Abstract
Background
Success in further reducing the burden of cardiovascular disease (CVD) is threatened by the increasing prevalence of obesity-related atherogenic dyslipidemia. HDL-cholesterol (HDL-C) level is inversely correlated with CVD risk; each 1 mg/dl decrease in HDL-C is associated with a 6% reduction in risk. We previously showed that a common CNR1 haplotype, H3 (frequency 20%), is protective against the reduction in HDL-C that typically accompanies weight gain. In the present study, we extend that observation by reporting the effect of CNR1 haplotype on HDL-C response to modification of dietary fat intake in weight maintenance and weight loss.
Methods
Six haplotype tagging SNPs that cover the CNR1 gene locus were genotyped in 590 adults of varying body mass index (cohort 1 is 411 males with BMI 18.5–30.0 kg/m2; cohort 2 is 71 females with BMI18.5–30.0 kg/m2; and cohort 3 is 108 females with BMI 30–39.9 kg/m2). Dietary intakes were modified so that fat intake in the “high fat” condition was 15–20% greater than in the “low fat” condition, and lipid profiles were compared between carriers versus noncarriers for each of the five commonly observed CNR1 haplotypes (H1–H5).
Results
In normal to overweight subjects on eucaloric diets, the H3 haplotype was significantly associated with short-term high fat diet induced changes in HDL-C level in females (carriers 5.9 mg/dl>noncarriers, p = 0.007). The H3 haplotype was also significantly associated with HDL-C level after 16 weeks on high fat calorie restricted diet in obese females (carriers 6.8 mg/dl>noncarriers, p = 0.009).
Conclusion
Variability within the CNR1 gene locus contributes to gender-related differences in the HDL-cholesterol response to change in dietary fat intake. Functional characterization of this relationship in vitro may offer insights that potentially yield therapeutic guidance targeting dietary macronutrient composition, a direction much needed in the current epidemic of obesity.
Introduction
Obesity is associated with an atherogenic lipoprotein profile characterized by increased very low-density lipoprotein triglycerides (VLDL-TG), small dense low density lipoprotein cholesterol (LDL-C), and decreased circulating high density lipoprotein cholesterol (HDL-C) [1]. The anti-inflammatory properties of HDL-C and its role in reverse cholesterol transport make it a significant modulator of risk for the development of cardiovascular disease (CVD). As event rate is inversely correlated with HDL-C concentration, each 1 mg/dl decrease in HDL-C is associated with a 6% reduction in CVD risk [2]. Decreased HDL-C is especially important in the current epidemic of obesity as higher body mass index (BMI) is associated with lower HDL-C levels in all racial/ethnic groups [3]. Notably, the prevalence of subthreshold (“at risk”) HDL-C levels has improved three-fold more in men than women (↓5.6% versus ↓1.8%) over the past 30 years [4]. In fact, the strongest clinically-relevant demographic predictors of plasma HDL-C concentration are BMI and gender [5], [6].
While modest weight loss of 5–10% improves plasma lipoprotein profile [7] and modifying dietary macronutrient intake can increase HDL-C level [8], scientific and clinical debate continues regarding the importance of the relative amount of fat in a calorically restricted diet [9], [10]. Low-fat (LF) diets can increase circulating VLDL-TG levels, reduce HDL-C, and thus, adversely affect atherogenic dyslipidemia [11], [12]. Although meta-analyses show greater improvements in HDL-C on high fat (HF) diets [13], [14], some studies yield conflicting results [15], [16]. Overall, the lipoprotein response to dietary macronutrient composition is a highly variable and complex trait [11], influenced by BMI [17], gender [18], and individual genetic characteristics [19], [20].
Preclinical and clinical studies targeting the endocannabinoid system (ECS) reveal potent effects on body weight, body fat distribution, and lipoprotein metabolism. The ECS consists primarily of two phospholipid-derived signaling molecules, N-arachidonoylethanolamide (anandamide) and 2-arachidonoylglycerol (2-AG), and two G protein-coupled receptors, CB1 and CB2 [21]. Most of the metabolic effects of ECS signaling appear to be mediated through CB1 receptors in central and peripheral tissues. In randomized clinical trials, CB1 receptor blockade resulted in weight loss, with increased HDL-C levels that exceeded the magnitude of change expected from weight loss alone [22], [23]. Collectively, these observations suggest that the ECS regulates lipid homeostasis via peripheral mechanisms that are at least partially independent of weight change.
Consistent with this hypothesis, we previously reported that sequence variation in CB1 (gene name CNR1) is associated with variability in circulating HDL-C levels independent of BMI [24], [25]. Importantly, a common CNR1 haplotype, H3, was further protective against the marked reduction in HDL-C that occurs in severe obesity (BMI≥40 kg/m2) [25]. However, it remains unknown whether the CNR1 H3 haplotype influences the effect of dietary fat intake on lipoprotein profiles. We tested this question in adults across a range of BMI who were prescribed various percentages of dietary fat intakes. Herein, we provide evidence that the response of HDL-C to increased dietary fat intake during weight maintenance is strongly influenced by the CNR1 H3 haplotype in women. We also report that this same H3 haplotype may protect women against low circulating HDL-C during voluntary weight loss.
Methods
Three distinct cohorts comprising 590 adults who participated in clinical trials designed to modify the percentage of dietary fat consumed were studied. Cohort #1 (normal to overweight males) and cohort #2 (normal to overweight females) were convenience samples from prior trials using eucaloric diets designed for weight maintenance. Because we had previously found an association between CNR1 and HDL-C in a population sample that was predominantly female [26], [27], we enrolled more males (cohort #1) than females (cohort #2) in the present study (at a ratio of 4∶1) to assure that we would not be missing a significant relationship in males. As the strength of the genotype-phenotype associations in cohorts #1 & #2 were consistent with our prior work, cohort #3 consisted of a sample of obese females on a calorically restricted diet, which not only broadened our range of BMI in females to better match current population demographics, but also allowed investigation of the effects of dietary fat intake within the context of weight loss.
For all subjects, the amount of fat prescribed in the “high fat” (HF) conditions were 15% higher than in the “low fat” (LF) conditions. Dietary intakes were assessed by Registered Dietitians (RDs) from 4-day food diaries and 24-hour diet recalls using multi-pass methodology [28]. Energy and macronutrient intake analysis was performed using Nutrition Data System for Research (NDS, Nutrition Coordinating Center, University of Minnesota, Minneapolis). Written informed consent was obtained and study protocols were approved by the Institutional Review Boards of Children's Hospital and Research Center of Oakland and the E.O. Lawrence Berkeley National Laboratory at the University of California for cohorts #1 & #2 and the Vanderbilt University Medical Center for cohort #3.
