Abstract
Background
Ganoderma lucidum (Reishi or Ling Zhi) is one of the most famous Traditional Chinese Medicines and has been widely used in the treatment of various human diseases in Asia countries. It is also a fungus with strong wood degradation ability with potential in bioenergy production. However, genes, pathways and mechanisms of these functions are still unknown.
Methodology/Principal Findings
The genome of G. lucidum was sequenced and assembled into a 39.9 megabases (Mb) draft genome, which encoded 12,080 protein-coding genes and ∼83% of them were similar to public sequences. We performed comprehensive annotation for G. lucidum genes and made comparisons with genes in other fungi genomes. Genes in the biosynthesis of the main G. lucidum active ingredients, ganoderic acids (GAs), were characterized. Among the GAs synthases, we identified a fusion gene, the N and C terminal of which are homologous to two different enzymes. Moreover, the fusion gene was only found in basidiomycetes. As a white rot fungus with wood degradation ability, abundant carbohydrate-active enzymes and ligninolytic enzymes were identified in the G. lucidum genome and were compared with other fungi.
Conclusions/Significance
The genome sequence and well annotation of G. lucidum will provide new insights in function analyses including its medicinal mechanism. The characterization of genes in the triterpene biosynthesis and wood degradation will facilitate bio-engineering research in the production of its active ingredients and bioenergy.
Introduction
Ganoderma lucidum (Leyss. ex. Fr) Karst., Ling-Zhi in Chinese and Reishi in Japanese, belonging to the Ganodermataceae of Aphyllophorales in Basidiomycetes [1], is a widely distributed fungus in the tropic and subtropics of Asia, Africa and America [2], with the most diversities in China. G. lucidum is one of the most famous Traditional Chinese Medicines and has been widely used as a tonic for longevity and overall health in China for thousands of years [3]. G. lucidum has been proved with remarkable pharmacological activities and therapeutic effects in immuno-modulation, anti-cancer, anti-radiation and detoxification for various human diseases [4]–[7]. However, the accumulation of its active ingredients and the pharmacological mechanisms are mainly unknown. The genome sequence and gene annotation of G. lucidum will provide key resources and may speed up the function research of G. lucidum to human health.
Ganoderic acids (GAs), one of the main active ingredients of G. lucidum, are a kind of triterpenoid secondary metabolites and shown the ability to participate in many biological activities including antitumor, antioxidant, etc. [8]. However, the content of GAs is very low and is suggested to be the quality indicator of G. lucidum in Japan [1], [9]. It is suggested that the triterpene backbone of GAs could be biosynthesized via the mevalonic acid (MVA) pathway. Several genes in this pathway have been cloned in G. lucidum, including 3-Hydroxy- 3- methylglutaryl- CoA reductase (HMGR) [10], Farnesyl diphosphate synthase (FPPs) [11], Squalene synthase (SQS) [12], and Lanosterol synthase (also namely 2, 3-oxidosqualene lanosterol cyclase, OSC) [13]. However, it rarely reported about the processes of decoration after the triterpene backbone biosynthesis, such as cyclization and glycosylation, which are very important for GAs synthesis. The genome sequence is expected to characterize the enzymes of these key steps in the GAs biosynthesis.
G. lucidum is one of the white-rot fungi that grow on the dead trees by degrading cellulose, hemicellulose and lignin. Lignin, one of the main polymeric components of plant cell wall, is highly resistant to chemical and biological degradation [14]. Although there are some reports mentioned ligninolytic enzymes, the mechanism of lignin degradation is still not fully understood [14]–[16]. In addition, different enzymatic systems are employed in different fungi [17]. As one of the dominant organisms decomposing lignocellulose, it would be interesting to figure out the enzymatic system and genes of G. lucidum in wood degradation.
With the development of next-generation DNA sequencing, several macrofungi have been sequenced and analyzed to illuminate different aspects. Ohm et al. [18] studied the fruiting bodies formation and lignocelluloses degradation of Schizophyllum commune. Stajich et al. [19] completed the chromosome assembly of Coprinopsis cinerea, and investigated the meiotic recombination, genes and gene families and so on. Martin et al. illustrated the different ways of genetic predisposition for symbiosis in basidiomycete Laccaria bicolor [20] and ascomycete Tuber melanosporum Vittad. [21]. With these fungi genomes, it is possible to make full annotation and comparison for G. lucidum genomes. The genome annotation of G. lucidum will provide important data to further function and mechanism research in G. lucidum and comparative genomics in fungi.
In this study, we sequenced the genome of monokaryotic G. lucidum strain isolated from China and assembled a 39.9 Mb genome. We made full annotations with the predicted genes in this genome and compared them with other fungi genomes. With integrated gene prediction and annotation, we illuminated the synthesis of GAs as a model system to study triterpenoid biosynthesis in fungi. Besides the importance of understanding the biosynthesis of this active ingredient, insights into the enzyme systems of lignocelluloses degradation in G. lucidum may speed up the process of understanding the lignocelluloses degradation mechanism for bioenergy applications.
Results
The genome characteristics of G. lucidum
The genome of monokaryotic G. lucidum was sequenced by whole genome shotgun strategy and produced 3,738 Mb clean data after filtering low quality and adapter contamination reads. The assembly was performed by SOAPdenovo genome assembler [22], firstly generated 1,724 contigs with N50 of 80,796 base pairs (bp) and then assembled into 634 scaffolds with N50 of 322,982 bp. The lengths of scaffolds ranged from 1,004 bp to 1,953,398 bp. Finally, we got a 39.9 Mb draft genome sequence for G. lucidum. Although we could not assemble these scaffolds into chromosomes, by using k-mer analysis, the expected genome size was 42.53 Mb, so these scaffolds covered 93.92% of the whole genome. The G+C content of the G. lucidum genome was 55.56%. The features of the assembled genome sequences are shown in Table 1.
Table 1. The characteristics of assembly scaffold and genome of G. lucidum.
| Scaffold characteristics | |
| Total number | 634 |
| Total length (bp) | 39,945,170 |
| N50 (bp) | 322,982 |
| N90 (bp) | 50,570 |
| Max length (bp) | 1,953,398 |
| Min length (bp) | 1,004 |
| Genome characteristics | |
| Genome assembly (Mb) | 39.9 |
| Whole GC content (%) | 55.56 |
| Coding sequence GC content (%) | 58.86 |
| Number of protein-coding genes | 12080 |
| Coding sequence > = 100 amino acids | 11522 |
| Coding sequences/genome | 43.31% |
| Average gene length (bp) | 1959 |
| Average coding sequence length (bp) | 1435 |
| Average exon length (nt) | 230 |
| Average intron length (nt) | 100 |
| Average number of exons per gene | 6.25 |
Repeat sequences in the genome
Five softwares were used to characterize transposons and the Tandem Repeat Finder was used to identify the tandem repeat sequences. Totally, we identified 2,025,242 bp repeat sequences, comprising 5.07% of the genome. No large scale dispersed segmental duplication was observed. Of them, tandem repeat sequences comprised 0.57% and transposable elements (TEs) were about 4.6% of the assembled genome. Among the TEs, long terminal repeats (LTR) and non-LTR transposons comprised 1.43% and 3.17% of the genome, respectively. Among the non-LTR transposons, DNA transposons (class II transposons) comprised 0.52% of the genome. The elements of DNA transposons mainly fell into four classes: Activator (hAT), Enhancer (En/spm), Harbinger and Mariner (Tc1).
Predicted Gene models
By combining several different gene predictors (see methods), we identified 12,080 protein-coding gene models, 245 tRNA, 1 rRNA and 15 snRNA with a total length of 17,343,729 bp, accounting for 43.41% of the genome (Table 1). The gene density was 3.34 genes/10 kilobases (kb) and the average size of protein coding genes was 1,435 bp. Genes were typically with small exons (average 230 bp) and introns (average 100 bp), which were similar with other basidiomycetes [20]. There were average 6.25 exons in one gene. Notably, the G+C content in protein coding gene regions was 58.86%, slightly higher than the whole genome (55.56%) and other basidiomycetes [20].
Among the 245 tRNA genes, 10 tRNAs were pseudogenes and 141 tRNAs contained an intron. Forty six out of the 61 possible anti-codon tRNA were found, corresponding to the codons of 20 amino acids. The anti-codon usage and codon usage were shown in File S1. Except for several codons, the usage frequencies of most codons were proportional to the numbers of anti-codon proportion (File S2). For lacking of the other 18 anti-codons, we speculated anticodon repertoire in this genome was consistent with the normal wobble rules [23], which allow the following anticodon and codon pairings: I/ANN:NNU/NNC; GNN:NNU/NNC; UNN:NNA and CNN:NNG.
