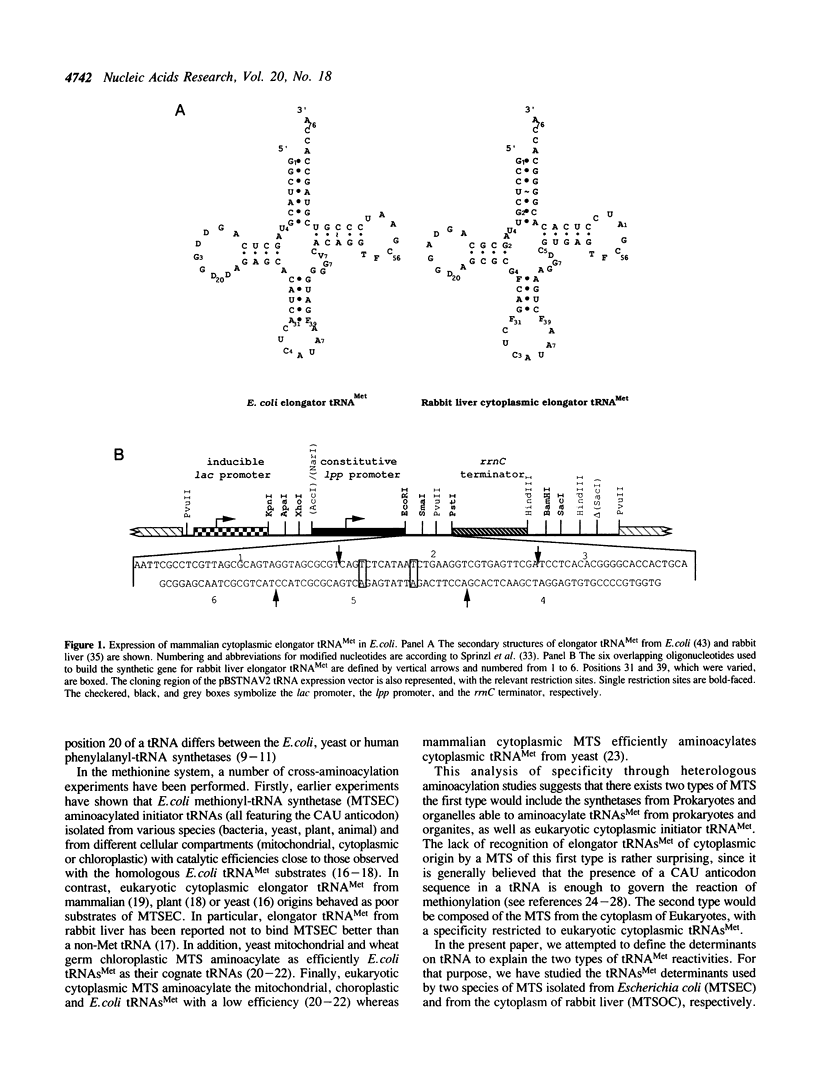
Abstract
The rates of the cross-aminoacylation reactions of tRNAs(Met) catalyzed by methionyl-tRNA synthetases from various organisms suggest the occurrence of two types of tRNA(Met)/methionyl-tRNA synthetase systems. In this study, the tRNA determinants recognized by mammalian or E. coli methionyl-tRNA synthetases, which are representative members of the two types, have been examined. Like its prokaryotic counterpart, the mammalian enzyme utilizes the anticodon of tRNA as main recognition element. However, the mammalian cytoplasmic elongator tRNA(Met) species is not recognized by the bacterial synthetase, and both the initiator and elongator E. coli tRNA(Met) behave as poor substrates of the mammalian cytoplasmic synthetase. Synthetic genes encoding variants of tRNAs(Met), including the elongator one from mammals, were expressed in E. coli. tRNAs(Met) recognized by a synthetase of a given type can be converted into a substrate of an enzyme of the other type by introducing one-base substitutions in the anticodon loop or stem. In particular, a reduction of the size of the anticodon loop of cytoplasmic mammalian elongator tRNA(Met) from 9 to 7 bases, through the creation of an additional Watson-Crick pair at the bottom of the anticodon stem, makes it a substrate of the prokaryotic enzyme and decreases its ability to be methionylated by the mammalian enzyme. Moreover, enlarging the size of the anticodon loop of E. coli tRNA(Metm) from 7 to 9 bases, by disrupting the base pair at the bottom of the anticodon stem, renders the resulting tRNA a good substrate of the mammalian enzyme, while strongly altering its reaction with the prokaryotic synthetase. Finally, E. coli tRNA(Metf) can be rendered a better substrate of the mammalian enzyme by changing its U33 into a C. This modification makes the sequence of the anticodon loop of tRNA(Metf) identical to that of cytoplasmic initiator tRNA(Met).
