Abstract
Background: Human leukocyte antigen-G (HLA-G) molecules play a prominent role in immune tolerance. Structurally similar to their classical HLA homologs, they are distinct by having high rate of polymorphism in the non-coding regions including a functionally relevant 14-base pair (bp) insertion/deletion (Ins/Del) allele in the 3′ untranslated region (3′UTR), rarely examined in a hematopoietic stem cell transplantation (HSCT) setting. Here, we analyzed the potential impact of HLA-G Ins/Del dimorphism on the incidence of acute graft-versus-host disease (aGvHD), transplant-related mortality (TRM), overall survival (OS), and incidence of relapse after HSCT using bone marrow (BM) as stem cell source from HLA-matched donors. Methods: One hundred fifty-seven sibling pairs, who had undergone HSCT, were studied for the distribution of the HLA-G 14 bp Ins/Del polymorphism using a polymerase chain reaction (PCR)-based technique. Potential genetic association with the incidence of aGvHD, TRM, and OS was analyzed by monovariate and multivariate analyses. Results: Monovariate analysis showed that the homozygous state for the 14-bp Ins allele is a risk factor for severe aGvHD (grade III and IV; P = 0.008), confirmed subsequently by multivariate analysis [hazard ratio (HR) = 3.5; 95% confidence interval (95%CI) = 1.3–9.5; P = 0.012]. We did not find any association between HLA-G polymorphism and the other studied complications. Conclusion: Our data suggest that the HLA-G low expressor 14 bp Ins allele constitutes a risk factor for the incidence of severe aGvHD in patients who received BM as stem cell source.
Keywords: hematopoietic stem cell transplantation, HLA-G polymorphism, acute GvHD
Introduction
In hematopoietic stem cell transplantation (HSCT), substantial advances in conditioning regimens, prophylactic measures and treatments of post-transplant complications have been made in combination with an improved donor/recipient (D/R) HLA genetic profiling. Despite such progresses, impending, and unpredictable severity of acute graft-versus-host disease (aGvHD), remains a major complication (Dickinson and Charron, 2005; Messina et al., 2008; Socié and Blazar, 2009). Consequently, much attention has been focused on genetic mechanisms that govern the transplant tolerance. Among the pivotal actors of such immuno-modulation, the human leukocyte antigen-G (HLA-G) molecules are of great interest because of their various immunosuppressive/immune tolerogenic properties such as, inhibition of both natural killer (NK) cytotoxicity (Rouas-Freiss et al., 1997) and antigen-specific cytotoxic CD8 + T cell (CTL) functions (Wiendl et al., 2002) as well as CD4 + T cell allogeneic proliferation (Bainbridge et al., 2000).
Belonging to the non-classical HLA-class Ib family and encoded by a locus mapped telomeric to HLA-A gene, the HLA-G molecules are structurally similar to their classical counterparts, yet are distinct by the following characteristics: limited tissue distribution in physiological conditions, diversity of isoforms generated by alternative splicing, i.e., four membrane-bound (HLA-G1 to -G4) and three soluble (HLA-G5 to -G7) isoforms (sHLA-G; Paul et al., 2000) and unique pattern of polymorphisms in the non-coding regions especially within the promoter and the 3′-untranslated region (3′UTR; Larsen and Hviid, 2009). As identified to date, HLA-G gene presents a limited number of exonic polymorphisms with 46 alleles accounting for 15 protein variants (IMGT/HLA Sequence Database1). Each of these alleles bears either a 14-base pair (bp) insertion (Ins) or deletion (Del) polymorphism in the 3′ UTR which influences the HLA-G expression. Indeed, the insertion allele (+14 bp), albeit initially described to improve HLA-G mRNA stability (O’Brien et al., 2001; Hviid et al., 2003; Rousseau et al., 2003), was subsequently demonstrated to be associated with low levels of HLA-G mRNA and serum sHLA-G isoforms (Chen et al., 2008; Rizzo et al., 2008) with likely consequences on functional properties of HLA-G molecules.