Gene annotation
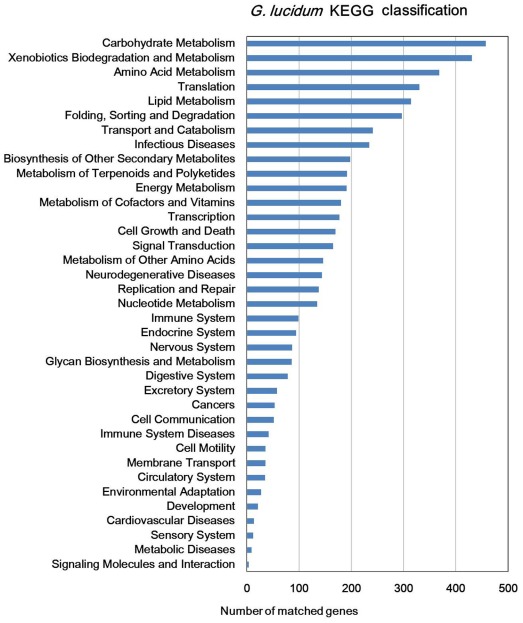
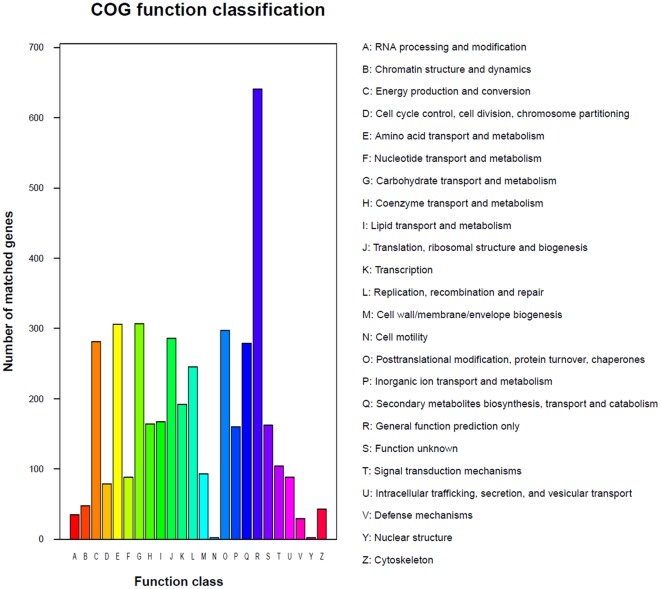
By homology search, we mapped our predicted proteins to Gene Ontology (GO), 5,893 (49%) of which were assigned to GO terms, including 5,410, 1,738 and 4,034 genes mapped to the molecular function, cellular component and biological process categories, respectively. We also assigned 4,737 proteins to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The annotations with KEGG, GO, InterPro, NCBI Clusters of Orthologous Groups of proteins (COG), NCBI non-redundant (nr), Pfam, SwissProt and TrEMBL protein databases were shown in File S1. KEGG function classification was shown in Figure 1, in which “Carbohydrate Metabolism”, “Xenobiotics Biodegradation and Metabolism” and “Amino Acid Metabolism” were the top 3 categories. Of these predicted genes in G. lucidum, up to 9,978, 6,436 and 9,981 showed a significant similarity (BLASTP, cut-off e-value<1e-7) to documented proteins in the NCBI nr database (Aug 2011), Swiss-Prot, and TrEMBL, respectively. As a result, about 83% of predicted proteins were similar to sequences in these public databases and only 2,094 genes were not similar to current public sequences, some of which might be G. lucidum specific genes. We further classed predicted genes into orthologous group (single-copy in G. lucidum and at least one other species ortholog), or paralogous group (multi-copy in G. lucidum). There were 4,689 orthologous genes and 5,510 paralogous genes by above definition. By the NCBI COG mapping, 3,509 (29%) proteins were assigned to COGs proteins (Figure 2). Similar to the KEGG annotation, some metabolisms and biosynthesis categories in COG were highly enriched.
Figure 1. The KEGG function annotaion of G. lucidum.
Distribution of Genes in different KEGG categories.
Figure 2. The COG function annotaion of G. lucidum.
Distribution of Genes in different COG function classification.
Comparing with other published basidiomycetes, G. lucidum has many different biological characteristics, such as saprophytism, multiple triterpenoids and polysaccharides metabolites. To compare and find genes for G. lucidum specific characteristics, we performed comprehensive comparisons among G. lucidum and other published fungi genomes in the follow sections.
Comparative genomics analysis of KEGG annotation
To make comprehensive comparison for KEGG annotations in fungi, KEGG pathway mapping was performed in 13 basidiomycetes and 5 ascomycetes (File S1). To facilitate comparison and show results in one table, we only showed the results of 8 basidiomycetes (7 in agaricomycotina and 1 in ustilaginomycotina) and 2 represented ascomycetes in the following analyses. In the second layer of KEGG pathway terms, we found fungi in agaricomycotina (including G. lucidum) had much more genes in each pathway than other fungi (Figure 1 and File S1). G. lucidum had relatively more genes in several pathways of metabolism and biosynthesis, such as “Metabolism of Terpenoids and Polyketides”, “Metabolism of Other Amino Acids” and “Xenobiotics Biodegradation and Metabolism”. In the third layer of KEGG, under the “Xenobiotics Biodegradation and Metabolism” pathway category, we found that G. lucidum and other Agaricomycotina fungi had relatively more proteins involved in several degradation pathways including pathways of aminobenzoate, bisphenol, dioxin and polycyclic aromatic hydrocarbon degradation (Table 2). There were about 190 genes involved in 3 of these degradation pathways in G. lucidum (Table 2). These results indicated that G. lucidum had strong ability of degradation. In addition, we also observed that “Metabolism of xenobiotics by cytochrome P450” and “Drug metabolism - cytochrome P450” sub-pathways had relative more genes in G. lucidum.
Table 2. The gene distribution of fungi in pathway “Xenobiotics Biodegradation and Metabolism”.
| Pathway in KEGG | Pathway annotation | Agaricomycotina | |||||||||
| G. luc | F. pin | P. chr | S. com | P. ost | L. bic | C. cin | M. glo | P. tri | S. cer | ||
| 00362 | Benzoate degradation | 33 | 47 | 29 | 31 | 30 | 21 | 26 | 7 | 73 | 16 |
| 00627 | Aminobenzoate degradation | 190* | 178 | 157 | 149 | 189 | 83 | 143 | 21 | 202 | 38 |
| 00364 | Fluorobenzoate degradation | 6 | 4 | 4 | 4 | 4 | 1 | 4 | 1 | 8 | 1 |
| 00625 | Chloroalkane and chloroalkene degradation | 71 | 85 | 78 | 108 | 67 | 43 | 39 | 14 | 79 | 27 |
| 00361 | Chlorocyclohexane and chlorobenzene degradation | 25 | 22 | 14 | 26 | 25 | 4 | 12 | 4 | 40 | 2 |
| 00623 | Toluene degradation | 13 | 14 | 8 | 12 | 15 | 4 | 12 | 4 | 28 | 1 |
| 00622 | Xylene degradation | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| 00633 | Nitrotoluene degradation | 0 | 0 | 1 | 0 | 1 | 3 | 1 | 1 | 1 | 0 |
| 00642 | Ethylbenzene degradation | 11 | 23 | 11 | 12 | 8 | 8 | 11 | 3 | 41 | 9 |
| 00643 | Styrene degradation | 14 | 11 | 14 | 8 | 9 | 5 | 7 | 3 | 23 | 4 |
| 00791 | Atrazine degradation | 3 | 3 | 3 | 1 | 2 | 1 | 1 | 0 | 7 | 1 |
| 00930 | Caprolactam degradation | 14 | 12 | 8 | 12 | 23 | 9 | 8 | 5 | 25 | 5 |
| 00351 | 1,1,1-Trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) degradation | 10 | 8 | 5 | 13 | 9 | 0 | 0 | 0 | 9 | 0 |
| 00363 | Bisphenol degradation | 196* | 183 | 165 | 194 | 186 | 84 | 129 | 18 | 162 | 27 |
| 00621 | Dioxin degradation | 35* | 21 | 27 | 24 | 12 | 9 | 12 | 0 | 19 | 0 |
| 00626 | Naphthalene degradation | 62 | 64 | 57 | 58 | 41 | 32 | 35 | 11 | 94 | 17 |
| 00624 | Polycyclic aromatic hydrocarbon degradation | 187* | 148 | 151 | 136 | 150 | 59 | 99 | 11 | 132 | 8 |
| 00980 | Metabolism of xenobiotics by cytochrome P450 | 44* | 41 | 45 | 30 | 35 | 22 | 31 | 5 | 38 | 9 |
| 00982 | Drug metabolism - cytochrome P450 | 38* | 31 | 32 | 25 | 26 | 27 | 26 | 6 | 32 | 10 |
| 00983 | Drug metabolism - other enzymes | 14 | 14 | 12 | 14 | 16 | 15 | 20 | 6 | 17 | 12 |
represents G. lucidum having relatively more genes than others. Abbreviations: G. lui, Ganoderma lucidum; F. pin: Fomitopsis pinicola; P. chr: Phanerochaete chrysosporium; S. com: Schizophyllum commune; P. ost: Pleurotus ostreatus; L. bic: Laccaria bicolor; C. cin: Coprinopsis cinerea; M. glo: Malassezia globosa; P. tri: Pyrenophora teres; S. cer: Saccharomyces cerevisiae.
