Abstract
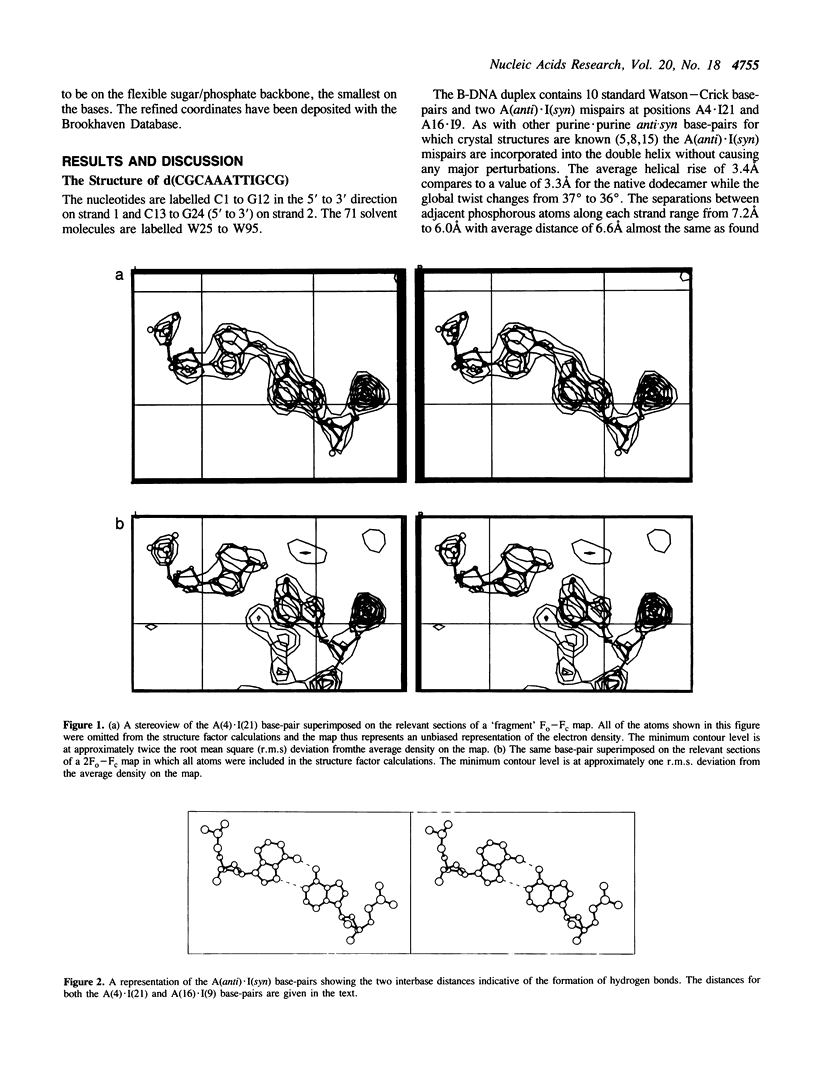
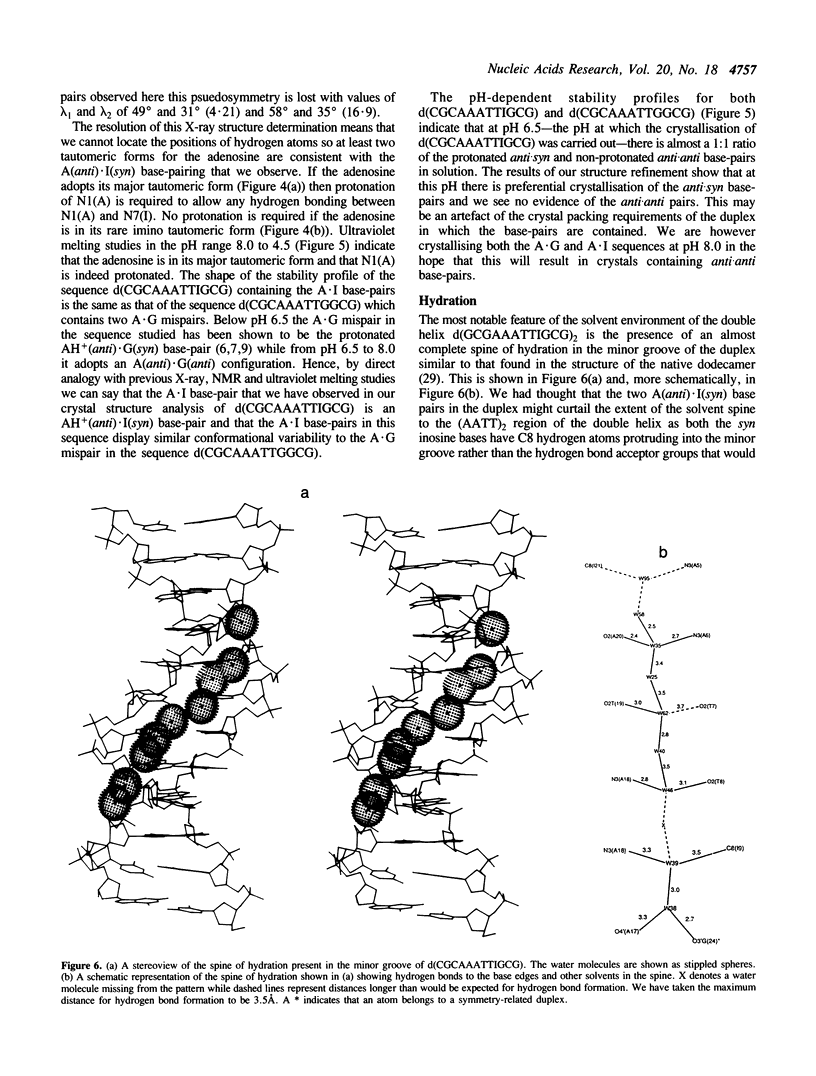
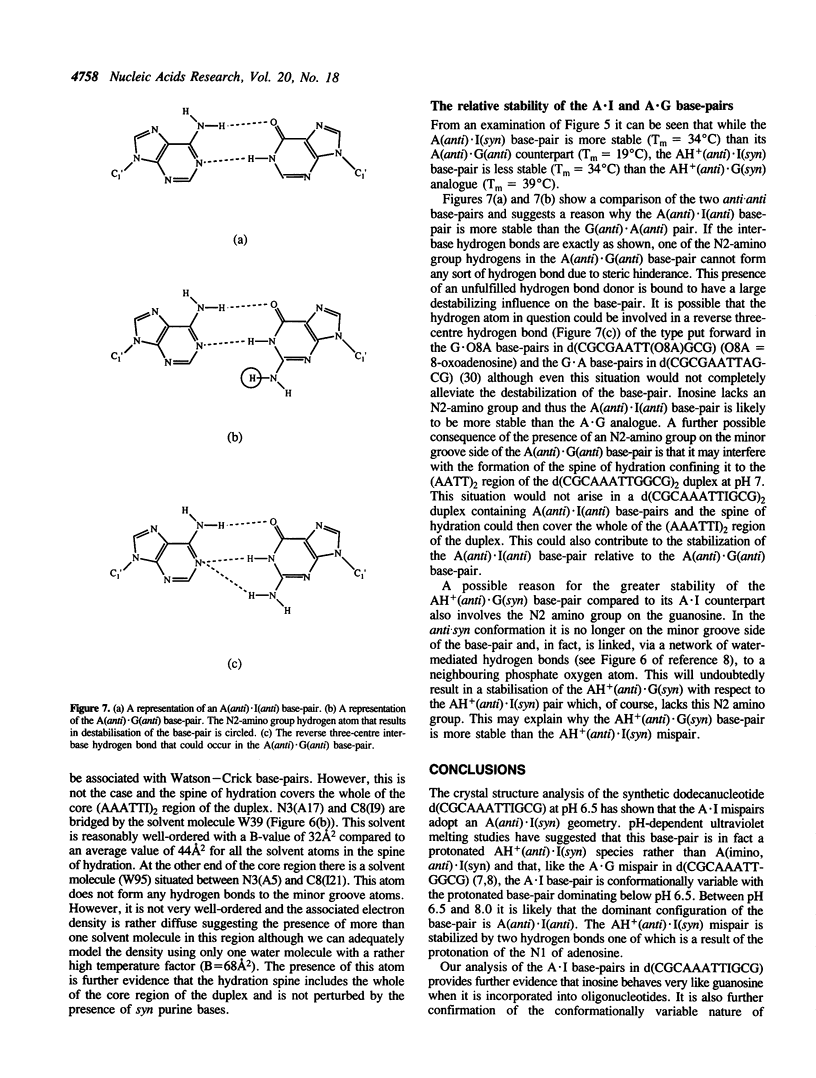
A crystal structure analysis of the synthetic deoxydodecamer d(CGCAAATTIGCG) which contains two adenosine.inosine (A.I) mispairs has revealed that, in this sequence, the A.I base-pairs adopt a A(anti).I(syn) configuration. The refinement converged at R = 0.158 for 2004 reflections with F greater than or equal to 2 sigma(F) in the range 7.0-2.5A for a model consisting of the DNA duplex and 71 water molecules. A notable feature of the structure is the presence of an almost complete spine of hydration spanning the minor groove of the whole of the (AAATTI)2 core region of the duplex. pH-dependent ultraviolet melting studies have suggested that the base-pair observed in the crystal structure is, in fact, a protonated AH+ (anti).I(syn) species and that the A.I base-pairs in the sequence studied display the same conformational variability as A.G mispairs in the sequence d(CGCAAATTGGCG). The AH+(anti).I(syn) base-pair predominates below pH 6.5 and an A(anti).I(anti) mispair is the major species present between pH 6.5 and 8.0. The protonated base-pairs are held together by two hydrogen bonds one between N6(A) and O6(I) and the other between N1(A) and N7(I). This second hydrogen bond is a direct result of the protonation of the N1 of adenosine. The ultraviolet melting studies indicate that the A(anti).I(anti) base-pair is more stable than the A(anti).G(anti) base-pair but that the AH+(anti).I(syn) base pair is less stable than its AH+(anti).G(syn) analogue. Possible reasons for this observation are discussed.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown T., Hunter W. N., Kneale G., Kennard O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T., Leonard G. A., Booth E. D., Chambers J. Crystal structure and stability of a DNA duplex containing A(anti).G(syn) base-pairs. J Mol Biol. 1989 May 20;207(2):455–457. doi: 10.1016/0022-2836(89)90268-4. [DOI] [PubMed] [Google Scholar]