Cohort #1: Males on Eucaloric Diet
Males age 21–60 years with BMI 18.9–30 kg/m2 who had participated in one of four trials [26], [29], [30], [31], [32] were included if they had no active chronic disease, did not take lipid altering medications, and were nonsmokers. Males were randomly assigned in a crossover design to eucaloric HF diet (prescribed as 35% fat [∼18% saturated, 11% monounsaturated, 6% polyunsaturated], 50% carbohydrate, 15% protein) and LF diet (prescribed as 20% fat [∼5% saturated, 11% monounsaturated, 4% polyunsaturated], 65% carbohydrate [half simple and half complex], 15% protein) for 6 weeks each. Cholesterol content of the prescribed diets was 125 mg/1000 kcal and fiber content was 5 g/1000 kcal. Subjects maintained habitual physical activity and weighed themselves daily. RDs adjusted menu energy content when necessary to maintain weight. Diet compliance was assessed from 4-day weighed food intake records and daily recording of menu items consumed. Height, weight and venous blood samples were collected at baseline and end of study in an overnight fasted state. Plasma total cholesterol and triglyceride levels were determined by enzymatic procedures on a Gilford Impact 400E or Express 550 Plus analyzer (Ciba Corning, Oberlin, OH). HDL-C was measured after dextran sulfate precipitation of plasma and LDL-C was calculated from the Friedewald formula [33].
Cohort #2: Females on Eucaloric Diet
Premenopausal females age 21–50 years with BMI 18.9–30 kg/m2 were included if they had no active chronic disease, did not take lipid altering medications, were nonsmokers, and were not pregnant, breastfeeding or taking oral contraceptives [27]. Although not randomly crossed over, females were prescribed similar eucaloric HF and LF diets as the males in cohort #1 (above) for 8 week periods. The same procedures as described for cohort #1 were followed for compliance and outcome measurements.
Cohort #3: Females on Calorie Restricted Diet
Premenopausal females age 21–50 years with BMI 30–39.9 kg/m2 were included if they had no active chronic disease, did not take medications for lipid metabolism, were nonsmokers, and were not pregnant, breastfeeding or taking oral contraceptives. After habituation on the lower fat diet condition (35% fat), subjects were prescribed a 15% calorically restricted HF diet (50% fat [1/3 saturated, 1/3 monounsaturated, 1/3 polyunsaturated], 30% carbohydrate [half simple and half complex], 20% protein) for 16 weeks. Cholesterol content of the HF diet was 150 mg/1000 kcal and fiber content was 7 g/1000 kcal. Standard assays were performed for triglyceride, total cholesterol, LDL-C and HDL-C by selective enzymatic hydrolysis at the Vanderbilt Department of Pathology Clinical Laboratory. Serum glucose was quantified by colorimetric timed endpoint method and serum insulin by chemiluminescent immunoassay. The homeostasis model assessment of insulin resistance (HOMA-IR) was calculated as [fasting glucose (mM) x fasting insulin (mU/L)]/22.5 [34].
Genotyping
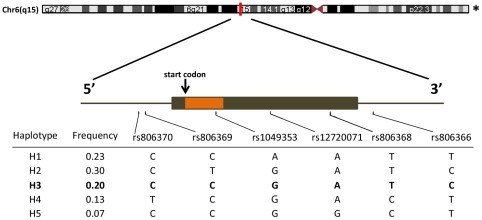
Genomic DNA was extracted from whole blood according to standard procedures. Based on our prior work [24], [25], we tagged the entire CNR1 gene plus 5 kilobases (kb) upstream and 5 kb downstream using Haploview (http://www.broad.mit.edu/mpg/haploview/) for linkage disequilibrium analysis [35] and selection of tagSNPs. Gene locus, haplotype structure and allele frequencies of the tagSNPs covering the variability within our locus of interest (Chromosome 6, physical position 88, 901, 306–88, 916, 775) are shown in Figure 1. These six tagSNPs include two promoter SNPs (rs806370, rs806369), one coding SNP (rs1049353), two SNPs in the 3′ untranslated region (3′-UTR: rs12720071, rs806368), and one SNP distal to the polyA tail (rs806366). The tagSNPs were genotyped for cohorts #1 & #2 using multiplex polymerase chain-reaction (PCR), single base primer extension (SBE), and generation of mass spectra (Sequenom, San Diego, CA). For cohort #3, genotyping was conducted using commercial Taqman Allelic discrimination assays (Applied Biosystems, Inc., Foster City, CA) [36]. Each assay contained two Taqman probes uniquely labeled to bind separate (major versus minor) alleles.
Figure 1. The CNR1 gene: chromosomal location, tagSNPs, haplotype structure and alleles.
*The CNR1 gene locus covers ∼15 kb on Chromosome 6 (chromosome 6 is shown in the reverse orientation, with arm q on the left). Six haplotype tagging SNPs were genotyped, based upon our prior work [25], and reference sequence numbers for the tag SNPs are presented. These SNPs are inherited in combination (haplotypes).
Statistical Analysis
Statistical analyses were conducted using SPSS version 17 (SPSS Inc., Chicago, IL), JMP version 7.0 (SAS Institute Inc., Cary, NC) and PLINK genetic analyses toolset version 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink) [37]. Data are expressed as means and standard deviations unless otherwise noted. Baseline lipid variables among cohorts were assessed using the Kruskal-Wallis test and relationships between demographics and lipids were examined using Spearman Rho correlations. Minor allele frequency and Hardy-Weinberg (H-W) equilibrium statistics were calculated for each SNP. As we have previously demonstrated that haplotypes provide greater power than single SNPs for identifying associations related to CNR1 [25], we performed haplotype analyses based on a dominant model (carrier versus noncarrier). Five CNR1 haplotypes (Figure 1) were identified at frequency over 5%. From 5′ to 3′, they are CCAATT (23%), CTGATC (30%), CCGATC (20%), TCGACT (13%), CCGGCT (7%). These five are the most common CNR1 haplotypes found in the Centre d'Etude du Polymorphisme Humain (CEPH) cohort within the International HapMap Consortium and in our prior cohorts [24]. Although the tagging SNPs were, by definition, in tight linkage dysequilibrium, we corrected for multiple hypothesis testing of the five haplotypes [38] and considered results statistically significant only with p value<0.01.
For cohorts #1 and #2, the five CNR1 haplotypes were estimated using PHASE software. As expected, the same five haplotypes were observed in cohort #3. However, the greater racial diversity of this cohort introduced more structural variability at the CNR1 gene locus, and thus, we identified two novel CNR1 haplotypes (CCGGTT, frequency 0.7%; and CCGATT, frequency 2%) in the African American (AA) females (data not presented). Importantly, these two new haplotypes were not significantly associated with HDL-C levels. For cohort #3, haplotypes were estimated by applying standard E-M algorithms located in the –hap option of PLINK.