In the fourth layer, G. lucidum had 27 KO terms with 1.5-fold genes more than other Agaricomycotina fungi (Table 3). Of them, the term K00490 (cytochrome P450) showed relatively more genes in G. lucidum. Since cytochrome P450 is a large group of enzymes involved in many important biosynthesis and metabolism pathways, we further identified and performed comparison about P450 genes at genome level in these fungi. We found that the numbers of P450 genes in agaricomycotina were much more than those in other subphylums of basidiomycota and in ascomycota (Table 4). G. lucidum had 222 putative P450 genes, which was the largest one in the 10 represented fungi and the top 3 in all the 18 fungi we analyzed.
Table 3. KO families showing relatively more genes in G. lucidum genome as compared to other Basidiomycota fungi.
| Pathway in KEGG | KO description | G. luc | F. pin | P. chr | S. com | P. ost | L. bic | C. cin |
| K00490 | CYP4F; cytochrome P450, family 4, subfamily F | 48 | 41 | 12 | 20 | 14 | 20 | 16 |
| K00480 | E1.14.13.1; salicylate hydroxylase | 34 | 20 | 27 | 23 | 11 | 8 | 11 |
| K01046 | E3.1.1.3; triacylglycerol lipase | 26 | 21 | 14 | 10 | 19 | 14 | 13 |
| K01279 | TPP1, CLN2; tripeptidyl-peptidase I | 24 | 31 | 10 | 5 | 7 | 7 | 2 |
| K04125 | E1.14.11.13; gibberellin 2-oxidase | 22 | 17 | 4 | 4 | 10 | 1 | 1 |
| K10866 | RAD50; DNA repair protein RAD50 | 22 | 14 | 21 | 12 | 7 | 6 | 8 |
| K01183 | E3.2.1.14; chitinase | 21 | 15 | 10 | 13 | 12 | 11 | 9 |
| K01423 | E3.4.-.-; | 19 | 8 | 17 | 26 | 12 | 5 | 12 |
| K00140 | malonate-semialdehyde dehydrogenase/methylmalonate-semialdehyde dehydrogenase | 18 | 1 | 45 | 1 | 1 | 2 | 1 |
| K01528 | DNM; dynamin GTPase | 11 | 4 | 3 | 4 | 4 | 3 | 3 |
| K00218 | E1.3.1.33; protochlorophyllide reductase | 8 | 6 | 4 | 2 | 2 | 3 | 5 |
| K01190 | lacZ; beta-galactosidase | 8 | 2 | 3 | 4 | 5 | 0 | 0 |
| K03942 | NDUFV1; NADH dehydrogenase (ubiquinone) flavoprotein 1 | 7 | 2 | 1 | 2 | 2 | 2 | 2 |
| K06148 | ABCC-BAC; ATP-binding cassette, subfamily C, bacterial | 7 | 2 | 5 | 3 | 2 | 4 | 6 |
| K01044 | E3.1.1.1; carboxylesterase | 6 | 0 | 2 | 4 | 4 | 3 | 5 |
| K02831 | RAD53; ser/thr/tyr protein kinase RAD53 | 6 | 1 | 1 | 0 | 2 | 2 | 1 |
| K00119 | E1.1.99.-; | 5 | 0 | 2 | 1 | 2 | 0 | 3 |
| K00129 | E1.2.1.5; aldehyde dehydrogenase (NAD(P)+) | 5 | 2 | 2 | 2 | 2 | 0 | 1 |
| K01082 | E3.1.3.7; 3′(2′), 5′-bisphosphate nucleotidase | 5 | 3 | 1 | 4 | 1 | 1 | 1 |
| K09202 | regulatory protein SWI5 | 5 | 2 | 1 | 1 | 3 | 2 | 1 |
| K09553 | STIP1; stress-induced-phosphoprotein 1 | 5 | 0 | 1 | 1 | 0 | 1 | 5 |
| K00135 | E1.2.1.16; succinate-semialdehyde dehydrogenase (NADP+) | 4 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01539 | ATP1A; sodium/potassium-transporting ATPase subunit alpha | 4 | 4 | 1 | 0 | 1 | 2 | 0 |
| K02133 | ATPeF1B, ATP5B; F-type H+-transporting ATPase subunit beta | 4 | 1 | 1 | 1 | 1 | 1 | 1 |
| K10590 | TRIP12; E3 ubiquitin-protein ligase TRIP12 | 4 | 1 | 1 | 1 | 1 | 1 | 1 |
| K12388 | SORT1; sortilin | 4 | 1 | 1 | 1 | 1 | 2 | 1 |
| K09753 | CCR; cinnamoyl-CoA reductase | 3 | 0 | 1 | 0 | 0 | 0 | 2 |
The abbreviations of species were the same with Table 2. The number of genes in G. lucidum of each KO of is 1.5 fold more than the average of the other Basidiomycota fungi.
Table 4. The gene distribution of fungi in P450 family and GST family.
| G. luc | F. pin | P. chr | S. com | P. ost | L. bic | C. cin | M. glo | P. tri | S. cer | ||
| P450 | 222* | 196 | 154 | 120 | 160 | 113 | 143 | 12 | 97 | 6 | |
| GST | |||||||||||
| EFBy | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 3 | |
| GTE | 4 | 10 | 5 | 9 | 7 | 3 | 14 | 0 | 2 | 0 | |
| GTT1 | 1 | 1 | 0 | 2 | 1 | 0 | 2 | 0 | 1 | 1 | |
| GTT2 | 8 | 4 | 3 | 6 | 3 | 11 | 6 | 0 | 0 | 1 | |
| MAK16 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | |
| omega | 18* | 7 | 8 | 8 | 8 | 3 | 5 | 1 | 4 | 3 | |
| URE2p | 6 | 8 | 9 | 0 | 1 | 1 | 2 | 1 | 2 | 1 | |
| TOTAL | 39* | 32 | 27 | 27 | 22 | 20 | 31 | 5 | 11 | 10 |
represents G. lucidum having the most genes than others. The abbreviations of species were the same with Table 2.
Under the “Metabolism of xenobiotics by cytochrome P450” pathway, we found the glutathione S-transferases (GST, EC 2.5.1.18), a kind of well-known detoxification enzymes [24], were greatly enriched in G. lucidum compared with other fungi. According to the classification of Morel et al. [25], we investigated the GSTs distribution in six known classes (GTT1, GTT2, URE2p, Omega, EFBγ, MAK16) and a new class (GTE) of all fungi in this study. Under the relatively strict cutoff (BLASTP e-value<1e-10 and identity>30%), we found 39 GST genes in G. lucidum, which was the highest GST gene numbers among all fungi we analysed. Notably, G. lucidum had 18 genes in the Omega subfamily, which were much more than other fungi (Table 4).
The pathway of triterpenes synthesis
The triterpenes have been reported of great importance in G. lucidum because of their significant roles in immune regulation and other biological activities [4]–[7]. In plants, there are two pathways to synthesize terpenoids: the Mevalonate (MVA) pathway and methylerythritol 4-phosphate/deoxyxylulose 5- phosphate (MEP/DOXP) pathway. It has been suggested that the MEP/DOXP pathway do not exist in fungi [8]. We checked the G. lucidum genes in the “terpenoid backbone biosynthesis (map00900)” pathway and found that the genes only distributed in MVA pathway, no gene existed on the MEP/DOXP pathway (File S2). The similar results were found in other basidiomycetes and ascomycetes. These observations verified that terpenoid backbone biosynthesis only could be through the MVA pathway in fungi at the genome level.
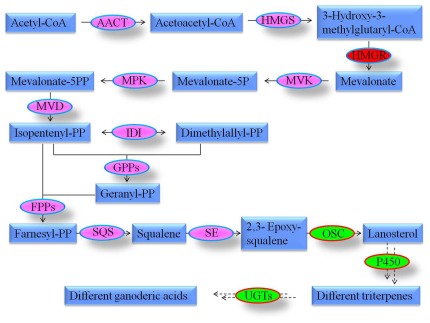
By integrating MVA pathway in KEGG and plant triterpenoid saponins biosynthesis from literatures, we summarized the potential triterpenoids biosynthesis pathway in G. lucidum (Figure 3). The pathway contained 14 steps catalyzed by different enzymes. The first 11 steps are the common steps for terpenoid skeleton biosynthesis and the last 3 steps may be specific for different triterpenes in different species. We identified and summarized the enzymes in the ganoderic acids (GAs) biosynthesis in table 5 from the G. lucidum genome, which includes 6 putative UDP-glycosyltransferases (UGTs) genes (Table 5).