Given such genotype-dependant phenotype expression status, studies have focused on the HLA-G dimorphism in various clinical settings including gestational complications, auto-immunity, infections, cancers, and solid organ transplantation (Larsen and Hviid, 2009). In a HSCT setting, three studies have addressed the potential role of HLA-G in the development of post-transplant complications (La Nasa et al., 2007; Le Maux et al., 2008; Chiusolo et al., 2011), two exploring the 14-bp dimorphism-related diversity and one evaluating the pre- and post-transplant level of sHLA-G molecules, both in relation with the incidence of aGvHD. However, conducted on small sample size of D/R cohorts, in different HSCT study design, these studies generated conflictory data regarding the role of HLA-G in HSCT outcomes. To clarify the issue, in this retrospective study, we have analyzed the HLA-G 14 bp Ins/Del polymorphism in a large cohort of patients who have undergone HSCT in relation with aGvHD, transplant-related mortality (TRM), relapse, and overall survival (OS).
Materials and Methods
Study population
One hundred fifty-seven consecutive patients who underwent non-T-cell depleted allogeneic HSCT were included in this study. All of them received bone marrow (BM) from HLA-identical sibling donors as stem cell source and were recruited between January 1995 and January 2004 in the BM Transplant Department of Saint-Louis Hospital, Paris, France. It is of interest to note that 136 patients (87%) were transplanted for hematological malignant disorders. The detailed characteristics of D/R pairs are given in Table 1. The supportive therapy as well as the criteria used to define the outcomes namely aGvHD, TRM, and OS were as previously detailed in a study involving a part of the present cohort (Rocha et al., 2002). It is important to precise that the main outcome, aGvHD, was defined according to modified Glucksberg’s criteria (Glucksberg et al., 1974; Przepiorka et al., 1995). The vast majority of patients (144/157, 92%) received the same GvHD prophylaxis protocol with cyclosporine A (CYA A) and pulsed doses of methotrexate (MTX). Informed consent was obtained according to the declaration of Helsinki.
Table 1.
Patients, disease, and transplant characteristics.
| Characteristics | n = 157 |
|---|---|
| RECIPIENTS | |
| Median age, years | 28.4 |
| Male (%) | 91 (58%) |
| Children, 15 year or younger (%) | 39 (25%) |
| Positive CMV serology (%) | 94 (60%) |
| HLA-G | |
| 14 Heterozygote | 76 (48%) |
| 14− Homozygote | 49 (31%) |
| 14+ Homozygote | 32 (21%) |
| Underlying diagnosis | |
| Chronic leukemia (%) | 31 (20%) |
| Acute leukemia (%) | 80 (51%) |
| Other malignant disorders (%) | 25 (16%) |
| Non-malignant disorder | 21 (13%) |
| Disease status for malignant disorders | |
| Early (%) | 107 (79%) |
| Intermediate (%) | 13 (10%) |
| Advanced (%) | 15 (11%) |
| DONORS | |
| Median age | 29.9 |
| Male (%) | 83 (53%) |
| Female donor to male recipient (%) | 43 (27%) |
| ABO major incompatibility (%) | 24 (15%) |
| Positive CMV serology (%) | 80 (51%) |
| HLA-G | |
| 14 Heterozygote | 76 (48%) |
| 14− Homozygote | 49 (31%) |
| 14+ Homozygote | 32 (21%) |
| TRANSPLANTATION | |
| GVHD prophylaxis | |
| CYA A | 5 (3%) |
| CYA A + MTX | 144 (92%) |
| CYA A + others | 3 (2%) |
| Conditioning | |
| TBI-based | 60 (38%) |
| Bu-based | 97 (62%) |
CMV, cytomegalovirus; CYA A, cyclosporin A; MTX, methotrexate; TBI, total body irradiation; Bu, Busulfan.
HLA-G genotyping
Both donors and recipients were tested for HLA-G genotype. Genomic DNA was extracted from EDTA-treated peripheral blood samples using the standard salting out procedure (John et al., 1991). The 14-bp Ins/Del polymorphism (rs 66554220) in the exon 8 encoding the HLA-G 3′ UTR was genotyped as previously described (Tripathi et al., 2004). Briefly, after polymerase chain reaction (PCR), amplified products were size-discriminated by agarose gel electrophoresis with appropriate controls of known HLA-G genotypes that had previously been characterized by nucleotide sequencing. Alleles having the 14-bp sequence in exon 8 was termed HLA-G 14pb Ins whereas those without, HLA-G 14pb Del.
