Abstract
Cytotoxic T lymphocytes (CTL) are important for controlling equine infectious anemia virus (EIAV). Because Gag matrix (MA) and capsid (CA) are the most frequently recognized proteins, the hypothesis that CTL from EIAV-infected horses with diverse MHC class I alleles recognize epitope clusters (EC) in these proteins was tested. Four EC were identified by CTL from 15 horses and 8 of these horses had diverse MHC class I alleles. Two of the eight had CTL to EC1, six to EC2, five to EC3, and four to EC4. Because EC2–4 were recognized by CTL from >50% of horses with diverse alleles, the hypothesis was accepted. EC1 and EC3 were the most conserved EC and these more conserved broadly recognized EC may be most useful for CTL induction, helping overcome MHC class I polymorphism and antigenic variation.
Keywords: Cytotoxic T lymphocyte, Equine infectious anemia virus, Gag, Matrix, Capsid, Epitope cluster, Horse MHC class I allele
Introduction
Equine infectious anemia virus (EIAV), a lentivirus, causes persistent infection in horses that is initially characterized by recurrent viremia with fever, thrombocytopenia, and anemia within the first 6–12 months of infection (Sellon et al., 1994). Disease episodes are associated with viral antigenic variants as defined by neutralizing antibody (Kono et al., 1973; Rwambo et al., 1990a) and, recently, by cytotoxic T lymphocytes (CTL) (Mealey et al., 2003). Immune responses to EIAV are critical for termination of the initial viremia, as shown by comparison of viremia between normal and severe combined immunodeficient (SCID) foals which lack functional T and B lymphocytes (Perryman et al., 1988). In addition, adoptive transfer of immune lymphocytes to an EIAV-infected SCID foal prevented continuous EIAV replication (Mealey et al., 2001). Disease episodes in most infected horses are eventually controlled and a lifelong carrier stage follows (Coggins, 1984). The mechanism used to control viremia during the carrier stage in EIAV-infected horses is not clear, but protective immune responses likely include neutralizing antibody and CTL (Hammond et al., 1997, 2000; Kono, 1969; Kono et al., 1970; Perryman et al., 1988; Zhang et al., 1998). The ability of horses to restrict plasma viremia to a very low level after a few disease episodes and become lifelong carriers provides an opportunity to define protective immune responses to a natural lentiviral infection.
CTL are critical for control of lentiviruses including EIAV. In EIAV, clearance of initial plasma viremia is associated with the presence of CTL (Hammond et al., 1997; McGuire et al., 1994) and occurs before the appearance of neutralizing antibody (Carpenter et al., 1987; Montelaro et al., 1984; O’Rourke et al., 1988). In addition, individual vaccinates in immunization trials were protected against homologous viral infection in the absence of detectable neutralizing antibody at the time of challenge (Issel et al., 1992). In HIV-1 and SIV infection, CTL responses correlate with virus clearance (Kuroda et al., 1999; Ogg et al., 1998). Eliminating CD8+ cells from monkeys during chronic SIV infection resulted in a rapid and marked increase in viremia that was again suppressed coincident with the reappearance of SIV-specific CD8+ T lymphocytes (Schmitz et al., 1999). This result demonstrates that CD8+ cells are necessary for controlling lentivirus infection and supports vaccine approaches that elicit CD8+ T lymphocytes.
Induction of effective CTL responses to lentiviruses is complicated by variation of CTL epitopes (Goulder et al., 2001; Kaur et al., 2001; Mealey et al., 2003; Zhang et al., 1999) and MHC class I polymorphism (Bailey et al., 2000; Bernoco et al., 1987; Ellis et al., 1995; Evans et al., 1999; Lazary et al., 1988). A comprehensive, yet difficult strategy, to overcome these complications is to use epitopes from multiple proteins from multiple viral variants presented by different MHC class I alleles to induce protective CTL responses. An easier approach is suggested in a report describing relatively conserved overlapping HIV-1 epitopes recognized by CTL from several individuals (Buseyne et al., 1993; Goulder et al., 2000a, 2000b; Wilson et al., 1997). One study of HIV-1-infected patients found three dominant CTL epitope clusters (EC) in Gag p17 [matrix (MA)] and p24 [capsid (CA)] that were recognized irrespective of the virus clade, ethnicity, or age group studied (Goulder et al., 2000a). However, those EC were defined with CTL from individuals that shared some MHC class I alleles (Goulder et al., 2000a). Therefore, EC need to be evaluated in individuals with more heterologous MHC class I alleles to determine if they could be used to induce broadly reactive CTL responses in populations with diverse MHC class I haplotypes.
