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Sentenac A., Fromageot P. Interaction of a new polypeptide with yeast RNA polymerase B. J Biol Chem. 1980 Jan 10;255(1):12–15. [PubMed] [Google Scholar]
- Scafe C., Martin C., Nonet M., Podos S., Okamura S., Young R. A. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol Cell Biol. 1990 Mar;10(3):1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich I., Riva M., Sentenac A. Zinc-binding subunits of yeast RNA polymerases. J Biol Chem. 1991 Nov 15;266(32):21971–21976. [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]