Statistical analysis
Differences in categorical variables between the two groups were evaluated by Chi-square analysis. Since HLA-G genotypes are identical among patients and donors, the predictive effect of recipient genotype (HLA-G 14 bp Ins/Ins, 14 bp Ins/Del, and 14 bp Del/Del) was assessed as variable. Cumulative incidence using competing risk method, as described by Fine and Gray (1999), was used for the assessment of prognostic factors of acute GvHD (grades II–IV or grades III–IV) with death as a competing event. The Cox regression (stepwise backward procedure) analysis and competing risk regression were used in multivariate analysis of independent risk factors of GvHD (Cox, 1972). Similar methodology was used to analyze the other outcomes namely TRM, relapse, and OS. All tests were two-sided, with type I error rate fixed at 0.05. Statistical analyses were performed with SPSS 15 software, Stata 11, and R packages “cmprsk” (competing risks). D/R pairs as well as transplant-related characteristics such as disease stage, D/R age, and sex as well as cytomegalovirus (CMV) serology status that could potentially influence the post-HSCT outcomes were evaluated in all pairs and were included in multivariate analysis.
Results
The sibling D/R pairs were fully matched for HLA-G genotypes with the following frequencies: HLA-G 14 bp Ins/Ins: 21%, HLA-G 14 bp Ins/Del: 48%, and HLA-G 14 bp Del/Del: 31% which were comparable to those previously reported2
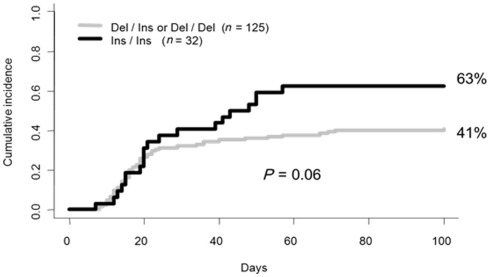
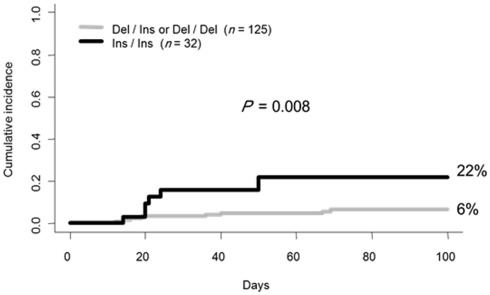
In the studied sample, 71 patients (45%) developed a grade II to IV acute GvHD at day 100. Among them, 15 patients (21%) experienced the severe form (grade III and IV). Monovariate analysis using competing risk showed that the homozygous state of the HLA-G 14 bp Ins/Ins genotype was more prevalent among patients who experienced aGvHD (grade 0, I versus II, III, IV), but failed to reach statistical significance (P = 0.06; Figure 1). Nevertheless additional monovariate analysis after patient stratification based on aGvHD severity (grade 0, I, II versus III, IV) revealed a significant association between the HLA-G 14 bp Ins/Ins genotype and severe aGvHD (HLA-G 14 bp Ins/Ins versus others: 22% versus 6%; P = 0.008; Figure 2). These data were further confirmed using two different multivariate analysis adjusted for confounding variables (gender, CMV status, age of recipient, and disease status). The only factor that remained statistically significant was HLA-G 14 bp Ins/Ins genotype; [hazard ratio (HR) = 3.26; 95% confidence interval (95%CI) = 1.16–9.14; P = 0.024 by Cox regression and HR = 3.5; (95%CI) = 1.3–9.5; P = 0.012 by competing risk regression]. Given the potential effect of methotrexate (MTX) on HSCT outcome in the context of HLA-G polymorphism (Chiusolo et al., 2011), we re-analyzed the sub-group who received this drug for aGvHD prophylaxis (n = 144). We found no changes in the above-mentioned association, but rather strengthened and this is despite a reduction in the sample size [P = 0.001 and HR = 4.7; (95%CI) = 1.5–3.15; P = 0.009 in monovariate and multivariate analyses respectively].
Figure 1.
Cumulative incidence for aGvHD following BMT based on HLA-G recipient genotype (grade 0, I versus II, III, IV).
Figure 2.