All genotype-phenotype association analyses were performed using the five CNR1 haplotypes. In cohorts #1 and #2, linear regression models were created to examine relationships between haplotypes and lipid variables. Data were adjusted for age and baseline BMI. To be consistent with this approach, for cohort #3 a linear regression model was applied to test each haplotype (versus all other haplotypes) using the –hap-linear option in PLINK. For cohort #3, which was obese by definition, baseline BMI and HOMA-IR were significantly associated with baseline serum HDL-C, and thus, included as covariates in the model using the –covar option. This approach allowed us to replicate our initial findings from cohorts #1  in a second independent cohort (cohort #3) to reduce the likelihood of a spurious association between CNR1 and HDL-C levels.
Results
Subject Characteristics
The combined sample included 411 males and 179 females with a mean age of 39.9±4.0 years (Table 1). As defined by eligibility criteria, subjects in cohorts #1 & #2 were normal to overweight (mean BMI 24.8±1.7 kg/m2), while cohort #3 subjects were obese (mean BMI 34.7±2.7 kg/m2). Ninety percent of subjects in cohorts #1 & #2 self-identified as being of European ancestry (EA). In cohort #3, 75 (69%) females self-identified as EA and 33 (31%) as AA. Baseline TG, total cholesterol, LDL-C and HDL-C were similar across cohorts. There were no significant associations between minor allele frequency and diet adherence (data not presented).
Table 1. Baseline characteristics of study subjects (n = 590).
| Characteristics | Cohort #1 | Cohort #2 | Cohort #3 |
| Sample Size | 411 | 71 | 108 |
| Male/Female | Male | Female | Female |
| Age, mean (SD), y | 45.0 (11.5) | 38.2 (6.5) | 37.5 (0.7) |
| BMI*, mean (SD) | 25.4 (3.0) | 23.2 (3.1) | 36.7 (1.1) |
| Total cholesterol, mean (SD), mg/dL | 197.9 (35.1) | 186.3 (31.0) | 168.1 (29.3) |
| Triglycerides, mean (SD), mg/dL | 129.6 (77.4) | 80.7 (44.2) | 85.1 (48.2) |
| HDL-C, mean (SD), mg/dL | 46.6 (12.2) | 60.3 (11.3) | 48.9 (12.8) |
| LDL-C, mean (SD), mg/dL | 125.7 (30.4) | 109.8 (29.1) | 102.1 (25.0) |
Abbreviations: SD, standard deviation; BMI, body mass index; HDL-C, high density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol.
Calculated as weight in kilograms divided by height in meters squared.
Cohort #1
As expected with eucaloric diets, no significant changes occurred in body weight in cohort #1 subjects. In these normal to overweight males, HF diet significantly decreased TG (−30.2±63.6 mg/dl, p<0.001), increased total cholesterol (+12.0±21.9 mg/dl, p<0.001), increased LDL-C (+11.5±20.6 mg/dl, p<0.001) and increased HDL-C (+6.4±5.7 mg/dl, p<0.001). Consistent with previous report in a predominantly female cohort [24], the reduction in TG was significantly greater in male noncarriers of the CNR1 H4 haplotype compared to male carriers (at least one copy), p = 0.005. Although prior work also indicated that the CNR1 H3 haplotype was associated with HDL-C in both genders, in the present study H3 was not significantly associated with HDL-C response to high fat diet in normal to overweight males (Table 2); male carriers of the H3 haplotype had only a 1.3 mg/dl higher HDL-C level (from 40.5±9.2 to 47.8±11.4 mg/dl) than male noncarriers (from 40.1±9.4 to 46.0±11.5 mg/dl).
Table 2. Effect of haplotype on triglycerides, LDL-cholesterol and HDL-cholesterol in 411 normal to overweight males (Cohort #1) on eucaloric diets.
| Triglyceride, mg/dl (SD) | LDL Cholesterol, mg/dl (SD) | HDL Cholesterol, mg/dl (SD) | |||||||
| Haplotype | Non-Carrier | Carrier | P-Value | Non-Carrier | Carrier | P-Value | Non-Carrier | Carrier | P-Value |
| H1 | |||||||||
| Sample Size | 241 | 170 | 241 | 170 | 241 | 170 | |||
| LF | 137.4 (100.2) | 145.5 (91.4) | 0.81 | 118.0 (32.7) | 109.6 (30.8) | 0.007 | 40.9 (9.0) | 39.2 (9.7) | 0.29 |
| HF | 112.3 (99.7) | 108.2 (72.9) | 0.19 | 129.3 (35.1) | 121.4 (34.8) | 0.02 | 47.2 (10.9) | 45.7 (12.3) | 0.65 |
| Difference | −25.1 (61.7) | −37.4 (65.7) | 0.09 | 11.3 (19.5) | 11.8 (22.2) | 0.72 | 6.3 (5.5) | 6.5 (6.0) | 0.45 |
| H2 | |||||||||
| Sample Size | 191 | 220 | 191 | 220 | 191 | 220 | |||
| LF | 146.3 (107.3) | 135.9 (86.3) | 0.67 | 112.7 (32.4) | 116.1 (32.0) | 0.19 | 39.7 (9.2) | 40.6 (9.4) | 0.51 |
| HF | 121.0 (117.2) | 101.6 (54.0) | 0.21 | 122.7 (34.1) | 128.9 (35.8) | 0.04 | 46.0 (11.8) | 47.1 (11.2) | 0.50 |
| Difference | −25.3 (67.9) | −34.3 (59.4) | 0.20 | 10.0 (21.1) | 12.8 (20.2) | 0.10 | 6.3 (5.8) | 6.5 (5.6) | 0.77 |
| H3 | |||||||||
| Sample Size | 284 | 127 | 284 | 127 | 284 | 127 | |||
| LF | 143.6 (95.6) | 134.5 (99.0) | 0.14 | 113.8 (31.1) | 116.2 (34.5) | 0.60 | 40.1 (9.4) | 40.5 (9.2) | 0.77 |
| HF | 111.6 (94.6) | 108.3 (77.5) | 0.42 | 126.4 (34.3) | 125.2 (37.1) | 0.56 | 46 (11.5) | 47.8 (11.4) | 0.16 |
| Difference | −31.9 (65.0) | −26.2 (60.4) | 0.58 | 12.6 (20.5) | 9.0 (20.7) | 0.05 | 6.0 (5.7) | 7.3 (5.6) | 0.03 |
| H4 | |||||||||
| Sample Size | 303 | 108 | 303 | 108 | 303 | 108 | |||
| LF | 139.9 (91.4) | 143.2 (110.4) | 0.97 | 114.4 (32.8) | 115.0 (30.3) | 0.65 | 39.8 (9.4) | 41.2 (9.3) | 0.20 |
| HF | 104.6 (62.2) | 127.4 (139.9) | 0.11 | 126.9 (35.8) | 123.7 (33.2) | 0.13 | 46.4 (11.2) | 47.0 (12.2) | 0.76 |
| Difference | −35.3 (60.4) | −15.8 (70.2) | 0.005 | 12.5 (21.0) | 8.7 (19.5) | 0.06 | 6.6 (5.6) | 5.8 (6.0) | 0.17 |
| H5 | |||||||||
| Sample Size | 328 | 63 | 328 | 63 | 328 | 63 | |||
| LF | 142.0 (98.2) | 133.7 (88.2) | 0.60 | 113.7 (31.7) | 119.2 (34.2) | 0.22 | 40.2 (9.5) | 40.2 (8.6) | 0.56 |
| HF | 112.7 (94.1) | 98.8 (57.7) | 0.49 | 124.9 (35.1) | 132.3 (35.2) | 0.14 | 46.7 (11.6) | 46.0 (10.8) | 0.27 |
| Difference | −29.3 (65.7) | −34.9 (50.4) | 0.44 | 11.2 (20.4) | 13.1 (21.9) | 0.56 | 6.5 (5.7) | 5.7 (5.4) | 0.23 |
Abbreviations: LF, low fat diet; HF, high fat diet.