Figure 3. Putative ganoderic acid biosynthesis pathway in G. lucidum.
Enzymes involved in this pathway are: AACT: acetyl-CoA acetyltransferase, [EC:2.3.1.9], K00626; HMGS: 3-hydroxy-3-methylglutaryl-CoA synthase, [EC:2.3.3.10], K01641; HMGR: 3-hydroxy-3-methylglutaryl-CoA reductase, [EC:1.1.1.34], K00021; MVK: mevalonate kinase, [EC:2.7.1.36], K00869; MPK: phosphomevalonate kinase, [EC:2.7.4.2], K00938; MVD: pyrophosphomevalonate decarboxylase, [EC:4.1.1.33], K01597; IDI: isopentenyl-diphosphate isomerase, [EC: 5.3.3.2], K01823; GPPs: geranyl diphosphate synthase, [EC: 2.5.1.1], K00787, K00804; FPPs: farnesyl diphosphate synthase, [EC: 2.5.1.10], K00787, K00804; SQS: squalene synthase, [EC: 2.5.1.21], K00801; SE: squalene monooxygenase, [EC: 1.14.99.7], K00511; OSC: 2, 3-oxidosqualene-lanosterol cyclase, [EC: 5.4.99.7], K01852; P450: cytochrome P450, [EC: 1.14.-.-]; UGTs: uridin diphosphate glycosyltransferases, [EC: 2.4.1.-]. Ingredients are in blue box. Limited enzymes are in red oval, key enzymes are in green oval while the other enzymes are in pink oval. Solid arrows and broken arrows represent single and putative multiple enzymatic steps respectively.
Table 5. The putative genes involved in triterpene biosynthesis.
| Gene full name | Abbr. | Enzyme | KO | Putative gene |
| acetyl-CoA acetyltransferase | AACT | EC:2.3.1.9 | K00626 | G_lucidum_10003032 |
| 3-hydroxy-3-methylglutaryl-CoA synthase | HMGS | EC:2.3.3.10 | K01641 | G_lucidum_10008701 |
| 3-hydroxy-3-methylglutaryl-CoA reductase | HMGR | EC:1.1.1.34 | K00021 | G_lucidum_10003589 |
| mevalonate kinase | MVK | EC:2.7.1.36 | K00869 | G_lucidum_10009892 |
| phosphomevalonate kinase | MPK | EC:2.7.4.2 | K00938 | G_lucidum_10010135 |
| pyrophosphomevalonate decarboxylase | MVD | EC:4.1.1.33 | K01597 | G_lucidum_10005090 |
| isopentenyl-diphosphate isomerase | IDI | EC:5.3.3.2 | K01823 | G_lucidum_10001705 |
| geranyl diphosphate synthase | GPPs | EC: 2.5.1.1 | K00787/K00804 | G_lucidum_10002724; |
| G_lucidum_ 10008471; | ||||
| G_lucidum_10004225 | ||||
| farnesyl diphosphate synthase | FPPs | EC: 2.5.1.10 | K00787/K00804 | G_lucidum_10002724; |
| G_lucidum_ 10008471; | ||||
| G_lucidum_10004225 | ||||
| squalene synthase | SQS | EC 2.5.1.21 | K00801 | G_lucidum_10005172 |
| squalene monooxygenase | SE | EC 1.14.99.7 | K00511 | G_lucidum_10007072 |
| 2, 3-oxidosqualene- lanosterol cyclase | OSC | EC 5.4.99.7 | K01852 | G_lucidum_10008645; |
| G_lucidum_10008646 | ||||
| cytochrome P450 | P450 | EC: 1.14.-.- | 222 putative genes | |
| UDP-glucosyl transferase | UGT | EC: 2.4.1.- | G_lucidum_10003239; | |
| G_lucidum_10003516; | ||||
| G_lucidum_10009503; | ||||
| G_lucidum_10009504; | ||||
| G_lucidum_10010093; | ||||
| G_lucidum_10010094 |
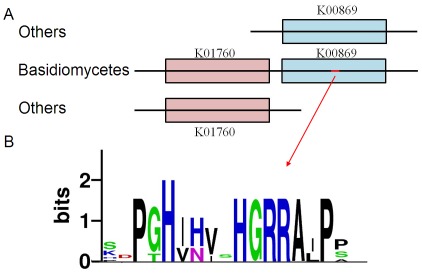
Interestingly, we observed a fusion gene in the triterpenes biosynthesis pathway in 12 Basidiomycete fungi except for L. bicolor. The N-terminal of the protein was similar to the enzyme K01760 (KEGG ID, cystathionine beta-lyase, metC), while the C-terminal was similar to another enzyme K00869 (KEGG ID, mevalonate kinase, MVK) which is an enzyme in the triterpenes biosynthesis pathway (Figure 4A). These proteins were referred as metC-MVKs in the following. In all other species except basidiomycetes, no such a protein matched the two enzymes at the same time. By multiple sequence alignment of the fusion protein in basidiomycetes, the average length of metC-MVKs was ∼886 aa and about half of it matched with K01760 and half matched with K00869. The metC-MVK protein was the only homologous protein with K00869 in our analyzed basidiomycetes, so they should be involved in terpenoid backbone biosynthesis functioning as K00869 in other species. We also noticed that this metC-MVK gene was the only gene which best hit K01760. In addition, seven of the 12 Basidiomycota fungi had a 16 amino acids conserved insertion sequence in the middle of the MVK regions of the metC-MVK gene (Figure 4B).
Figure 4. The features of metC-MVK in basidiomycetes.
(A) The metC-MVK matched two enzymes at the same time and some of them have an addition sequence in the middle of K00869 (red line). (B) The added conservative sequence in 7 of 13 Basidiomycota fungi.
Phylogeny of G. lucidum and multigene families
The phylogenetic tree constructed by concatenated sequences alignments showed that G. lucidum was close to another polyporale fungus Fomitopsis pinicola in the evolutionary relationship among all our analyzed fungi (Figure 5A). In the all-to-all BLASTP analysis, 9,278 predicted proteins of G. lucidum showed high sequence similarity with that of F. pinicola (BLASTP, cut-off e-value<1e-7). Following, 9,013 and 8,872 predicted proteins showed significant sequence similarity to that of Gloeophyllum trabeum and Stereum hirsutum, which were all in polyporales.
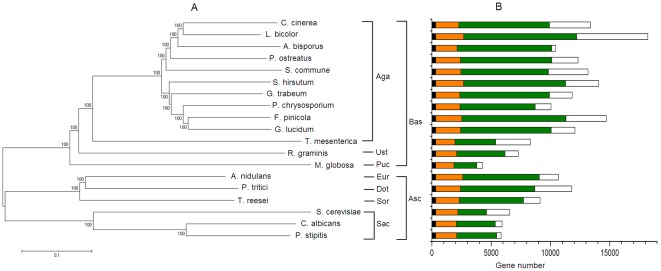
Figure 5. Phylogeny tree of 19 fungi and genes in their genomes.
(A) The Neighbor Joining tree (NJ) was constructed with 1,000 bootstrap replications from a concatenated alignment of 323 single-copy proteins. (B) Bars represent a comparison of the gene content of all these species in the corresponding position in NJ tree. Bars are subdivided to indicate different types of homology relationships. Black: genes that are found only one copy in all these fungi (323 genes); Orange: genes that were in all species but maybe more copies in some species; Green: genes presenting in more than one fungus but not in all these fungi; White: species-specific genes with no detectable homologs in other species genes. Abbreviations: Aga, Agaricomycotina; Ust, Ustilaginomycotina; Puc, Pucciniomycotina; Eur, Eurotiomycetes; Dot, Dothideomycetes; Sor, Sordariomycetes; Sac, Saccharomycotina; Bas, Basidiomycota; Asc, Ascomycota.
In order to investigate the gene family expansion in G. lucidum, we performed analyses for multi-gene families, which were generated from proteins in 8 Agaricomycotina species. In total, 10,720 gene families (File S1) containing at least two members were generated using the Tribe-MCL tool, of which 5,947 families had at least one G. lucidum gene and 1,487 families had at least two G. lucidum genes. The largest gene family had 517 genes and 126 of them were G. lucidum genes. In 3,540 lineage specific gene families, 287 families were G. lucidum specific (File S1). The number was very similar to that of G. trabeum and much lower than other basidiomycetes. L. bicolor had the largest (947) lineage-specific gene families, which may be related to its biggest genome size among our analyzed basidiomycetes. The distributions of genes with different copies or species-specific are shown in Figure 5B.
Besides the lineage specific gene families, the evolutionary changes in the size of each gene family were performed using CAFE program. As a result, we found that among the 7,180 non-lineage specific gene families for G. lucidum, 636 of them were expanded and 994 of them had undergone contraction. The function of the most abundant gene family was uncharacterized for lack of available annotation, while genes in the second most abundant gene family encoding proteins with a P450 domain (File S1). The expanded and contracted gene families and their annotations were shown in File S1.