Cumulative incidence for aGvHD based on HLA-G genotype in patients stratified for severity (grade 0, I, II versus III, IV).
We did not find any other relationship between HLA-G genotype and the other complications explored in this study (TRM, relapse, and OS; data not shown).
Discussion
Inappropriate immune responses are often the substratum of immune disorders including those related to the prevention/treatment of the underlying disease per se. A typical example of such process is represented by the post-HSCT aGvHD. GvHD results from a sequential activation of both D/R immune cells allowing overwhelming of inflammation with consequent tissue damage (Reddy and Ferrara, 2003; Ferrara et al., 2009; Socié and Blazar, 2009). It is evident that understanding the molecular mechanisms that could lead to induction of immune tolerance may provide clues for novel therapeutic management. In such context, given the prominent role of HLA-G molecules in immune tolerance, we focused our attention on the genetics of this locus to clarify the inter-patient differences in the incidence and severity of aGvHD in a HSCT setting.
Indeed, studying a functionally relevant HLA-G gene polymorphism, namely the HLA-G 14 bp Ins/Del variation, we found that the presence of the HLA-G 14 Ins/Ins genotype increased the risk of developing severe aGvHD in patients transplanted with BM as the stem cell source. These findings are in line with the data provided by other studies in different clinical settings including gestational complications, auto-immunity, infections, cancers as well as solid organ transplantation (Larsen and Hviid, 2009). This polymorphic variation in HLA-G seems to influence the RNA splicing stability by mechanisms that are yet to be understood. The presence of 14-bp insertion, introducing an additional splice site, results in the removal of the first 92 bp of exon 8 generating more stable HLA-G mRNAs species than the complete mRNA (O’Brien et al., 2001; Hviid et al., 2003; Rousseau et al., 2003). In fact, the 3′ UTR 14 bp insertion have been consistently associated with low expression of HLA-G mRNA and low serum sHLA-G (Chen et al., 2008; Rizzo et al., 2008). Such a discrepancy between stability and output, termed “the 14-bp polymorphism paradox” (Veit and Chies, 2009), predicts complex mechanisms of regulation of HLA-G expression. Recent studies implicate the potential role of microRNAs which by interacting with the HLA-G 3′ mRNA region, may regulate its phenotype expression (Veit and Chies, 2009; Castelli et al., 2010). Indeed, a C to G polymorphic change at position + 3142 of the 3′ UTR (rs1063320) has been identified as a putative binding site for three microRNAs (miR-148a, miR-148b, and miR-152) and functional studies demonstrate that the 3′ UTR + 3142 G allele favors binding of these microRNAs with consequent down regulation of HLA-G expression (Tan et al., 2007). In addition, presence of an adenine at position + 3187 (rs 9380142), in the close vicinity of an (AU)-rich motif, was also found to be associated with diminished stability of HLA-G mRNA species both in in silico and in vitro experiments (Yie et al., 2008). Interestingly, the two above-mentioned variant alleles, one affecting the mRNA translation and the other affecting the mRNA stability, are found to be in strong linkage disequilibrium with the HLA-G 14 bp Ins allele in independent studies involving various population groups (Tan et al., 2007; Castelli et al., 2010; Larsen et al., 2010). The observed effect of Ins allele on aGvHD could be either due to the above-mentioned combined effect (haplotype effect) or due to another yet to be identified linked functional variant. A recent study associated the 14-bp Ins/ + 3142G/ + 3187A haplotype with the risk to develop pre-eclampsia, a multisystemic complication of pregnancy (Larsen et al., 2010).
Our data on the effect of the insertion allele in the incidence of aGvHD fit perfectly within the concept that a downregulated expression of HLA-G molecules could decrease their immunosuppressive/tolerogenic properties with consequent aGvHD development. They are also in concordance with the data showing that high pre- and post-transplantation levels of sHLA-G molecules correlate not only with a decreased incidence of aGvHD but also with a high frequency of circulating regulatory T-cells (Treg; Le Maux et al., 2008). Similar correlation between sHLA-G and Treg cells was also observed in in vitro mixed leukocyte reaction assay (Le Maux et al., 2008), in the context of liver transplantation (Le Rond et al., 2006) and in a transgenic murine model system (Ristich et al., 2007).