Studies of CTL responses to HIV-1 and EIAV demonstrated that Gag proteins were recognized by CTL from the most individuals. HIV-1 subtype C isolates had low aa diversity in Gag CA protein, and infected patients had an immunodominant CTL response to this protein (Novitsky et al., 2002). Analysis of CTL recognition of EIAV proteins using retroviral vectors expressing Gag p9, p11, p15 (MA), p26 (CA), Pol, SU, TM, Tat, Rev, and S2 found that CTL recognition of Gag MA and CA proteins was most frequent (McGuire et al., 2000). Gag MA and CA proteins were recognized by CTL from 100% to 86% of seven EIAV-infected horses, respectively. The results suggested that the 359 aa comprising MA and CA proteins were capable of stimulating CTL in most horses, although these results need to be confirmed using horses with defined differences in MHC class I alleles. A previous study dissecting CTL responses from five EIAV-infected horses found recognition of overlapping peptides in EIAV Gag indicating a possible EC in the MA protein and another in the CA protein; however, the horses had limited diversity of MHC class I haplotypes (Zhang et al., 1998).
In this study, horses with heterologous MHC class I alleles defined by sequence-based typing were used to rigorously test the hypothesis that CTL from EIAV-infected horses with diverse MHC class I alleles recognize EC in Gag MA and CA proteins. Additionally, the use of CTL from 17 EIAV-infected carrier horses enhanced the probability of identifying EC involved in virus control. Four EC were identified and three of these were recognized by 50% or more infected horses in a subset with diverse MHC class I alleles. The identification of these EC will allow preparation of small polypeptide antigens containing multiple CTL epitopes for evaluation in outbred horses.
Results
CTL from EIAV-infected horses recognize Gag EC
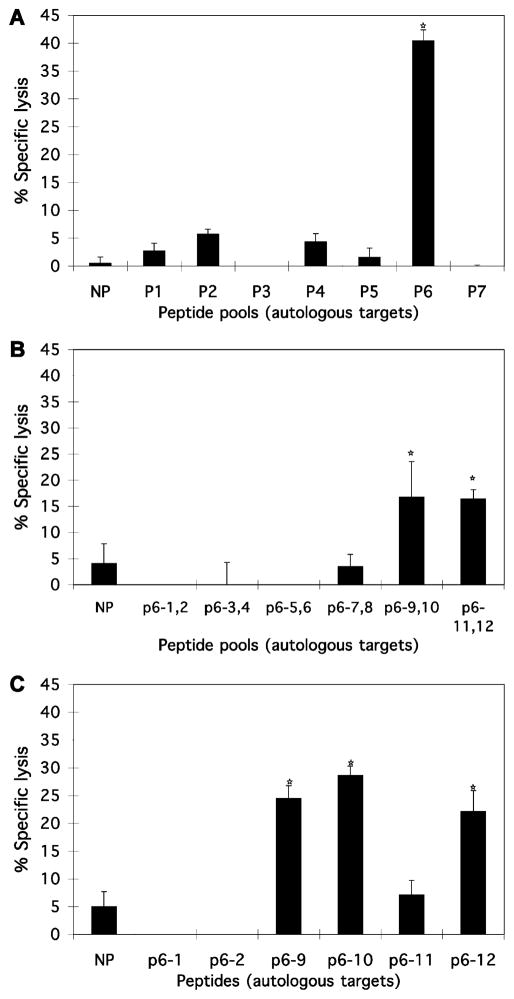
To identify EC, CTL from 17 EIAV-infected horses (Table 1) were evaluated. Peptide mapping of CTL from 12 horses was done in this study and data were used from five other horses in a previous publication (Zhang et al., 1998). CTL recognition of peptides was assessed using EK target cells pulsed with seven pools of MA and CA peptides. These