G. lucidum has multiple copy het-like genes
Among the 287 G. lucidum specific gene families (File S1), the largest G. lucidum specific gene family had 101 genes and 89 of them had the HET (heterokaryon incompatibility protein) domain, which is related to vegetative (or heterokaryon) incompatibility (VI). It is surprised that so many het-like genes were found in G. lucidum, while few het genes were reported in other fungi. In PFAM database, there are three vegetative incompatibility related domains, which are HET, Het-c and HET-s. Since the HET related studies were mostly reported in fungi P. anserina and N. crassa, we added them in our analyzed fungi list to identify the HET genes. Thus, in total, we scanned 7 ascomycetes and 13 basidiomycetes for genes with the three het related domains. The results were shown in File S1 and only P. anserina had one HET-s domain. The number of Het-c genes in each species was always 0–2 and the highest one was four. While the number of genes with HET domain varied from 0 to 126. It seems that het-c and HET-s are comparatively conserved. G. lucidum had two genes with Het-c domain and 96 genes with HET domain, which was much more than other basidiomycetes and most ascomycetes. In the comparison, we also observed that there were 62 and 126 HET-like genes in N. crassa and P. anserine in which the number of het genes were reported for 11 and 9, respectively [26]. Thus, some of the het-like genes may play roles in other function not for VI, such as mat a/A for mating in N. crassa and het c for ascospore formation in P. anserina [26], [27]. Therefore, it may be a complex system not only one locus affect the VI. Since one of het-c loci in P. anserina is similar to the glycolipid transfer protein (GLTP) [27], the GLTP domain was also scanned in this study. We found two genes G_lucidum_10005152 and G_lucidum_10009654 with a GLTP domain, which also might be het-c genes.
These HET genes in G. lucidum encoded proteins with an average length of 2,686 amino acids and did not uniformly spread across the genome. The 98 genes were located on 45 scaffolds (total 634 scaffolds). Of them, 13 scaffolds had more than two HET genes and three scaffolds had more than 10 HET genes, suggesting the expansion of HET genes might have undergone tandem duplications. Except for the HET domain, some HET genes also had other domains, such as, adh_short,Aldo_ket_red, ICMT,Nup96, p450, SUR7, and WD40.
Function annotation of putative CAZymes
CAZy is a carbohydrate-active enzymes (CAZymes) database (http://www.cazy.org/) [28], which classifies the CAZymes into 5 major modules: Glycoside Hydrolases (GH), Glycosyl Transferases (GT), Polysaccharide Lyases (PL), Carbohydrate Esterases (CE), and Carbohydrate-Binding Modules (CBM). We mapped our analyzed fungi genomes to CAZy to study the members and features of these Carbohydrate-active enzymes. The results revealed that the gene numbers in the 5 major modules of CAZymes were similar in Agaricomycotina fungi, while much fewer in Ustilaginomycotina and Ascomycota fungi. G. lucidum possessed a wide spectrum of CAZymes responsible for the biosynthesis, degradation and modification of oligo- and polysaccharides, and of glycoconjugates (Table 6). The GHs and CEs in G. lucidum showed a little more than average count, while GTs, CBMs and PLs showed less than the Agaricomycotina average (Table 6).
Table 6. The gene distribution of different fungi in CAZymes family.
| CAZY | GH | GT | CBM | CE | PL | Total | |
| Agaricomycotina | G. lucidum | 216* | 56 | 34 | 40* | 3 | 349 |
| F. pinicola | 169 | 61 | 13 | 33 | 1 | 277 | |
| P. chrysosporium | 149 | 54 | 42 | 23 | 1 | 269 | |
| S. commune | 204 | 65 | 38 | 42 | 13 | 362 | |
| P. ostreatus | 221 | 57 | 64 | 47 | 13 | 402 | |
| L. bicolor | 105 | 59 | 12 | 27 | 1 | 204 | |
| C. cinerea | 189 | 65 | 56 | 49 | 9 | 368 | |
| Above average | 179 | 60 | 37 | 37 | 6 | 319 | |
| Ustilaginomycotina | M. globosa | 13 | 34 | 1 | 6 | 1 | 55 |
| Dothideomycetes | P. tritici | 221 | 91 | 43 | 60 | 10 | 425 |
| Saccharomycotina | S. cerevisiae | 46 | 68 | 14 | 4 | 0 | 132 |
represents G. lucidum having relatively more genes than the average of 7 Agaricomycotina fungi. Abbreviations: GH, Glycoside Hydrolases; GT, GlycosylTransferases; CBM, Carbohydrate-Binding Modules; CE, Carbohydrate Esterases; PL, Polysaccharide Lyases.
Function annotation of putative FOLymes
To assess the degradation in genomic level, proteins of G. lucidum were aligned to proteins in the FOLy (Fungal Oxidative Lignin enzymes) database, which collects and classifies enzymes involved in lignin catabolism. The FOLymes mainly comprise two families, lignin oxidases (LO families) and lignin-degrading auxiliary enzymes (LDA families) that generate H2O2 for peroxidases. G. lucidum contained a total of 48 members in FOLymes (24 genes in LO families and 24 genes in LDA families, Table 7) which was more than brown-rot fungi F. pinicola,G. trabeum and the fungi without ligninolytic activity, such as Malassezia globosa, Pyrenophora teres and Saccharomyces cerevisiae. In contrast, it had fewer FOLymes than the coprophilic fungus Coprinopsis cinerea (59 FOLymes) and the white-rot fungus Pleurotus ostreatus (72 FOLymes). While G. lucidum had the largest number of lignin oxidases (LO families). The LO families can further divided into 3 subfamilies, which are laccases (LO1), lignin peroxidases, manganese peroxidases, versatile peroxidases (LO2) and cellobiose dehydrogenases (CDHs; LO3). G. lucidum contained 16 laccase genes (LO1), 7 peroxidase (LO2) and 1 cellobiose dehydrogenase (LO3). For the seven peroxidases (LO2) genes in G. lucidum, two of them located at scaffold 10 and 3 located at scaffold 79 which maybe form a gene cluster. While LDA families have 7 sub families, G. lucidum contained 10 aryl-alcohol oxidase (LDA1), 9 copper radical oxidase (LDA3), 3 glucose oxidase (LDA6) and 2 benzoquinone reductase (LDA7). Similar to most other fungi, no LDA2, LDA4 or LDA5 gene was found in G. lucidum. The major fungi contained multi-copy genes in LO1 except P. chrysosporium, which had only 1 LO1 gene but 16 LO2 genes.
Table 7. The gene distribution of FOLymes in G. lucidum and other fungi.
| FOLYmes | G. luc | F. pin | P. chr | S. com | P. ost | L. bic | C. cin | M. glo | P. tri | S. cer |
| LDA1 | 10 | 7 | 4 | 3 | 29 | 4 | 27 | 1 | 8 | 0 |
| LDA2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| LDA3 | 9 | 4 | 7 | 2 | 16 | 5 | 6 | 1 | 2 | 0 |
| LDA4 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| LDA6 | 3 | 1 | 3 | 11 | 4 | 2 | 4 | 3 | 4 | 0 |
| LDA7 | 2 | 1 | 4 | 4 | 2 | 2 | 3 | 0 | 1 | 3 |
| LO1 | 16 | 6 | 1 | 5 | 12 | 11 | 17 | 2 | 7 | 2 |
| LO2 | 7 | 1 | 16 | 0 | 8 | 1 | 1 | 0 | 0 | 0 |
| LO3 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 0 |
| total | 48 | 20 | 37 | 27 | 72 | 25 | 59 | 7 | 25 | 5 |
The abbreviations of species were the same with Table 2. LO1, laccases; LO2, peroxidases; LO3, cellobiose dehydrogenases; LDA1, aryl alcohol oxidases; LDA2, vanillyl-alcohol oxidases; LDA3, glyoxal oxidases; LDA4, pyranose oxidases; LDA5, galactose oxidases; LDA6, glucose oxidases; LDA7, benzoquinone reductases.
Discussion
G. lucidum is one of the most famous traditional medicines in China and it is also an important fungus in cellulose and lignin degradation with potential ability in energy production. The genome sequencing and annotation of G. lucidum are crucial for its function and comparative genomics research. Here we selected the most commonly used G. lucidum in China and sequenced its genome sequence by Solexa technology. We assembled the sequences into 634 scaffolds in 39.9 Mb sequences represented about 93.92% of the whole genome and annotated 12,080 gene models at genome level. We noticed that JGI (DOE Joint Genome Institute) also have sequenced and annotated the genome of a North American isolate G. lucidum. The predicted gene models of our sequenced genome were very similar with JGI genome annotation, which indicated the quality of our sequence and annotation were reliable.