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blanquet S., Iwatsubo M., Waller J. P. The mechanism of action of methionyl-tRNA synthetase from Escherichia coli. 1. Fluorescence studies on tRNAMet binding as a function of ligands, ions and pH. Eur J Biochem. 1973 Jul 2;36(1):213–226. doi: 10.1111/j.1432-1033.1973.tb02903.x. [DOI] [PubMed] [Google Scholar]
- Blanquet S., Petrissant G., Waller J. P. The mechanism of action of methionyl-tRNA synthetase. 2. Interaction of the enzyme with specific and unspecific tRNAs. Eur J Biochem. 1973 Jul 2;36(1):227–233. doi: 10.1111/j.1432-1033.1973.tb02904.x. [DOI] [PubMed] [Google Scholar]
- Carias J. R., Mouricout M., Julien R. Chloroplastic methionyl-tRNA synthetase from wheat. Biochem Biophys Res Commun. 1981 Feb 12;98(3):735–742. doi: 10.1016/0006-291x(81)91174-8. [DOI] [PubMed] [Google Scholar]
- Cigan A. M., Feng L., Donahue T. F. tRNAi(met) functions in directing the scanning ribosome to the start site of translation. Science. 1988 Oct 7;242(4875):93–97. doi: 10.1126/science.3051379. [DOI] [PubMed] [Google Scholar]
- Cory S., Marcker K. A. The nucleotide sequence of methionine transfer RNA-M. Eur J Biochem. 1970 Jan;12(1):177–194. doi: 10.1111/j.1432-1033.1970.tb00836.x. [DOI] [PubMed] [Google Scholar]
- Cummins C. M., Culbertson M. R., Knapp G. Frameshift suppressor mutations outside the anticodon in yeast proline tRNAs containing an intervening sequence. Mol Cell Biol. 1985 Jul;5(7):1760–1771. doi: 10.1128/mcb.5.7.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987 Dec 11;238(4833):1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- Edwards H., Schimmel P. A bacterial amber suppressor in Saccharomyces cerevisiae is selectively recognized by a bacterial aminoacyl-tRNA synthetase. Mol Cell Biol. 1990 Apr;10(4):1633–1641. doi: 10.1128/mcb.10.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards H., Schimmel P. An E. coli aminoacyl-tRNA synthetase can substitute for yeast mitochondrial enzyme function in vivo. Cell. 1987 Nov 20;51(4):643–649. doi: 10.1016/0092-8674(87)90133-4. [DOI] [PubMed] [Google Scholar]
- Edwards H., Trézéguet V., Schimmel P. An Escherichia coli tyrosine transfer RNA is a leucine-specific transfer RNA in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1153–1156. doi: 10.1073/pnas.88.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon J. M., Meinnel T., Mechulam Y., Lazennec C., Blanquet S., Fayat G. Nucleotides of tRNA governing the specificity of Escherichia coli methionyl-tRNA(fMet) formyltransferase. J Mol Biol. 1992 Mar 20;224(2):359–367. doi: 10.1016/0022-2836(92)91000-f. [DOI] [PubMed] [Google Scholar]
- Hasegawa T., Himeno H., Ishikura H., Shimizu M. Discriminator base of tRNA(Asp) is involved in amino acid acceptor activity. Biochem Biophys Res Commun. 1989 Sep 29;163(3):1534–1538. doi: 10.1016/0006-291x(89)91154-6. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry. 1989 Aug 22;28(17):6800–6804. doi: 10.1021/bi00443a003. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y., Himeno H., Miura K., Watanabe K. Unilateral aminoacylation specificity between bovine mitochondria and eubacteria. J Biochem. 1991 Mar;109(3):421–427. doi: 10.1093/oxfordjournals.jbchem.a123397. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y., Yokogawa T., Hasegawa E., Miura K., Watanabe K. The aminoacylation of structurally variant phenylalanine tRNAs from mitochondria and various nonmitochondrial sources by bovine mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 1989 Aug 5;264(22):13005–13011. [PubMed] [Google Scholar]