peptides were 15–16 aa long and overlapped by 11–12 aa (Tables 2 and 3). Effectors were PBMC from EIAV-infected horses stimulated with EIAVWSU5. Fig. 1 contains peptide-mapping data derived with CTL from horse H596 that was representative of data from the other horses. The initial mapping with larger peptide pools (Fig. 1A) was followed by a second mapping with pools of two peptides (Fig. 1B) and a third mapping with individual peptides (Fig. 1C). It was found that CTL from 16 of 17 horses recognized at least one peptide from Gag MA and CA proteins (Fig. 2). The exception was PBMC from horse A2153 that did not recognize any of the peptides. Further analysis of data in Fig. 2 resulted in the identification of four EC based on regions with the most peptides recognized by CTL from the most horses. These were designated EC1–4 by their position in the linear sequence of Gag MA and CA proteins (Fig. 2).
Table 1.
Recognition of Gag EC by CTL from 17 EIAV-infected horses
| Horse no. | MHC class I haplotype
|
EC recognized
|
||
|---|---|---|---|---|
| Alleles identified by sequencing (Chung et al., 2003) | ELA-A type | Virus stimulation | EC peptide stimulation | |
| A2147 | 7-1, 7-4 | A4a | 2, 3 | 2, 3, 4 |
| A2150 | 104, 105, 106, 141 | A1/W11 | 2 | 2 |
| H593 | 113, 114 | A1a | 3 | 1, 3 |
| H596 | 115, 116, 117, 118, 119 | A4/W11 | 2, 3 | 2, 3 |
| H610 | 111, 121, 122 | A6a | 2, 4 | 2, 4 |
| H614 | 123, 124, 125, 126, 127 | A9a | 3 | 2, 3, 4 |
| H629 | Not done | A3/A5 | 4 | 4 |
| H631 | 133, 134, 135, 136 | Undeterminedb | 1, 2 | 1, 2, 3 |
| A2140 | Not done | A1/W11 | 1 | 1, 2, 3, 4 |
| H601 | Not done | A1/W11 | 1 | 1 |
| H507c | Not done | A7/W11 | 1, 3 | Not done |
| H513c | Not done | A5/A8 | 1 | Not done |
| H521c | Not done | A1/W11 | 1, 3 | Not done |
| H529c | Not done | A1/A5 | 1, 2 | Not done |
| H532c | Not done | A5/A6 | 4 | Not done |
| H540 | 109, 111 | A5/A6 | None | None |
| A2153 | 107, 7-1, 7-4 | A4a | None | None |
Only one ELA-A type was detected with the antisera used (A1–A10 and W11).
Undetermined indicates that none of the available antisera reacted with lymphocytes from this horse.
Gag epitope mapping with CTL from these five horses was previously reported (Zhang et al., 1998).
Table 2.