Our findings are also in agreement with the majority of expression studies of HLA-G in solid organ transplantation evidencing the beneficial effect of HLA-G molecules against acute rejection/chronic dysfunction of the transplanted heart and kidney (Lila et al., 2000, 2002; Crispim et al., 2008a) and also with those establishing correlations between the presence of the HLA-G 14 bp Ins allele and organ rejection (Crispim et al., 2008b; Piancatelli et al., 2009; Twito et al., 2011).
In the overall context of transplantation, the only controversy noted in the literature has been in association with HSCT outcome (La Nasa et al., 2007; Chiusolo et al., 2011). Indeed, in one study, the authors found that the aGvHD risk was associated with the HLA-G 14 bp Del/Del genotype (La Nasa et al., 2007) which appears to be in contradiction with the established fact that the 14-bp Del allele is associated with higher levels of sHLA-G than the 14-bp Ins allele. Nevertheless, it must be noted that this study concerned a relatively small cohort of D/R pairs (n = 53) and moreover HLA-matched unrelated-donor (UD) HSCT in the context of a non-malignant disorder, namely beta-thalassemia. The mechanism of aGvHD could however be different in this situation. For example, since the HLA-matched unrelated D/R pairs are not identical by descent, it could be possible that the HLA-G 14pb Del/Del genotype may reflect the MHC non-HLA haplotype disparity (Petersdorf et al., 2007). More recently, another work involving 47 HSCT patients with a variety of hematological malignancies failed to observe any significant association between the HLA-G 14 bp Ins/Del polymorphism and aGvHD (Chiusolo et al., 2011), but found that patients homozygous for the 14-bp Ins allele were characterized by a lower survival rate and disease free survival. The authors related their findings to the possible relationship between the HLA-G 14 bp dimorphism, methotrexate-based aGvHD prophylaxis, and HLA-G expression. Possibly in line with this, our finding of positive association with aGvHD was not altered by the stratification of our patients based on their aGvHD prophylaxis.
It must also be stressed that the herein studied cohort of HSCT patients was homogeneous with respect to the donor stem cell source, namely BM derived stem cells. In this context, it is of interest to note that the main soluble HLA-G isoform, HLA-G5, a recognized major inducer of regulatory T cell expansion, is specifically expressed by erythroid progenitors residing within the BM (Menier et al., 2004).
HSCT is a sensitive in vivo setting capable of revealing fine immune response traits, undetectable in physiological situations. This is the plausible reason why the effect of Ins/Ins HLA-G genotype on conferring low immune tolerance has been uncovered in this study.
Contribution
Wahid Boukouaci designed the research, analyzed, and interpreted the data and wrote the manuscript. Marc Busson collected data, performed statistical analysis, analyzed, and interpreted the data. Catherine Fortier and Kahina Amokrane designed and performed experiments. Régis Peffault de Latour and Marie Robin collected and organized the clinical data. Rajagopal Krishnamoorthy designed the research and critically reviewed the manuscript. Antoine Toubert participated in the research design and critically reviewed the manuscript. Dominique Charron participated in the research design and critically reviewed the manuscript. Gérard Socié analyzed and interpreted the data and critically reviewed the manuscript. Ryad Tamouza designed the research, analyzed, and interpreted the data and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by institutional funding to Assistance Publique-Hôpitaux de Paris (AP-HP) and INSERM UMRS 940.
Footnotes
Abbreviations
95%CI, 95% confidence interval; BMT, bone marrow transplantation; bp, base pair; CMV, cytomegalovirus; CTL, cytotoxic T lymphocyte; CYA A, cyclosporine A; Del, deletion; D/R, donor/recipient; EBV, epstein-barr-virus; GvHD, graft-versus-host disease; HLA, human leukocyte antigen; HR, hazard ratio; HSCT, hematopoietic stem cell transplantation; Ins, insertion; MHC, major histocompatibility complex; MTX, methotrexate; NK, natural killer; OS, overall survival; PBSC, peripheral blood stem cells; PCR, polymerase chain reaction; TRM, transplant-related mortality; UD-BMT, unrelated-donor bone marrow transplantation.