P-values are presented after adjusting for age and baseline BMI; statistical significance level established at α = 0.01.
Cohort #2
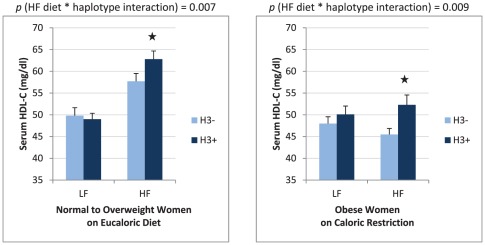
Like the males in cohort #1, females in cohort #2 experienced no significant changes in body weight. In this cohort, HF diet significantly decreased TG (−17.5±34.1 mg/dl, p<0.001), increased total cholesterol (+12.2±19.8 mg/dl, p<0.001), increased LDL-C (+4.8±17.0 mg/dl, p = 0.02) and increased HDL-C (+10.9±8.7 mg/dl, p<0.001). In contrast to the males in cohort #1, the CNR1 H3 haplotype was strongly associated (p = 0.007) with high fat diet induced changes in serum HDL-C levels in these normal to overweight females (Table 3). The increase in HDL-C level after HF diet in normal to overweight female carriers of the H3 haplotype (from 49±8 to 62.8±11.2 mg/dl) was 5.9 mg/dl greater than the increase in noncarriers (from 49.8±10.9 to 57.7±10.9 mg/dl) (Figure 2a).
Table 3. Effect of haplotype on triglycerides, LDL-cholesterol and HDL-cholesterol in 71 normal to overweight females (cohort #2) on eucaloric diet.
| Triglyceride, mg/dl (SD) | LDL Cholesterol, mg/dl (SD) | HDL Cholesterol, mg/dl (SD) | |||||||
| Haplotype | Non-Carrier | Carrier | P-Value | Non-Carrier | Carrier | P-Value | Non-Carrier | Carrier | P-Value |
| H1 | |||||||||
| Sample Size | 42 | 29 | 42 | 29 | 42 | 29 | |||
| LF | 93.5 (47.5) | 105.0 (47.4) | 0.18 | 104.5 (25.5) | 105.8 (39.7) | 0.91 | 48.3 (9.8) | 50.9 (8.8) | 0.26 |
| HF | 74.1 (37.1) | 90.2 (52.0) | 0.08 | 110.5 (28.3) | 108.7 (30.6) | 0.82 | 60.5 (11.5) | 59.9 (11.2) | 0.74 |
| Difference | −19.4 (32.7) | −14.8 (36.3) | 0.53 | 6.1 (15.7) | 2.9 (18.8) | 0.56 | 12.2 (8.1) | 9.0 (9.2) | 0.14 |
| H2 | |||||||||
| Sample Size | 42 | 29 | 42 | 29 | 42 | 29 | |||
| LF | 100.9 (54.1) | 94.3 (36.4) | 0.75 | 104 (34.5) | 106.5 (28.0) | 0.71 | 49.5 (8.6) | 49.2 (10.8) | 0.89 |
| HF | 86.5 (51.9) | 72.3 (28.4) | 0.31 | 108.7 (28.5) | 111.4 (30.4) | 0.68 | 59.8 (10.1) | 61 (12.9) | 0.68 |
| Difference | −14.4 (35.5) | −22.0 (32.0) | 0.37 | 4.7 (15.0) | 4.9 (19.8) | 0.97 | 10.3 (7.7) | 11.7 (10.0) | 0.52 |
| H3 | |||||||||
| Sample Size | 35 | 36 | 35 | 36 | 35 | 36 | |||
| LF | 107.3 (54.9) | 89.4 (37.7) | 0.61 | 108.1 (37.7) | 102 (25.0) | 0.93 | 49.8 (10.9) | 49.0 (8.0) | 0.13 |
| HF | 88.9 (53.2) | 72.7 (32.0) | 0.95 | 108.9 (32.4) | 110.6 (25.9) | 0.42 | 57.7 (10.9) | 62.8 (11.2) | 0.33 |
| Difference | −18.4 (32.1) | −16.7 (36.3) | 0.60 | 0.8 (19.2) | 8.6 (13.8) | 0.11 | 7.9 (6.2) | 13.8 (9.7) | 0.007 |
| H4 | |||||||||
| Sample Size | 51 | 20 | 51 | 20 | 51 | 20 | |||
| LF | 91.0 (35.2) | 116.5 (67.5) | 0.81 | 101.5 (26.7) | 113.9 (41.7) | 0.55 | 50.1 (9.3) | 47.6 (9.8) | 0.78 |
| HF | 71.2 (27.5) | 104.9 (65.9) | 0.58 | 106.8 (26.1) | 117.4 (35.2) | 0.44 | 62 (11.6) | 55.8 (9.4) | 0.37 |
| Difference | −19.8 (33.2) | −11.7 (36.3) | 0.57 | 5.3 (15.2) | 3.5 (21.3) | 0.77 | 12 (9.7) | 8.2 (4.3) | 0.20 |
| H5 | |||||||||
| Sample Size | 68 | 3 | 68 | 3 | 68 | 3 | |||
| LF | 97.6 (47.3) | 112.7 (60.9) | 0.33 | 105.3 (31.2) | 98 (52.4) | 0.97 | 49.4 (9.5) | 48.3 (10.1) | 0.54 |
| HF | 81.1 (45.0) | 71.7 (14.4) | 0.61 | 110.2 (29.0) | 101.7 (37.1) | 0.78 | 60.4 (11.4) | 57.7 (9.0) | 0.36 |
| Difference | −16.5 (33.5) | −41.0 (47.1) | 0.25 | 4.8 (17.2) | 3.7 (15.5) | 0.58 | 11.0 (8.9) | 9.3 (1.2) | 0.68 |
Abbreviations: LF, low fat diet; HF, high fat diet.