The G. lucidum genome was characteristics of a relatively high GC content and less TEs compared with other sequenced fungi. In G. lucidum genome, we only identified 4.6% TE sequences of the genome, which was much less than other reported fungi, for example, Laccaria bicolor (21%) [20] and Tuber melanosporum (58%) [29]. To check whether the lower repeat percentage was caused by methodology or not, we had used our approaches to identify the repeat sequences in L. bicolor and found a similar percentage of repeat sequences with reported [20]. This confirmed that the TEs in G. lucidum were really much less than other fungi. Although the repeat sequence percentage was different, the genome size, gene length, and gene annotation of G. lucidum were similar by comparing with other Basidiomycota fungi.
Ganoderic acids biosynthesis pathway
Ganoderic acid (GA), a kind of triterpenoids, is the main medicinal component of G. lucidum with function of anti-tumor, immuno-reglulation, and anti-oxidant et al. [8]. Currently, studies on triterpenoid biosynthesis are mostly performed in plants; the detailed biosynthesis pathways in fungi are still unclear. Shiao [30] proved that GAs were synthesized via MVA pathway by using isotopic tracer experiments. In G. lucidum genome, we found genes only involved in MVA pathway but not MEP pathway, which is another terpenoids biosynthesis pathway in plants. Thus, at the genome level, we confirmed that the terpenoids biosynthesis of G. lucidum was only via MVA pathway, not MEP pathway. Interestingly, we identified a fusion gene (metC-MVK) in basidiomycetes, half of which was homologous to MVK enzyme in terpenoids biosynthesis and half of which was similar to metC enzyme, a cystathionine beta-lyase. In animal and even Ascomycota fungi, they were two separate genes, implying the appearance of metC-MVK occurred in the ancestor of Basidiomycota fungi. The study of the reason and function of the gene fusion is going on.
We have characterized all the enzymes in the terpenoid backbone biosynthesis in G. lucidum (Table 5). In triterpenoid synthesis, 2, 3-epoxysqualene is the precursor and the difference of cyclization, oxidation, hydroxylation and glycosylation leads to different triterpenoids [31]. However, there is little knowledge about the pathway that lies in downstream of cyclization. It has been reported that these modification are carried out by cytochrome P450, glycosyltransferases and other enzymes [31]. P450 are speculated to be involved in a wide range of modification, including oxidation and hydroxylation in the synthesis of triterpenoids [32]. Due to the large number and diversity, it is difficult to identify their specific functions based on homology. There were only four P450 genes reported to involve in the biosynthesis of triterpenoid saponins [32]–[34]. So far no P450 genes have been cloned in GAs biosynthesis of G. lucidum, we characterized 222 genes encoding P450 enzymes and 21 of them were very similar with the four known P450 genes. The plentiful of putative P450 genes provided the potential of different oxidation and hydroxylation, thus formed plentiful GAs in G. lucidum.
Glycosylation, which transfers the active saccharides to the triterpenoid backbones and alters its physiological activity [35], is the last and key modification in GAs biosynthesis. UGTs are reported to contribute the glycosylation in triterpenoid biosynthesis and so far only six UGTs are experimentally identified in the triterpenoid biosynthesis [33], [34]. By searching against known UGTs sequences [32], we found six putative UGTs in G. lucidum genome, which might be responsible for the glycosylation modification in the GAs biosynthesis in G. lucidum. Among these six putative UGTs, the gene (G_lucidum_10009504) was highly similar with UGT73K1, which glycosylated both triterpenoids and (iso)flavones in Medicago truncatula [36]. Extended searching other fungi against known UGTs sequences, we found that there were several UGT homologs in the Agaricomycotina fungi but none of them in the Ascomycota fungi. This might suggest that other glycosylation enzymes instead of UGTs were applied in the triterpenoid synthesis of ascomycetes.
Genes related to biodegradation in G. lucidum
Besides its medicinal and economic value, limited studies were performed regarding the function of biodegradation of G. lucidum so far. JGI sequenced its genome due to its ability in wood degradation and potential value in bioenergy production. In our analysis, we found that many genes might be involved in bio-degradation in G. lucidum genome. G. lucidum could degrade the major components of plant cell walls including cellulose, hemicelluloses and lignin. After predicted the CAZymes in G. lucidum, 216 putative GH genes and 56 putative GT genes were found. The number of GHs was comparatively larger than GTs. This may be related to its lifestyle, in which its survival depends on decomposed lignocelluloses, thus decomposing polysaccharides is more important than constructed. Similar phenomenon was observed in P. chrysosporium [29].
Lignin is the second most abundant renewable organic polymer and its degradation has great potential value to reproductive energy. Thus the research about lignin degradation especially in white rot fungi is increasing. We checked the FOLymes enzymes in G. lucidum and found 16 laccase (LO1), 7 peroxidases (LO2) and 9 glyoxal oxidases (LDA3) genes. Laccase is one of the most applied ligninolytic enzymes, for its broad substrate specificity and the generation of water as by-product [37]. Except P. chrysosporium had single copy of LO1 genes, all of other fungi contained more than one LO1 genes suggesting LO1 played an important role in lignin degradation of fungi. Besides its essential for wood and lignin decomposition, LO1 is also involved in other functions, such as pigments synthesis, fruiting bodies and spores formation [38]. Makela et al. reported that multiple lip, mnp and lac transcripts were coexpressed in P. radiate, indicating the potential synergy of the fungal LO1 and LO2 upon white rot fungal decay of wood [39].
LO2 mainly existed in white rot and wood-colonising basidiomycetes (P. chrysosporium, P. ostreatus, and G. lucidum). It has been reported that glyoxal oxidases (LDA3) were inactive unless they coupled to LO2 reaction [40]. Here, we identified 9 LDA3 in G. lucidum. Considering the total number of LO2 and LDA3 genes in all fungi we analyzed, P. ostreatus (24 genes) and P. chrysosporium (23 genes) were the top two fungi and were reported with strong ligninolytic ability. G. lucidum had the third largest number of LO2 and LDA3 genes (16 genes), which may suggest its strong ligninolytic ability. In addition, in G. lucidum genome we also found 10 LDA1 gene, which could reduce the level of radical compounds and quinonoids produced by LO1, leading an oxidative enzyme system with LO1 [41].
Moreover, G. lucidum showed rich P450 family, abundant GST enzymes (mainly Omega class) and abundant genes involved in “Xenobiotics Biodegradation and Metabolism” pathways. Some of these pathways are related to the degradation of refractory compounds, such as dioxin and naphthalene. These enzymes have potential to degrade various industrial pollutants [42]. In view of the extensive contamination of the environment by persistent and toxic chemical pollutants, the utility of the degradation of fungi including G. lucidum may be an attractive and effective approach on pollution controlling.
Materials and Methods
Strains and culture conditions
A fruiting body of G. lucidum was collected from oak at Hengshan, Hunan province, China, on May 21th, 2001. No specific permits were required for the described field studies. We confirmed that the location was not privately-owned or protected in any way. The G. lucidum was deposited at the edible fungi institute of Hunan agricultural university (Changsha, Hunan, China). Basidiospores from the fruiting body of G. lucidum were collected by hood. Single spores were separated by micro-manipulation of basidiospores and allowed to geminate on PDA (Potato Dextrose Agar) enrichment medium (20% potato, 2% dextrose, 0.2% yeast extract, 0.2% peptone,0.3% monopotassium phosphate, 0.15% magnesium sulfate and 2% agar) at 25°C, dark. Germination started 4–8 days after plating. After 3–5 days growth, clamp connection was observed by using optical microscopy. Forty-six single basidiospores with no clamp connection were isolated and transferred individually to fresh dishes, sealed and stored at 4°C after 10–15 days growth. For genomic DNA isolation, single basidiospore named P9 which grew well was cultured in potato dextrose agar enrichment medium without agar at 25°C, dark, shaken at 120 r/min, for 8–10 days.
DNA isolation, genome sequencing and assembly
Genomic DNA of G. lucidum was isolated by improved cetyl trimethylammonium bromide (CTAB) method [43] and sequenced using a whole-genome shotgun strategy. All data were generated by paired-end sequencing of cloned inserts with two different insert sizes (200 bp, 6000 bp) using Illumina Hiseq2000 Sequencer at BGI-Shenzhen. After removing the low complexity, low quality, adapter and duplication contamination raw reads, the clean reads were assembled using the whole-genome do novo assembler SOAPdenovo [22].
Annotation methods
Protein coding gene models were predicted using de novo prediction tools Genscan [44],Augustus [45] and GeneMark-ES [46] and homology based gene prediction tool Genewise [47] with the default parameters. The homology-based and de novo gene sets were merged to form a comprehensive and non-redundant reference gene set by Glean [48].