References
- Bainbridge D. R., Ellis S. A., Sargent I. L. (2000). HLA-G suppresses proliferation of CD4(+) T-lymphocytes. J. Reprod. Immunol. 48, 17–26 10.1016/S0165-0378(00)00070-X [DOI] [PubMed] [Google Scholar]
- Castelli E. C., Mendes-Junior C. T., Deghaide N. H., de Albuquerque R. S., Muniz Y. C., Simões R. T., Carosella E. D., Moreau P., Donadi E. A. (2010). The genetic structure of 3′ untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun. 11, 134–141 10.1038/gene.2009.74 [DOI] [PubMed] [Google Scholar]
- Chen X. Y., Yan W. H., Lin A., Xu H. H., Zhang J. G., Wang X. X. (2008). The 14 bp deletion polymorphisms in HLA-G gene play an important role in the expression of soluble HLA-G in plasma. Tissue Antigens 72, 335–341 10.1111/j.1399-0039.2008.01107.x [DOI] [PubMed] [Google Scholar]
- Chiusolo P., Bellesi S., Piccirillo N., Giammarco S., Marietti S., De Ritis D., Metafuni E., Stignani M., Baricordi O. R., Sica S., Leone G., Rizzo R. (2011). The role of HLA-G 14-bp polymorphism in allo-HSCT after short-term course MTX for GvHD prophylaxis. Bone Marrow Transplant. [Epub ahead of print]. 10.1038/bmt.2011.40 [DOI] [PubMed] [Google Scholar]
- Cox D. R. (1972). Regression models and life tables (with discussion). J. R. Stat. Soc. Series B Stat. Methodol. 34, 187 [Google Scholar]
- Crispim J. C., Duarte R. A., Soares C. P., Costa R., Silva J. S., Mendes-Júnior C. T., Wastowski I. J., Faggioni L. P., Saber L. T., Donadi E. A. (2008a). Human leukocyte antigen-G expression after kidney transplantation is associated with a reduced incidence of rejection. Transpl. Immunol. 18, 361–367 10.1016/j.trim.2007.10.010 [DOI] [PubMed] [Google Scholar]
- Crispim J. C., Mendes-Junior C. T., Wastowski I. J., Costa R., Castelli E. C., Saber L. T., Donadi E. A. (2008b). Frequency of insertion/deletion polymorphism in exon 8 of HLA-G and kidney allograft outcome. Tissue Antigens 71, 35–41 [DOI] [PubMed] [Google Scholar]
- Dickinson A. M., Charron D. (2005). Non-HLA immunogenetics in hematopoietic stem cell transplantation. Curr. Opin. Immunol. 17, 517–525 10.1016/j.coi.2005.07.017 [DOI] [PubMed] [Google Scholar]
- Ferrara J. L., Levine J. E., Reddy P., Holler E. (2009). Graft-versus-host, disease. Lancet 373, 1550. 10.1016/S0140-6736(09)60237-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine J. P., Gray R. J. (1999). A proportional hazards model for the sub distribution of a competing risk. JASA 94, 496 [Google Scholar]
- Glucksberg H., Storb R., Fefer A., Buckner C. D., Neiman P. E., Clift R. A., Lerner K. G., Thomas E. D. (1974). Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation 18, 295–304 10.1097/00007890-197410000-00001 [DOI] [PubMed] [Google Scholar]
- Hviid T. V., Hylenius S., Rørbye C., Nielsen L. G. (2003). HLA-G allelic variants are associated with differences in the HLA-G mRNA isoform profile and HLA-G mRNA levels. Immunogenetics 55, 63–79 [DOI] [PubMed] [Google Scholar]
- John S. W., Weitzner G., Rozen R., Scriver C. R. (1991). A rapid procedure for extracting genomic DNA from leukocytes. Nucleic Acids Res. 19, 408. 10.1093/nar/19.2.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Nasa G., Littera R., Locatelli F., Lai S., Alba F., Caocci G., Lisini D., Nesci S., Vacca A., Piras E., Bernardo M. E., Di Cesare-Merlone A., Orrù S., Carcassi C. (2007). The human leucocyte antigen-G 14-basepair polymorphism correlates with graft-versus-host disease in unrelated bone marrow transplantation for thalassaemia. Br. J. Haematol. 139, 284–288 10.1111/j.1365-2141.2007.06779.x [DOI] [PubMed] [Google Scholar]