P-values are presented after adjusting for age and baseline BMI; statistical significance level established at α = 0.01.
Figure 2. Effect of H3 haplotype on serum HDL-cholesterol levels in women.
Values shown are mean+standard error. In normal to overweight females, changing from low fat (LF) to high fat (HF) diet increased HDL-C from 49.0±8.0 mg/dl to 62.8±11.2 mg/dl in H3 haplotype carriers and from 49.8±10.9 mg/dl to 57.7±10.9 mg/dl in H3 non-carriers. In obese females, calorically restricted (−15% kcal) HF diet increased HDL-C from 50.11±11.60 mg/dl to 52.29±10.38 mg/dl in H3 haplotype carriers and decreased HDL-C from 47.99±12.90 mg/dl to 45.49±8.95 mg/dl in H3 non-carriers.
Cohort #3
The obese females experienced a significant weight loss of 6.2±3.5 kg over the 16-week intervention (p<0.001) due to the 15% caloric restriction prescribed in the HF diet condition. Overall, cohort #3 experienced significantly decreased BMI (−2.5±1.5 units, p<0.001), total cholesterol (−9.6±19.2 mg/dl, p<0.001), LDL-C (−8.0±17.6 mg/dl, p<0.001), plasma insulin (−2.03±4.79 mg/dl, p = 0.001), and had non-significant changes in TG (−5.58±33.6 mg/dl, p = 0.178), HDL-C (0.30±7.90 mg/dl, p = 0.758), and plasma glucose (−1.16±10.3 mg/dl, p = 0.360). As with normal to overweight females, the CNR1 H3 haplotype was strongly associated with serum HDL-C levels in obese females, p = 0.009 (Table 4). After 16 weeks of high fat diet, obese female carriers of the H3 haplotype had significantly higher HDL-C levels (52.3±2.3 mg/dl) than noncarriers (45.5±1.4 mg/dl) (Figure 2b). In re-testing the EA obese females alone, the H3 haplotype remained significantly associated (p = 0.005) with HDL-C level, and thus, the relationship was independent of race. The relationship between H3 haplotype and HDL-C was also independent of circulating TG level (p = 0.06 for the association between H3 haplotype and TG/HDL-C ratio).
Table 4. Effect of haplotype on triglycerides, LDL-cholesterol and HDL-cholesterol in 108 obese females (cohort #3) on calorie restricted diet.a .
| Triglyceride, mg/dl (SD) | LDL Cholesterol, mg/dl (SD) | HDL Cholesterol, mg/dl (SD) | ||||||||
| Haplotype | Sample Sizeb | Non-Carrier | Carrier | P-Valuec | Non-Carrier | Carrier | P-Valueb | Non-Carrier | Carrier | P-Valuec |
| H1 | ||||||||||
| LF | 66/40 | 78.6 (42.6) | 96.8 (55.8) | 0.06 | 98.7 (23.8) | 106.6 (25.9) | 0.11 | 50.3 (11.8) | 46.2 (10.8) | 0.10 |
| HF | 39/25 | 75.6 (39.8) | 78.2 (39.7) | 0.80 | 98.8 (21.6) | 94.7 (23.7) | 0.47 | 49.1 (10.0) | 45.6 (9.6) | 0.17 |
| H2 | ||||||||||
| LF | 54/52 | 82.5 (51.7) | 88.6 (45.4) | 0.52 | 98.9 (25.1) | 104.6 (24.4) | 0.24 | 49.8 (11.9) | 47.6 (13.1) | 0.38 |
| HF | 31/33 | 68.1 (37.8) | 84.6 (39.9) | 0.09 | 93.3 (23.2) | 100.6 (21.4) | 0.18 | 48.9 (9.8) | 46.6 (10.0) | 0.37 |
| H3 | ||||||||||
| LF | 69/37 | 89.2 (53.3) | 78.5 (37.8) | 0.28 | 102.6 (24.5) | 99.9 (25.6) | 0.60 | 48.0 (12.9) | 50.1 (11.6) | 0.41 |
| HF | 43/21 | 79.6 (41.2) | 70.5 (35.9) | 0.39 | 100.8 (20.6) | 89.3 (24.6) | 0.05 | 45.5 (8.9) | 52.3 (10.4) | 0.009 |
| H4 | ||||||||||
| LF | 86/20 | 86.4 (50.2) | 81.4 (41.7) | 0.68 | 103.7 (25.0) | 93.1 (22.6) | 0.08 | 48.9 (12.6) | 48.1 (12.1) | 0.80 |
| HF | 52/12 | 76.9 (40.3) | 75.3 (37.5) | 0.90 | 97.2 (22.8) | 97.1 (21.4) | 0.99 | 48.8 (10.4) | 43.2 (6.0) | 0.08 |
| H5 | ||||||||||
| LF | 96/10 | 87.3 (50.0) | 68.0 (27.3) | 0.23 | 102.1 (25.1) | 97.9 (22.4) | 0.61 | 48.9 (11.8) | 47.1 (18.0) | 0.67 |
| HF | 58/6 | 75.6 (38.2) | 86.5 (53.7) | 0.52 | 96.7 (22.9) | 102.2 (17.6) | 0.58 | 47.9 (9.8) | 46.2 (11.9) | 0.69 |
Abbreviations: LF, low fat diet; HF, high fat diet; TG, triglycerides; HDL-C, high density lipoprotein cholesterol.
Number of Non Carriers/number of Carriers.
Statistical significance set at α = 0.01.
Subjects were prescribed high fat diet within the context of caloric restriction for weight loss, however, the effects observed between haplotype and HDL-C was independent of weight loss.
Discussion
The present findings demonstrate that variability in the CNR1 gene influences HDL-C response to changes in dietary fat intake. In normal to overweight adults on eucaloric diets, subjects expressing the CNR1 H3 haplotype had a more robust increase in HDL-C level during high fat intake and the magnitude of the effect of the H3 haplotype on HDL-C was four-fold greater in females compared to males. We observed a similar response to high fat intake in females with class I and II obesity who were prescribed caloric restriction. After 16 weeks of high fat diet, obese female carriers of the H3 haplotype had HDL-C levels that were ∼7 mg/dl greater than noncarriers (52.3±2.3 mg/dl in carriers versus 45.5±1.4 mg/dl in noncarriers, p = 0.009). The current findings are consistent with prior evidence that the CNR1 H3 haplotype was strongly associated with HDL-C levels in individuals with class III obesity [25].