The functionally annotation of predicted gene models were mainly based on homology to known annotated genes and BLAST was the mainly used tool in our analyses. We aligned all protein models by BLASTP to SwissProt, TrEMBL, and NCBI nr, InterPro, Pfam [49] and also mapped them onto functional terms, including GO [50], COGs [51] and KEGG pathways [52] (BLASTP cut-off e value<1e-7). Since each gene mapped to different database sequences, there may be multiple aligned results meeting the cut-off, the annotations of the sequences with the best score were chosen to be the annotation of the gene in G. lucidum.
Transposons were identified by aligning the assembled results with the known sequences of the transposon library. The specific method was made through the RepeatMasker software (http://www.repeatmasker.org, using Repbase database [53]), RepeatProteinMasker software (using the transposon protein library that comes with RepeatMasker) and 3 other tools LTR-FINDER [54], RepeatScout [55] and PILER [56] with default parameters. Tandem Repeat Finder [57] software was used to predict tandem repeats.
rRNAs were identified by BLAST against the rRNA libraries or predicted by using rRNAmmer [58] software. tRNAscan-SE [59] software was used to detect tRNA regions and its secondary structures. Other non-coding RNAs such as miRNA, sRNA and snRNA were predicted by Rfam.
KEGG pathway analysis
To compare the pathway annotation of G. lucidum with other fungi, we also mapped genes in other fungal genomes to KEGG database. The Basidiomycota comprises three taxa (agaricomycotina, ustilaginomycotina and pucciniomycotina). The Ascomycota includes four taxa– eurotiomycetes, dothideomycetes, sordariomycetes and saccharomycotina. For comparative genome analysis, we selected 12 basidiomycetes, 5 ascomycota and downloaded their genomic gene models [20], [29], [60] (File S1). Then, we compared the number of genes in each KEGG terms among all these genomes. Terms in KEGG are divided into four layers. The first layer consists of seven sections, including “Metabolism”, “Genetic information processing”, “Environmental information processing” and so on. Each section is further divided into several small entries, which are the second layers. The third layer is the specific pathway map and the fourth layer includes specific genes in each pathway. We compared the gene distribution of these species in the second layer and investigated the sub-terms (e.g. KO in KEGG pathway) if the term had significant different gene numbers in the second layer.
Because many P450 genes were involved in KEGG pathway, we compared P450 genes at the genomic level. All genome sequences were aligned to fungi P450 sequences in Cytochrome P450 database (cut-off e-value: 1e-10 and identity>30%). For glutathione S-transferase (GST) gene, we mapped all genomic genes to GSTs genes described in literature [25], which classified fungal GSTs into different sub families based on phylogenetic analysis (BLASTP cut-off e-value:1e-10 and identity>30%).
Phylogenetic tree construction and gene families
First, an all-to-all BLASTP alignment was programmed for G. lucidum and other 18 species (BLASTP cut-off e-value<1e-7) and core genes which were single-copy in all species were extracted. Then, following a certain order, all core genes of each species were made multiple sequence alignment using the MUSCLE Software and then were concatenated into a super sequence. The Neighbor Joining tree (NJ) was constructed with 1,000 bootstrap replications from aligned sequences by MEGA-5.05 [61]. Based on the all-to-all BLASTP results, Tribe-MCL tools [62] were used to generate the multigene families with default parameters (inflation parameter = 3) in G. lucidum and other 7 Agaricomycotina fungi (F. pinicol, G. trabeum, S. hirstutum, P. chrysosporium, S. commune, L. bicolor, and C. cinerea). The gene families containing genes from only one species were considered as lineage specific. For other gene families (> = 2 members), the evolutionary changes in the protein family size were analyzed using the CAFE program [63], which assesses the protein family expansion or contraction based on the topology of the phylogenetic tree.
Carbohydrate-related enzymes (CAZymes) and Lignin oxidative enzymes (FOLymes) annotation
As abundant putative genes involved in carbohydrate metabolites and xenobiotics biodegradation in our KEGG analysis, annotations of CAZymes (http://www.cazy.org/) [28] and FOLymes (http://foly.esil.univ-mrs.fr/) [17] were performed using BLASTP analyisis (e-value<1e-10) in G. lucidum and other fungi against libraries of CAZy and FOLy database. Refer to [64], a protein was identified as a CAZyme/FOLyme when it showed a similarity score above 50% with sequences of biochemically characterized enzymes. Because FOLyme database is under construction currently, we used genes provided in its publication as seed sequences. In addition, because of the update of database and different parameter chose, the predicted numbers of CAZymes/FOLymes may have a few differences with previous reports.
Data availability and accession numbers
Data from this Whole Genome Shotgun project have been deposited at DDBJ/EMBL/GenBank (http://www.ncbi.nlm.nih.gov/) under the accession no. AHGX00000000. The version described in this paper is the first version, AHGX01000000. Raw sequencing data have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRA048091 and study accession no. SRP009345.
Supporting Information
Table S1. The codon and anti-codon usage of G. lucidum . Table S2. G. lucidum gene annotation. Table S3. Fungi used in the study and their download websites. Table S4. The gene distribution of each fungus in each pathway. Table S5. The number of genes in each gene family and each organism. Table S6. G. lucidum specific gene families as compared to other fungi. Table S7. Protein families showing the highest rate of expansion in G. lucidum genome as compared to other fungi. Table S8. Protein families showing lower size in G. lucidum genome as compared to other fungi. Table S9. The number of genes related vegetative incompatibility in each fungus.
(XLS)
Figure S1. The frequencies of codon usage and anti-codon usage. Figure S2. “Terpenoid backbone biosynthesis” pathway of G. lucidum .
(DOC)
Acknowledgments
We thank the Edible Fungi Institute of Hunan Agricultural University for kind help with samples. We thank the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the user community and Broad Institute of Harvard and MIT (http://www.broadinstitute.org/) for providing most of the sequencing data of other fungi used in this study. We also acknowledge helpful comments, suggestions and manuscript reading and English corrections provided by anonymous reviewers.
Footnotes
Competing Interests: The authors have the following interests to declare: JG, HMZ, and WL are interns of the Nextomics Biosciences company and DW is an employee of this company. ZH is an employee of the Hunan Wanyuan Bio-tech company. There are no patents, products in development or marketed products to declare. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: Funding of this project was provided by the programs National Program on Key Basic Research Project (973 Program, 2012CB723000 to DBL), Key Projects in the National Science & Technology Program (2012BAD33B00 to DBL), and National Natural Science Foundation of China (31171271 to AYG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lin Z. The Modern Research of Ganoderma Lucidum. Beijing: Peking University Medical Press; 2007. [Google Scholar]
- 2.JiDing Z. Chinese Ganoderma lucidum new edition. Beijing: Science Press; 1989. 32 [Google Scholar]
- 3.Lin ZB. Cellular and molecular mechanisms of immuno-modulation by Ganoderma lucidum. J Pharmacol Sci. 2005;99:144–153. doi: 10.1254/jphs.crj05008x. [DOI] [PubMed] [Google Scholar]
- 4.Yuen JW, Gohel MD. Anticancer effects of Ganoderma lucidum: a review of scientific evidence. Nutr Cancer. 2005;53:11–17. doi: 10.1207/s15327914nc5301_2. [DOI] [PubMed] [Google Scholar]
- 5.Lin ZB, Zhang HN. Anti-tumor and immunoregulatory activities of Ganoderma lucidum and its possible mechanisms. Acta Pharmacol Sin. 2004;25:1387–1395. [PubMed] [Google Scholar]
- 6.Lee SY, Rhee HM. Cardiovascular effects of mycelium extract of Ganoderma lucidum: inhibition of sympathetic outflow as a mechanism of its hypotensive action. Chem Pharm Bull (Tokyo) 1990;38:1359–1364. doi: 10.1248/cpb.38.1359. [DOI] [PubMed] [Google Scholar]
- 7.Chu TT, Benzie IF, Lam CW, Fok BS, Lee KK, et al. Study of potential cardioprotective effects of Ganoderma lucidum (Lingzhi): results of a controlled human intervention trial. Br J Nutr. 2011:1–11. doi: 10.1017/S0007114511003795. [DOI] [PubMed] [Google Scholar]
- 8.Shi L, Ren A, Mu D, Zhao M. Current progress in the study on biosynthesis and regulation of ganoderic acids. Appl Microbiol Biotechnol. 2010;88:1243–1251. doi: 10.1007/s00253-010-2871-1. [DOI] [PubMed] [Google Scholar]
- 9.Sijia Zhao XC, Chen Ping. Comparison of quality of cultured Ganoderma lucidum from six leading producer. 2011. Proceedings of the 2011 International Meeting on Ganoderma Research.