- Larsen M. H., Hviid T. V. (2009). Human leukocyte antigen-G polymorphism in relation to expression, function, and disease. Hum. Immunol. 70, 1026–1034 10.1016/j.humimm.2009.07.015 [DOI] [PubMed] [Google Scholar]
- Larsen M. H., Hylenius S., Andersen A. M., Hviid T. V. (2010). The 3′-untranslated region of the HLA-G gene in relation to pre-eclampsia: revisited. Tissue Antigens 75, 253–261 10.1111/j.1399-0039.2009.01435.x [DOI] [PubMed] [Google Scholar]
- Le Maux A., Noël G., Birebent B., Grosset J. M., Vu N., De Guibert S., Bernard M., Semana G., Amiot L. (2008). Soluble human leucocyte antigen-G molecules in peripheral blood haematopoietic stem cell transplantation: a specific role to prevent acute graft-versus-host disease and a link with regulatory T cells. Clin. Exp. Immunol. 152, 50–56 10.1111/j.1365-2249.2008.03598.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rond S., Azéma C., Krawice-Radanne I., Durrbach A., Guettier C., Carosella E. D., Rouas-Freiss N. (2006). Evidence to support the role of HLA-G5 in allograft acceptance through induction of immunosuppressive/regulatory T cells. J. Immunol. 176, 3266–3276 [DOI] [PubMed] [Google Scholar]
- Lila N., Amrein C., Guillemain R., Chevalier P., Latremouille C., Fabiani J. N., Dausset J., Carosella E. D., Carpentier A. (2002). Human leukocyte antigen-G expression after heart transplantation is associated with a reduced incidence of rejection. Circulation 105, 194–1954 10.1161/hc0202.102238 [DOI] [PubMed] [Google Scholar]
- Lila N., Carpentier A., Amrein C., Khalil-Daher I., Dausset J., Carosella E. D. (2000). Implication of HLA-G molecule in heart-graft acceptance. Lancet 355, 2138. 10.1016/S0140-6736(00)02386-2 [DOI] [PubMed] [Google Scholar]
- Menier C., Rabreau M., Challier J. C., Le Discorde M., Carosella E. D., Rouas-Freiss N. (2004). Erythroblasts secrete the nonclassical HLA-G molecule from primitive to definitive hematopoiesis. Blood 104, 3153–3160 10.1182/blood-2004-03-0809 [DOI] [PubMed] [Google Scholar]
- Messina C., Faraci M., de Fazio V., Dini G., Calò M. P., Calore E. (2008). Prevention and treatment of acute GvHD. Bone Marrow Transplant. 41, S65. 10.1038/bmt.2008.57 [DOI] [PubMed] [Google Scholar]
- O’Brien M., McCarthy T., Jenkins D., Paul P., Dausset J., Carosella E. D., Moreau P. (2001). Altered HLA-G transcription in pre-eclampsia is associated with allele specific inheritance: possible role of the HLA-G gene in susceptibility to the disease. Cell. Mol. Life Sci. 58, 1943–1949 10.1007/PL00000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P., Cabestre F. A., Ibrahim E. C., Lefebvre S., Khalil-Daher I., Vazeux G., Quiles R. M., Bermond F., Dausset J., Carosella E. D. (2000). Identification of HLA-G7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6, and -G7 transcripts in human transfected cells. Hum. Immunol. 61, 1138–1149 10.1016/S0198-8859(00)00154-3 [DOI] [PubMed] [Google Scholar]
- Petersdorf E. W., Malkki M., Gooley T. A., Martin P. J., Guo Z. (2007). MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med. 4, e8. 10.1371/journal.pmed.0040008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piancatelli D., Maccarone D., Liberatore G., Parzanese I., Clemente K., Azzarone R., Pisani F., Famulari A., Papola F. (2009). HLA-G 14-bp insertion/deletion polymorphism in kidney transplant patients with metabolic complications. Transplant. Proc. 41, 1187–1188 10.1016/j.transproceed.2009.03.028 [DOI] [PubMed] [Google Scholar]
- Przepiorka D., Weisdorf D., Martin P., Klingemann H. G., Beatty P., Hows J., Thomas E. D. (1995). 1994 Consensus conference on acute GVHD grading. Bone Marrow Transplant. 15, 825–828 [PubMed] [Google Scholar]