It is important to recognize that the frequency of the H3 haplotype does not differ by gender and is common (20% frequency) in the general population. Given that women disproportionately seek dietary approaches for weight loss [39], along with the increasing recognition that women are at significant risk for cardiovascular disease - particularly in the current obesogenic environment, it is imperative to understand factors that determine favorable lipoprotein responses to dietary weight loss interventions. Prior investigation indicates that women may be more sensitive to diet induced changes in HDL-C during weight loss than men [40]. For example, a recent comparison of popular weight loss diets showed women had markedly increased HDL-C on 50% dietary fat intake (Atkins diet: ∼5 mg/dl increase in HDL-C), moderately increased HDL-C on 30% fat intake (Zone and Learn diets: ∼2.5 mg/dl increase in HDL-C) and no change in HDL-C on 10% fat intake (Ornish diet) [41]. The gender distinction observed in HDL-C response to popular weight loss diets may in part be attributable to underlying variability in the CNR1 gene.
The specific physiological mechanism linking the CNR1 gene product to lipid homeostasis is not completely understood. The CB1 receptor modulates food intake through an effect on central neuronal circuits [42]. Central CB1 receptor signaling is also known to directly influence peripheral metabolic processes involved in VLDL-TG secretion and clearance [43], [44]. Although changes in the abundance of VLDL could alter circulating HDL-C levels, by shifting the flux of free fatty acids between larger and smaller lipoproteins [45], our data do not support such a conclusion. In the present study, it appears that CNR1 modulated HDL-C response to high fat diet independent of BMI and fasting TG level. Peripherally, the CB1 receptor is expressed in the liver [46], pancreas, gastrointestinal tract [47] and adipose tissue [48]. HDL particle composition, and therefore HDL clearance rate, is modified within these tissues through the action of lipolytic enzymes [49]. Studies conducted using conditional (hepatic) CB1 knockout mice have shown that the CB1 receptor directly regulates hepatic lipid metabolism [50]. Lipoprotein lipase (LPL) and hepatic lipase (HL) convert larger HDL particles into smaller HDL remnants, resulting in HDL particles that are cleared more rapidly from the circulation. Endothelial lipase (EL) is also a strong predictor of circulating HDL-C concentration [51], [52], and EL expression is modulated by adipocytokines [53]. Because CB1 antagonists modulate the synthesis and secretion of adiponectin [54] and pro-inflammatory cytokines like TNFα [55], [56], genetic variability in CNR1 may be affecting HDL-C level through a mechanism involving its intravascular modeling. While our study was limited to assessment of HDL-C concentration, future investigation is needed to address more mechanistic hypotheses regarding the distribution of particle subfraction and changes in inflammatory biomarkers within the HDL proteome. In addition, large datasets are needed to condition the genetic effect of CNR1 on LPL, HL and EL, as well as known functional variants in the ATP-binding cassette transporter A1 (ABCA1), cholesteryl ester transfer protein (CETP), and scavenger receptor class B type 1 (SRB1). While all are known to influence HDL-C level [5], [51], [57], combinatorial models assessing interactions [58] will require targeted genotyping within cohorts of enormous size.
As these studies proceed, it is important to recognize that the causative allele linking CNR1 to HDL-C level remains undetermined. It is likely that the H3 haplotype examined in the present study is only in partial linkage (ie, D′<1.0) with the actual functional allele, a group of common and rare variants with the capacity to alter CNR1 gene function [59], [60]. To fully characterize these variants, future investigation must target deep re-sequencing of the CNR1 gene in individuals expressing H3, from cohorts of racially diverse populations enriched for the traits of interest. In conclusion, we have demonstrated that the common H3 haplotype at the CNR1 gene locus potently influences HDL-C response to change in dietary fat intake in a gender-specific manner. Functional characterization of this relationship may further our understanding of the biology underlying lipid homeostasis, and potentially yield therapeutic guidance to improve public health by optimizing dietary macronutrient modifications, a direction much needed in the current epidemic of obesity.
Acknowledgments
The authors thank Taren B Spence, R.D. for study implementation and Will Bush, Ph.D. for assistance with manuscript preparation. In addition, we thank the following for donating study food supplies: Atkins Nutritionals, Inc; Oregon Hazelnut Commission; National Peanut Board; Setton Farms; National Sunflower Association; Almond Board of California.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by The Dr. Robert C. and Veronica Atkins Foundation (HJS), the Vanderbilt CTSA grant 1 UL1 RR024975 from the National Center for Research Resources, National Institutes of Health (HJS), the Tennessee Valley Healthcare System, NIH grants DK064857 and DK069927, and the Vanderbilt Diabetes Research and Training Center grant DK020593 (KDN), NIH Grant U01HL069757 (RMK), the Department of Atherosclerosis Research, the Children's Hospital Oakland Research Institute (RMK), and NIH Grant R01DK080007 (RAW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. Jama. 2002;287:356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 2.Ashen MD, Blumenthal RS. Clinical practice. Low HDL cholesterol levels. N Engl J Med. 2005;353:1252–1260. doi: 10.1056/NEJMcp044370. [DOI] [PubMed] [Google Scholar]
- 3.Brown CD, Higgins M, Donato KA, Rohde FC, Garrison R, et al. Body mass index and the prevalence of hypertension and dyslipidemia. Obes Res. 2000;8:605–619. doi: 10.1038/oby.2000.79. [DOI] [PubMed] [Google Scholar]
- 4.Kim JK, Alley D, Seeman T, Karlamangla A, Crimmins E. Recent changes in cardiovascular risk factors among women and men. J Womens Health (Larchmt) 2006;15:734–746. doi: 10.1089/jwh.2006.15.734. [DOI] [PubMed] [Google Scholar]