- Martinis S. A., Schimmel P. Enzymatic aminoacylation of sequence-specific RNA minihelices and hybrid duplexes with methionine. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):65–69. doi: 10.1073/pnas.89.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Blanquet S., Fayat G. Binding of the anticodon domain of tRNA(fMet) to Escherichia coli methionyl-tRNA synthetase. J Mol Biol. 1991 Jul 20;220(2):205–208. doi: 10.1016/0022-2836(91)90003-o. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Fayat G. Fast purification of a functional elongator tRNAmet expressed from a synthetic gene in vivo. Nucleic Acids Res. 1988 Aug 25;16(16):8095–8096. doi: 10.1093/nar/16.16.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Le Corre D., Panvert M., Blanquet S., Fayat G. Selection of suppressor methionyl-tRNA synthetases: mapping the tRNA anticodon binding site. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):291–295. doi: 10.1073/pnas.88.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellot P., Mechulam Y., Le Corre D., Blanquet S., Fayat G. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J Mol Biol. 1989 Aug 5;208(3):429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- Mirande M., Cirakoğlu B., Waller J. P. Seven mammalian aminoacyl-tRNA synthetases associated within the same complex are functionally independent. Eur J Biochem. 1983 Mar 1;131(1):163–170. doi: 10.1111/j.1432-1033.1983.tb07244.x. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988 Nov 10;336(6195):179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Nazarenko I. A., Peterson E. T., Zakharova O. D., Lavrik O. I., Uhlenbeck O. C. Recognition nucleotides for human phenylalanyl-tRNA synthetase. Nucleic Acids Res. 1992 Feb 11;20(3):475–478. doi: 10.1093/nar/20.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrissant G., Boisnard M., Puissant C. Purification d'un tRNA accepteur de la méthionine dans le foie de lapin. Biochim Biophys Acta. 1970 Jul 16;213(1):223–225. [PubMed] [Google Scholar]
- Pétrissant G., Boisnard M. Particularités structurales du méthionin-tRNAmMet de foie de lapin. Biochimie. 1974;56(5):787–790. doi: 10.1016/s0300-9084(74)80053-2. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- RajBhandary U. L., Ghosh H. P. Studies on polynucleotides. XCI. Yeast methionine transfer ribonucleic acid: purification, properties, and terminal nucleotide sequences. J Biol Chem. 1969 Mar 10;244(5):1104–1113. [PubMed] [Google Scholar]
- Roe B., Sirover M., Dudock B. Kinetics of homologous and heterologous aminoacylation with yeast phenylalanyl transfer ribonucleic acid synthetase. Biochemistry. 1973 Oct 9;12(21):4146–4154. doi: 10.1021/bi00745a018. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Behlen L. S., DiRenzo A. B., Uhlenbeck O. C. Recognition of yeast tRNA(Phe) by its cognate yeast phenylalanyl-tRNA synthetase: an analysis of specificity. Biochemistry. 1992 May 5;31(17):4161–4167. doi: 10.1021/bi00132a002. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989 Mar 10;243(4896):1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- Schneller J. M., Schneider C., Stahl A. J. Distinct nuclear genes for yeast mitochondrial and cytoplasmic methionyl-tRNA synthetases. Biochem Biophys Res Commun. 1978 Dec 29;85(4):1392–1399. doi: 10.1016/0006-291x(78)91158-0. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon loop size and sequence requirements for recognition of formylmethionine tRNA by methionyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6755–6759. doi: 10.1073/pnas.80.22.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Schulman L. H. Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1991;41:23–87. [PubMed] [Google Scholar]
- Schwob E., Sanni A., Fasiolo F., Martin R. P. Purification of the yeast mitochondrial methionyl-tRNA synthetase. Common and distinctive features of the cytoplasmic and mitochondrial isoenzymes. Eur J Biochem. 1988 Dec 1;178(1):235–242. doi: 10.1111/j.1432-1033.1988.tb14448.x. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trézéguet V., Edwards H., Schimmel P. A single base pair dominates over the novel identity of an Escherichia coli tyrosine tRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1991 May;11(5):2744–2751. doi: 10.1128/mcb.11.5.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Woo N. H., Rich A. Initiator tRNAs have a unique anticodon loop conformation. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3289–3293. doi: 10.1073/pnas.76.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]