- Reddy P., Ferrara J. L. (2003). Immunobiology of acute graft-versus-host disease. Blood Rev. 17, 187–194 10.1016/S0268-960X(03)00009-2 [DOI] [PubMed] [Google Scholar]
- Ristich V., Zhang W., Liang S., Horuzsko A. (2007). Mechanisms of prolongation of allograft survival by HLA-G/ILT4-modified dendritic cells. Hum. Immunol. 68, 264–271 10.1016/j.humimm.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Rizzo R., Hviid T. V., Govoni M., Padovan M., Rubini M., Melchiorri L., Stignani M., Carturan S., Grappa M. T., Fotinidi M., Ferretti S., Voss A., Laustrup H., Junker P., Trotta F., Baricordi O. R. (2008). HLA-G genotype and HLA-G expression in systemic lupus erythematosus: HLA-G as a putative susceptibility gene in systemic lupus erythematosus. Tissue Antigens 71, 520–529 10.1111/j.1399-0039.2008.01037.x [DOI] [PubMed] [Google Scholar]
- Rocha V., Franco R. F., Porcher R., Bittencourt H., Silva W. A., Jr., Latouche A., Devergie A., Esperou H., Ribaud P., Socie G., Zago M. A., Gluckman E. (2002). Host defense and inflammatory gene polymorphisms are associated with outcomes after HLA-identical sibling bone marrow transplantation. Blood 100, 3908–3912 10.1182/blood-2002-04-1033 [DOI] [PubMed] [Google Scholar]
- Rouas-Freiss N., Marchal R. E., Kirszenbaum M., Dausset J., Carosella E. D. (1997). The alpha 1 domain of HLA-G1 and HLA-G2 inhibits cytotoxicity induced by natural killer cells: is HLA-G the public ligand for natural killer cell inhibitory receptors? Proc. Natl. Acad. Sci. U.S.A. 94, 5249–5254 10.1073/pnas.94.21.11520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau P., Le Discorde M., Mouillot G., Marcou C., Carosella E. D., Moreau P. (2003). The 14 bp deletion-insertion polymorphism in the 3′ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum. Immunol. 64, 1005–1010 10.1016/j.humimm.2003.08.347 [DOI] [PubMed] [Google Scholar]
- Socié G., Blazar B. R. (2009). Acute graft-versus-host disease; from the bench to the bedside. Blood 114, 4327–4336 10.1182/blood-2009-06-204669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., Randall G., Fan J., Camoretti-Mercado B., Brockman-Schneider R., Pan L., Solway J., Gern J. E., Lemanske R. F., Nicolae D., Ober C. (2007). Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am. J. Hum. Genet. 81, 829–834 10.1086/521200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P., Abbas A., Naik S., Agrawal S. (2004). Role of 14-bp deletion in the HLA-G gene in the maintenance of pregnancy. Tissue Antigens 64, 706–710 10.1111/j.1399-0039.2004.00308.x [DOI] [PubMed] [Google Scholar]
- Twito T., Joseph J., Mociornita A., Rao V., Ross H., Delgado D. H. (2011). The 14-bp deletion in the HLA-G gene indicates a low risk for acute cellular rejection in heart transplant recipients. J. Heart Lung Transplant. 30, 778–782 10.1016/j.healun.2011.01.726 [DOI] [PubMed] [Google Scholar]
- Veit T. D., Chies J. A. (2009). Tolerance versus immune response – microRNAs as important elements in the regulation of the HLA-G gene expression. Transpl. Immunol. 20, 229–231 10.1016/j.trim.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Wiendl H., Mitsdoerffer M., Hofmeister V., Wischhusen J., Bornemann A., Meyermann R., Weiss E. H., Melms A., Weller M. (2002). A functional role of HLA-G expression in human gliomas: an alternative strategy of immune escape. J. Immunol. 168, 4772–4780 [DOI] [PubMed] [Google Scholar]
- Yie S. M., Li L. H., Xiao R., Librach C. L. (2008). A single base-pair mutation in the 3′-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol. Hum. Reprod. 14, 649–653 10.1093/molehr/gan059 [DOI] [PubMed] [Google Scholar]