- 5.Wilke RA. High-density lipoprotein (HDL) cholesterol: leveraging practice-based biobank cohorts to characterize clinical and genetic predictors of treatment outcome. Pharmacogenomics J. 2011;11:162–173. doi: 10.1038/tpj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilke RA, Berg RL, Linneman JG, Peissig P, Starren J, et al. Quantification of the clinical modifiers impacting high-density lipoprotein cholesterol in the community: Personalized Medicine Research Project. Prev Cardiol. 2010;13:63–68. doi: 10.1111/j.1751-7141.2009.00055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanders B, Peterson FJ, Lavin P. Long term health effects associated with significant weight loss: A study of the dose response effect. In: Kanders GLBaBS., editor. Obesity Pathophysiology, Psychology and Treatment. New York: Chapman & Hall; 1994. pp. 167–181. [Google Scholar]
- 8.Stein O, Stein Y. Atheroprotective mechanisms of HDL. Atherosclerosis. 1999;144:285–301. doi: 10.1016/s0021-9150(99)00065-9. [DOI] [PubMed] [Google Scholar]
- 9.Bray GA. Is dietary fat important? Am J Clin Nutr. 2011;93:481–482. doi: 10.3945/ajcn.110.011114. [DOI] [PubMed] [Google Scholar]
- 10.Willett WC. Dietary fat and obesity: an unconvincing relation. Am J Clin Nutr. 1998;68:1149–1150. doi: 10.1093/ajcn/68.6.1149. [DOI] [PubMed] [Google Scholar]
- 11.Siri-Tarino PW, Sun Q, Hu FB, Krauss RM. Meta-analysis of prospective cohort studies evaluating the association of saturated fat with cardiovascular disease. Am J Clin Nutr. 2010;91:535–546. doi: 10.3945/ajcn.2009.27725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sieri S, Krogh V, Berrino F, Evangelista A, Agnoli C, et al. Dietary glycemic load and index and risk of coronary heart disease in a large italian cohort: the EPICOR study. Arch Intern Med. 2010;170:640–647. doi: 10.1001/archinternmed.2010.15. [DOI] [PubMed] [Google Scholar]
- 13.Sacks FM, Katan M. Randomized clinical trials on the effects of dietary fat and carbohydrate on plasma lipoproteins and cardiovascular disease. Am J Med. 2002;113(Suppl 9B):13S–24S. doi: 10.1016/s0002-9343(01)00987-1. [DOI] [PubMed] [Google Scholar]
- 14.Nordmann AJ, Nordmann A, Briel M, Keller U, Yancy WS, Jr, et al. Effects of low-carbohydrate vs low-fat diets on weight loss and cardiovascular risk factors: a meta-analysis of randomized controlled trials. Arch Intern Med. 2006;166:285–293. doi: 10.1001/archinte.166.3.285. [DOI] [PubMed] [Google Scholar]
- 15.Mensink RP, Zock PL, Kester AD, Katan MB. Effects of dietary fatty acids and carbohydrates on the ratio of serum total to HDL cholesterol and on serum lipids and apolipoproteins: a meta-analysis of 60 controlled trials. Am J Clin Nutr. 2003;77:1146–1155. doi: 10.1093/ajcn/77.5.1146. [DOI] [PubMed] [Google Scholar]
- 16.Volek JS, Sharman MJ, Gomez AL, DiPasquale C, Roti M, et al. Comparison of a very low-carbohydrate and low-fat diet on fasting lipids, LDL subclasses, insulin resistance, and postprandial lipemic responses in overweight women. J Am Coll Nutr. 2004;23:177–184. doi: 10.1080/07315724.2004.10719359. [DOI] [PubMed] [Google Scholar]
- 17.Jansen S, Lopez-Miranda J, Salas J, Castro P, Paniagua JA, et al. Plasma lipid response to hypolipidemic diets in young healthy non-obese men varies with body mass index. J Nutr. 1998;128:1144–1149. doi: 10.1093/jn/128.7.1144. [DOI] [PubMed] [Google Scholar]
- 18.Weggemans RM, Zock PL, Urgert R, Katan MB. Differences between men and women in the response of serum cholesterol to dietary changes. Eur J Clin Invest. 1999;29:827–834. doi: 10.1046/j.1365-2362.1999.00524.x. [DOI] [PubMed] [Google Scholar]
- 19.Denke MA, Adams-Huet B, Nguyen AT. Individual cholesterol variation in response to a margarine- or butter-based diet: A study in families. JAMA. 2000;284:2740–2747. doi: 10.1001/jama.284.21.2740. [DOI] [PubMed] [Google Scholar]
- 20.Ordovas JM. Nutrigenetics, plasma lipids, and cardiovascular risk. J Am Diet Assoc. 2006;106:1074–1081. doi: 10.1016/j.jada.2006.04.016. quiz 1083. [DOI] [PubMed] [Google Scholar]
- 21.Piomelli D. The molecular logic of endocannabinoid signalling. Nat Rev Neurosci. 2003;4:873–884. doi: 10.1038/nrn1247. [DOI] [PubMed] [Google Scholar]
- 22.Despres JP, Golay A, Sjostrom L. Effects of rimonabant on metabolic risk factors in overweight patients with dyslipidemia. N Engl J Med. 2005;353:2121–2134. doi: 10.1056/NEJMoa044537. [DOI] [PubMed] [Google Scholar]
- 23.Ruilope LM, Despres JP, Scheen A, Pi-Sunyer X, Mancia G, et al. Effect of rimonabant on blood pressure in overweight/obese patients with/without co-morbidities: analysis of pooled RIO study results. J Hypertens. 2008;26:357–367. doi: 10.1097/HJH.0b013e3282f2d625. [DOI] [PubMed] [Google Scholar]
- 24.Baye TM, Zhang Y, Smith E, Hillard CJ, Gunnell J, et al. Genetic variation in cannabinoid receptor 1 (CNR1) is associated with derangements in lipid homeostasis, independent of body mass index. Pharmacogenomics. 2008;9:1647–1656. doi: 10.2217/14622416.9.11.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Q, Jiang L, Berg RL, Antonik M, MacKinney E, et al. A common CNR1 (cannabinoid receptor 1) haplotype attenuates the decrease in HDL cholesterol that typically accompanies weight gain. PLoS One. 2010;5:e15779. doi: 10.1371/journal.pone.0015779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krauss RM, Dreon DM. Low-density-lipoprotein subclasses and response to a low-fat diet in healthy men. Am J Clin Nutr. 1995;62:478S–487S. doi: 10.1093/ajcn/62.2.478S. [DOI] [PubMed] [Google Scholar]
- 27.Dreon DM, Fernstrom HA, Williams PT, Krauss RM. LDL subclass patterns and lipoprotein response to a low-fat, high-carbohydrate diet in women. Arterioscler Thromb Vasc Biol. 1997;17:707–714. doi: 10.1161/01.atv.17.4.707. [DOI] [PubMed] [Google Scholar]
- 28.Moshfegh AJ, Borrud L, Perloff B, LaComb R. Improved Method for the 24-hour Dietary Recall for Use in National Surveys. FASEB Journal. 1999;13:A603. [Google Scholar]
- 29.Dreon DM, Fernstrom HA, Campos H, Blanche P, Williams PT, et al. Change in dietary saturated fat intake is correlated with change in mass of large low-density-lipoprotein particles in men. Am J Clin Nutr. 1998;67:828–836. doi: 10.1093/ajcn/67.5.828. [DOI] [PubMed] [Google Scholar]