EIAV Gag MA protein synthetic peptides used for EC identification
| Pool | Amino acid sequence | Peptide name |
|---|---|---|
| P1 | MGDPLTWSKALKKLE | P1-1 |
| LTWSKALKKLEKVTV | P1-2 | |
| KALKKLEKVTVQGSQ | P1-3 | |
| KLEKVTVQGSQKLTT | P1-4 | |
| VTVQGSQKLTTGNCN | P1-5 | |
| GSQKLTTGNCNWALS | P1-6 | |
| LTTGNCNWALSLVDL | P1-7 | |
| NCNWALSLVDLFHDT | P1-8 | |
| ALSLVDLFHDTNFVK | P1-9 | |
| VDLFHDTNFVKEKDW | P1-10 | |
| HDTNFVKEKDWQLRD | P1-11 | |
| FVKEKDWQLRDVIPL | P1-12 | |
| KDWQLRDVIPLLEDV | P1-13 | |
| LRDVIPLLEDVTQTL | P1-14 | |
| P2 | IPLLEDVTQTLSGQE | P2-1 |
| EDVTQTLSGQEREAF | P2-2 | |
| QTLSGQEREAFERTW | P2-3 | |
| GQEREAFERTWWAIS | P2-4 | |
| EAFERTWWAISAVKM | P2-5 | |
| RTWWAISAVKMGLQI | P2-6 | |
| AISAVKMGLQINNVV | P2-7 | |
| VKMGLQINNVVDGKA | P2-8 | |
| LQINNVVDGKASFQL | P2-9 | |
| NVVDGKASFQLLRAK | P2-10 | |
| GKASFQLLRAKYEKK | P2-11 | |
| FQLLRAKYEKKTANK | P2-12 | |
| RAKYEKKTANKKQSE | P2-13 | |
| EKKTANKKQSEPSEEY | P2-14 |
Table 3.
EIAV Gag CA protein synthetic peptides used for EC identification
| Pool | Amino acid sequence | Peptide name |
|---|---|---|
| P3 | PIMIDGAGNRNFRPL | P3-1 |
| DGAGNRNFRPLTPRG | P3-2 | |
| NRNFRPLTPRGYTTW | P3-3 | |
| RPLTPRGYTTWVNTI | P3-4 | |
| PRGYTTWVNTIQTNG | P3-5 | |
| TTWVNTIQTNGLLNE | P3-6 | |
| NTIQTNGLLNEASQN | P3-7 | |
| TNGLLNEASQNLFGI | P3-8 | |
| LNEASQNLFGILSVD | P3-9 | |
| SQNLFGILSVDCTSE | P3-10 | |
| FGILSVDCTSEEMNA | P3-11 | |
| P4 | SVDCTSEEMNAFLDV | P4-1 |
| TSEEMNAFLDVVPGQ | P4-2 | |
| MNAFLDVVPGQAGQK | P4-3 | |
| LDVVPGQAGQKQILL | P4-4 | |
| PGQAGQKQILLDAID | P4-5 | |
| GQKQILLDAIDKIAD | P4-6 | |
| ILLDAIDKIADDWDN | P4-7 | |
| AIDKIADDWDNRHPL | P4-8 | |
| IADDWDNRHPLPNAP | P4-9 | |
| WDNRHPLPNAPLVAP | P4-10 | |
| HPLPNAPLVAPPQGP | P4-11 | |
| P5 | NAPLVAPPQGPIPMT | P5-1 |
| VAPPQGPIPMTARFI | P5-2 | |
| QGPIPMTARFIRGLG | P5-3 | |
| PMTARFIRGLGVPRE | P5-4 | |
| RFIRGLGVPRERQME | P5-5 | |
| GLGVPRERQMEPAFD | P5-6 | |
| PRERQMEPAFDQFRQ | P5-7 | |
| QMEPAFDQFRQTYRQ | P5-8 | |
| AFDQFRQTYRQWIIE | P5-9 | |
| FRQTYRQWIIEAMSE | P5-10 | |
| P6 | YRQWIIEAMSEGIKV | P6-1 |
| IIEAMSEGIKVMIGK | P6-2 | |
| MSEGIKVMIGKPKAQ | P6-3 | |
| GIKVMIGKPKAQNIRQ | P6-4 | |
| MIGKPKAQNIRQGAKE | P6-5 | |
| PKAQNIRQGAKEPYPE | P6-6 | |
| NIRQGAKEPYPEFVDR | P6-7 | |
| GAKEPYPEFVDRLLSQ | P6-8 | |
| PYPEFVDRLLSQIKSE | P6-9 | |
| FVDRLLSQIKSEGHPQ | P6-10 | |
| LLSQIKSEGHPQEISK | P6-11 | |
| IKSEGHPQEISKFLTD | P6-12 | |
| P7 | GHPQEISKFLTDTLTI | P7-1 |
| EISKFLTDTLTIQNAN | P7-2 | |
| FLTDTLTIQNANEECR | P7-3 | |
| TLTIQNANEECRNAMR | P7-4 | |
| QNANEECRNAMRHLRP | P7-5 | |
| EECRNAMRHLRPEDTL | P7-6 | |
| NAMRHLRPEDTLEEKM | P7-7 | |
| HLRPEDTLEEKMYACR | P7-8 | |
| EDTLEEKMYACRDIGT | P7-9 | |
| EEKMYACRDIGTTKQK | P7-10 | |
| YACRDIGTTKQKMMLL | P7-11 | |
| DIGTTKQKMMLLAKAL | P7-12 |
Fig. 1.