- Carbonnaux C., van der Marel G. A., van Boom J. H., Guschlbauer W., Fazakerley G. V. Solution structure of an oncogenic DNA duplex containing a G.A mismatch. Biochemistry. 1991 Jun 4;30(22):5449–5458. doi: 10.1021/bi00236a018. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield P. W., Hunter W. N., Brown T., Robinson P., Kennard O. Inosine.adenine base pairs in a B-DNA duplex. Nucleic Acids Res. 1987 Oct 12;15(19):7935–7949. doi: 10.1093/nar/15.19.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse W. B., Aymani J., Kennard O., Brown T., Jack A. G., Leonard G. A. Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs. Nucleic Acids Res. 1989 Jan 11;17(1):55–72. doi: 10.1093/nar/17.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. D., Anderson P., Sparling P. F. Pairing of inosine with adenosine in codon position two in the translation of polyinosinic acid. J Mol Biol. 1973 May 15;76(2):223–232. doi: 10.1016/0022-2836(73)90386-0. [DOI] [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W., Tsui W. C. Kinetic basis of spontaneous mutation. Misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerases of Escherichia coli. J Mol Biol. 1982 Mar 25;156(1):37–51. doi: 10.1016/0022-2836(82)90457-0. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Quigley G. J., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. Methylation of the EcoRI recognition site does not alter DNA conformation: the crystal structure of d(CGCGAm6ATTCGCG) at 2.0-A resolution. J Biol Chem. 1988 Nov 25;263(33):17872–17879. doi: 10.2210/pdb4dnb/pdb. [DOI] [PubMed] [Google Scholar]