- 10.Shang CH, Zhu F, Li N, Ou-Yang X, Shi L, et al. Cloning and characterization of a gene encoding HMG-CoA reductase from Ganoderma lucidum and its functional identification in yeast. Biosci Biotechnol Biochem. 2008;72:1333–1339. doi: 10.1271/bbb.80011. [DOI] [PubMed] [Google Scholar]
- 11.Ding YX, Ou-Yang X, Shang CH, Ren A, Shi L, et al. Molecular cloning, characterization, and differential expression of a farnesyl-diphosphate synthase gene from the basidiomycetous fungus Ganoderma lucidum. Biosci Biotechnol Biochem. 2008;72:1571–1579. doi: 10.1271/bbb.80067. [DOI] [PubMed] [Google Scholar]
- 12.Zhao MW, Liang WQ, Zhang DB, Wang N, Wang CG, et al. Cloning and characterization of squalene synthase (SQS) gene from Ganoderma lucidum. J Microbiol Biotechnol. 2007;17:1106–1112. [PubMed] [Google Scholar]
- 13.Shang CH, Shi L, Ren A, Qin L, Zhao MW. Molecular cloning, characterization, and differential expression of a lanosterol synthase gene from Ganoderma lucidum. Biosci Biotechnol Biochem. 2010;74:974–978. doi: 10.1271/bbb.90833. [DOI] [PubMed] [Google Scholar]
- 14.Martinez AT, Speranza M, Ruiz-Duenas FJ, Ferreira P, Camarero S, et al. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int Microbiol. 2005;8:195–204. [PubMed] [Google Scholar]
- 15.Wang HX, Ng TB. A laccase from the medicinal mushroom Ganoderma lucidum. Appl Microbiol Biotechnol. 2006;72:508–513. doi: 10.1007/s00253-006-0314-9. [DOI] [PubMed] [Google Scholar]
- 16.Murugesan K, Yang IH, Kim YM, Jeon JR, Chang YS. Enhanced transformation of malachite green by laccase of Ganoderma lucidum in the presence of natural phenolic compounds. Appl Microbiol Biotechnol. 2009;82:341–350. doi: 10.1007/s00253-008-1819-1. [DOI] [PubMed] [Google Scholar]
- 17.Levasseur A, Piumi F, Coutinho PM, Rancurel C, Asther M, et al. FOLy: an integrated database for the classification and functional annotation of fungal oxidoreductases potentially involved in the degradation of lignin and related aromatic compounds. Fungal Genet Biol. 2008;45:638–645. doi: 10.1016/j.fgb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 19.Stajich JE, Wilke SK, Ahren D, Au CH, Birren BW, et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci U S A. 2010;107:11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin F, Aerts A, Ahren D, Brun A, Danchin EG, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 21.Martin F, Kohler A, Murat C, Balestrini R, Coutinho PM, et al. Perigord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature. 2010;464:1033–1038. doi: 10.1038/nature08867. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Zhu H, Ruan J, Qian W, Fang X, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crick FH. Codon–anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 24.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 25.Morel M, Ngadin AA, Droux M, Jacquot JP, Gelhaye E. The fungal glutathione S-transferase system. Evidence of new classes in the wood-degrading basidiomycete Phanerochaete chrysosporium. Cell Mol Life Sci. 2009;66:3711–3725. doi: 10.1007/s00018-009-0104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loubradou G, Turcq B. Vegetative incompatibility in filamentous fungi: a roundabout way of understanding the phenomenon. Res Microbiol. 2000;151:239–245. doi: 10.1016/s0923-2508(00)00145-5. [DOI] [PubMed] [Google Scholar]
- 27.Saupe S, Descamps C, Turcq B, Begueret J. Inactivation of the Podospora anserina vegetative incompatibility locus het-c, whose product resembles a glycolipid transfer protein, drastically impairs ascospore production. Proc Natl Acad Sci U S A. 1994;91:5927–5931. doi: 10.1073/pnas.91.13.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 30.Shiao MS. Natural products of the medicinal fungus Ganoderma lucidum: occurrence, biological activities, and pharmacological functions. Chem Rec. 2003;3:172–180. doi: 10.1002/tcr.10058. [DOI] [PubMed] [Google Scholar]
- 31.Haralampidis K, Trojanowska M, Osbourn AE. Biosynthesis of triterpenoid saponins in plants. Adv Biochem Eng Biotechnol. 2002;75:31–49. doi: 10.1007/3-540-44604-4_2. [DOI] [PubMed] [Google Scholar]
- 32.Augustin JM, Kuzina V, Andersen SB, Bak S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 2011;72:435–457. doi: 10.1016/j.phytochem.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Sun C, Li Y, Wu Q, Luo H, Sun Y, et al. De novo sequencing and analysis of the American ginseng root transcriptome using a GS FLX Titanium platform to discover putative genes involved in ginsenoside biosynthesis. BMC Genomics. 2010;11:262. doi: 10.1186/1471-2164-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Q, Ma X, Mo C, Wilson IW, Song C, et al. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis. BMC Genomics. 2011;12:343. doi: 10.1186/1471-2164-12-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hefner T, Arend J, Warzecha H, Siems K, Stockigt J. Arbutin synthase, a novel member of the NRD1beta glycosyltransferase family, is a unique multifunctional enzyme converting various natural products and xenobiotics. Bioorg Med Chem. 2002;10:1731–1741. doi: 10.1016/s0968-0896(02)00029-9. [DOI] [PubMed] [Google Scholar]
- 36.Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, et al. Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 2005;41:875–887. doi: 10.1111/j.1365-313X.2005.02344.x. [DOI] [PubMed] [Google Scholar]
- 37.Canas AI, Camarero S. Laccases and their natural mediators: biotechnological tools for sustainable eco-friendly processes. Biotechnol Adv. 2010;28:694–705. doi: 10.1016/j.biotechadv.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Lundell TK, Makela MR, Hilden K. Lignin-modifying enzymes in filamentous basidiomycetes–ecological, functional and phylogenetic review. J Basic Microbiol. 2010;50:5–20. doi: 10.1002/jobm.200900338. [DOI] [PubMed] [Google Scholar]
- 39.Makela MR, Hilden KS, Hakala TK, Hatakka A, Lundell TK. Expression and molecular properties of a new laccase of the white rot fungus Phlebia radiata grown on wood. Curr Genet. 2006;50:323–333. doi: 10.1007/s00294-006-0090-1. [DOI] [PubMed] [Google Scholar]
- 40.Kersten PJ. Glyoxal oxidase of Phanerochaete chrysosporium: its characterization and activation by lignin peroxidase. Proc Natl Acad Sci U S A. 1990;87:2936–2940. doi: 10.1073/pnas.87.8.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marzullo L, Cannio R, Giardina P, Santini MT, Sannia G. Veratryl alcohol oxidase from Pleurotus ostreatus participates in lignin biodegradation and prevents polymerization of laccase-oxidized substrates. J Biol Chem. 1995;270:3823–3827. doi: 10.1074/jbc.270.8.3823. [DOI] [PubMed] [Google Scholar]
- 42.Asgher M, Bhatti HN, Ashraf M, Legge RL. Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation. 2008;19:771–783. doi: 10.1007/s10532-008-9185-3. [DOI] [PubMed] [Google Scholar]
- 43.Kang XC, Liu DB, Xia ZL, Chen F. Study on Genomic genomic DNA Extraction extraction from Cordyceps Militaris. Journal of Hunan Agricultural University. 2011;37:147–149. [Google Scholar]
- 44.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 46.Borodovsky M, Lomsadze A, Ivanov N, Mills R. Eukaryotic gene prediction using GeneMark.hmm. Curr Protoc Bioinformatics. 2003;Chapter 4:Unit4 6. doi: 10.1002/0471250953.bi0406s01. [DOI] [PubMed] [Google Scholar]
- 47.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elsik CG, Mackey AJ, Reese JT, Milshina NV, Roos DS, et al. Creating a honey bee consensus gene set. Genome Biol. 2007;8:R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 54.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 56.Edgar RC, Myers EW. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21(Suppl 1):i152–158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- 57.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, et al. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 2010;38:D420–427. doi: 10.1093/nar/gkp751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 64.Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The codon and anti-codon usage of G. lucidum . Table S2. G. lucidum gene annotation. Table S3. Fungi used in the study and their download websites. Table S4. The gene distribution of each fungus in each pathway. Table S5. The number of genes in each gene family and each organism. Table S6. G. lucidum specific gene families as compared to other fungi. Table S7. Protein families showing the highest rate of expansion in G. lucidum genome as compared to other fungi. Table S8. Protein families showing lower size in G. lucidum genome as compared to other fungi. Table S9. The number of genes related vegetative incompatibility in each fungus.
(XLS)
Figure S1. The frequencies of codon usage and anti-codon usage. Figure S2. “Terpenoid backbone biosynthesis” pathway of G. lucidum .
(DOC)
Data Availability Statement
Data from this Whole Genome Shotgun project have been deposited at DDBJ/EMBL/GenBank (http://www.ncbi.nlm.nih.gov/) under the accession no. AHGX00000000. The version described in this paper is the first version, AHGX01000000. Raw sequencing data have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRA048091 and study accession no. SRP009345.