- 30.Dreon DM, Krauss RM. Diet-gene interactions in human lipoprotein metabolism. J Am Coll Nutr. 1997;16:313–324. doi: 10.1080/07315724.1997.10718692. [DOI] [PubMed] [Google Scholar]
- 31.Krauss RM. Genetic, metabolic, and dietary influences on the atherogenic lipoprotein phenotype. World Rev Nutr Diet. 1997;80:22–43. doi: 10.1159/000059581. [DOI] [PubMed] [Google Scholar]
- 32.Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 33.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 34.Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care. 2004;27:1487–1495. doi: 10.2337/diacare.27.6.1487. [DOI] [PubMed] [Google Scholar]
- 35.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 36.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′----3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russo P, Strazzullo P, Cappuccio FP, Tregouet DA, Lauria F, et al. Genetic variations at the endocannabinoid type 1 receptor gene (CNR1) are associated with obesity phenotypes in men. J Clin Endocrinol Metab. 2007;92:2382–2386. doi: 10.1210/jc.2006-2523. [DOI] [PubMed] [Google Scholar]
- 39.Serdula MK, Mokdad AH, Williamson DF, Galuska DA, Mendlein JM, et al. Prevalence of attempting weight loss and strategies for controlling weight. Jama. 1999;282:1353–1358. doi: 10.1001/jama.282.14.1353. [DOI] [PubMed] [Google Scholar]
- 40.Sharman MJ, Gomez AL, Kraemer WJ, Volek JS. Very low-carbohydrate and low-fat diets affect fasting lipids and postprandial lipemia differently in overweight men. J Nutr. 2004;134:880–885. doi: 10.1093/jn/134.4.880. [DOI] [PubMed] [Google Scholar]
- 41.Gardner CD, Kiazand A, Alhassan S, Kim S, Stafford RS, et al. Comparison of the Atkins, Zone, Ornish, and LEARN diets for change in weight and related risk factors among overweight premenopausal women: the A TO Z Weight Loss Study: a randomized trial. Jama. 2007;297:969–977. doi: 10.1001/jama.297.9.969. [DOI] [PubMed] [Google Scholar]
- 42.Di Marzo V, Matias I. Endocannabinoid control of food intake and energy balance. Nat Neurosci. 2005;8:585–589. doi: 10.1038/nn1457. [DOI] [PubMed] [Google Scholar]
- 43.Ruby MA, Nomura DK, Hudak CS, Mangravite LM, Chiu S, et al. Overactive endocannabinoid signaling impairs apolipoprotein E-mediated clearance of triglyceride-rich lipoproteins. Proc Natl Acad Sci U S A. 2008;105:14561–14566. doi: 10.1073/pnas.0807232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stafford JM, Yu F, Printz R, Hasty AH, Swift LL, et al. Central nervous system neuropeptide Y signaling modulates VLDL triglyceride secretion. Diabetes. 2008;57:1482–1490. doi: 10.2337/db07-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ginsberg HN. Lipoprotein physiology. Endocrinol Metab Clin North Am. 1998;27:503–519. doi: 10.1016/s0889-8529(05)70023-2. [DOI] [PubMed] [Google Scholar]
- 46.Osei-Hyiaman D, DePetrillo M, Pacher P, Liu J, Radaeva S, et al. Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. J Clin Invest. 2005;115:1298–1305. doi: 10.1172/JCI23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyler K, Hillard CJ, Greenwood-Van Meerveld B. Inhibition of small intestinal secretion by cannabinoids is CB1 receptor-mediated in rats. Eur J Pharmacol. 2000;409:207–211. doi: 10.1016/s0014-2999(00)00843-8. [DOI] [PubMed] [Google Scholar]
- 48.Bensaid M, Gary-Bobo M, Esclangon A, Maffrand JP, Le Fur G, et al. The cannabinoid CB1 receptor antagonist SR141716 increases Acrp30 mRNA expression in adipose tissue of obese fa/fa rats and in cultured adipocyte cells. Mol Pharmacol. 2003;63:908–914. doi: 10.1124/mol.63.4.908. [DOI] [PubMed] [Google Scholar]
- 49.Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ Res. 2005;96:1221–1232. doi: 10.1161/01.RES.0000170946.56981.5c. [DOI] [PubMed] [Google Scholar]
- 50.Osei-Hyiaman D, Liu J, Zhou L, Godlewski G, Harvey-White J, et al. Hepatic CB1 receptor is required for development of diet-induced steatosis, dyslipidemia, and insulin and leptin resistance in mice. J Clin Invest. 2008;118:3160–3169. doi: 10.1172/JCI34827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yasuda T, Ishida T, Rader DJ. Update on the role of endothelial lipase in high-density lipoprotein metabolism, reverse cholesterol transport, and atherosclerosis. Circ J. 2010;74:2263–2270. doi: 10.1253/circj.cj-10-0934. [DOI] [PubMed] [Google Scholar]
- 53.Badellino KO, Wolfe ML, Reilly MP, Rader DJ. Endothelial lipase is increased in vivo by inflammation in humans. Circulation. 2008;117:678–685. doi: 10.1161/CIRCULATIONAHA.107.707349. [DOI] [PubMed] [Google Scholar]
- 54.Gary-Bobo M, Elachouri G, Scatton B, Le Fur G, Oury-Donat F, et al. The cannabinoid CB1 receptor antagonist rimonabant (SR141716) inhibits cell proliferation and increases markers of adipocyte maturation in cultured mouse 3T3 F442A preadipocytes. Mol Pharmacol. 2006;69:471–478. doi: 10.1124/mol.105.015040. [DOI] [PubMed] [Google Scholar]
- 55.Gary-Bobo M, Elachouri G, Gallas JF, Janiak P, Marini P, et al. Rimonabant reduces obesity-associated hepatic steatosis and features of metabolic syndrome in obese Zucker fa/fa rats. Hepatology. 2007;46:122–129. doi: 10.1002/hep.21641. [DOI] [PubMed] [Google Scholar]
- 56.Jourdan T, Djaouti L, Demizieux L, Gresti J, Verges B, et al. CB1 antagonism exerts specific molecular effects on visceral and subcutaneous fat and reverses liver steatosis in diet-induced obese mice. Diabetes. 2010;59:926–934. doi: 10.2337/db09-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knoblauch H, Bauerfeind A, Toliat MR, Becker C, Luganskaja T, et al. Haplotypes and SNPs in 13 lipid-relevant genes explain most of the genetic variance in high-density lipoprotein and low-density lipoprotein cholesterol. Hum Mol Genet. 2004;13:993–1004. doi: 10.1093/hmg/ddh119. [DOI] [PubMed] [Google Scholar]
- 58.Turner SD, Berg RL, Linneman JG, Peissig PL, Crawford DC, et al. Knowledge-driven multi-locus analysis reveals gene-gene interactions influencing HDL cholesterol level in two independent EMR-linked biobanks. PLoS One. 2011;6:e19586. doi: 10.1371/journal.pone.0019586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 60.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]