- Kouchakdjian M., Bodepudi V., Shibutani S., Eisenberg M., Johnson F., Grollman A. P., Patel D. J. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry. 1991 Feb 5;30(5):1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kumar V. D., Harrison R. W., Andrews L. C., Weber I. T. Crystal structure at 1.5-A resolution of d(CGCICICG), an octanucleotide containing inosine, and its comparison with d(CGCG) and d(CGCGCG) structures. Biochemistry. 1992 Feb 11;31(5):1541–1550. doi: 10.1021/bi00120a035. [DOI] [PubMed] [Google Scholar]
- Lane A. N., Jenkins T. C., Brown D. J., Brown T. N.m.r. determination of the solution conformation and dynamics of the A.G mismatch in the d(CGCAAATTGGCG)2 dodecamer. Biochem J. 1991 Oct 1;279(Pt 1):269–281. doi: 10.1042/bj2790269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard G. A., Booth E. D., Brown T. Structural and thermodynamic studies on the adenine.guanine mismatch in B-DNA. Nucleic Acids Res. 1990 Oct 11;18(19):5617–5623. doi: 10.1093/nar/18.19.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Ohtsuka E., Matsuki S., Ikehara M., Takahashi Y., Matsubara K. An alternative approach to deoxyoligonucleotides as hybridization probes by insertion of deoxyinosine at ambiguous codon positions. J Biol Chem. 1985 Mar 10;260(5):2605–2608. [PubMed] [Google Scholar]
- Privé G. G., Heinemann U., Chandrasegaran S., Kan L. S., Kopka M. L., Dickerson R. E. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987 Oct 23;238(4826):498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Kato K., Hayashizaki Y., Wakabayashi T., Ohtsuka E., Matsuki S., Ikehara M., Matsubara K. Molecular cloning of the human cholecystokinin gene by use of a synthetic probe containing deoxyinosine. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1931–1935. doi: 10.1073/pnas.82.7.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]