Identification of peptides containing CTL epitopes. PBMC from EIAV-infected horse H596 were stimulated with EIAVWSU5 and evaluated. Target cells were pulsed first with peptide pools (A), then with two peptides (B), and finally with individual peptides (C). Asterisks on top of columns indicate a % specific lysis that was significantly different (defined in Materials and methods) from target cells with no peptide (NP). The error bars on the columns are 1 SE.
Fig. 2.
Gag MA (A) and CA (B) peptides recognized by CTL from EIAV-infected horses. All peptides recognized by CTL from horses H507, H513, H521, H529 and H540 are from a previous study (Zhang et al., 1998), whereas the remaining peptides are from this study. EC1–4 is shadowed. Epitope-lacking regions are shadowed and boxed. Peptide sequences and horse numbers in boxes indicate fine-mapped epitopes. The underlined peptides were recognized by CTL from eight horses with unique MHC class I alleles.
EC1 was 35 aa including MA 1–35 and was recognized by CTL from 7 of the total 17 horses (Fig. 2). MA 77–119 (43 aa), designated EC2, was recognized by CTL from six horses. CA 156–187 (32 aa), designated EC3, was also recognized by CTL from six horses. Finally, CA 192–232 (41 aa), designated EC4, was recognized by CTL from seven horses. Horse H540 had CTL that recognized peptides in a non-EC region, but not in an EC. EIAV-stimulated PBMC from horse A2153 did not recognize either EC or other MA or CA peptides as indicated above. The remaining 15 horses had CTL to Gag MA and CA proteins that recognized one or more of the identified EC.
Expansion of EC1–4 recognition by stimulation of CTL with EC peptide pools
Once EC1–4 were identified using CTL stimulated with EIAVWSU5, it was of interest to determine if CTL stimulation with EC peptides would result in expanded EC1–4 recognition. PBMC from all but the five horses (H507, H513, 521, H529, and H532) used for CTL epitope mapping in a previous study (Zhang et al., 1998) were available for stimulation with each of four peptide pools containing peptides from each EC. PBMC stimulated with a particular EC peptide pool were tested on targets pulsed with the same EC peptide pool. There was still no EC recognition by stimulated PBMC from horses H540 and A2153; however, five horses had CTL that recognized additional EC. CTL from four of these horses including H593, H614, H631, and A2147 recognized from one to two additional EC that were not recognized in mapping with EIAVWSU5-stimulated PBMC (Table 1, compare last two columns). CTL from A2140 recognized three additional EC (Table 1).
EC recognition by EIAV-infected horses with diverse MHC class I alleles
To determine if EC were recognized by horses with diverse MHC class I alleles, a subset of horses from Table 1 was evaluated. This subset consisted of the first eight horses listed in Table 1 because these horses had unique MHC class I alleles in comparison to the remaining horses. MHC class I alleles were determined by sequence-based typing except for H629, which had a different ELA-A type. In a previous study, horses with different ELA-A haplotypes did not share alleles by sequence-based typing (Chung et al., 2003). CTL from A2147 and H596, which shared an ELA-A4 haplotype, but did not share the same MHC class I alleles determined by sequencing, both recognized peptides P6-9 and P6-10 in EC3 (Fig. 2). However, fine mapping demonstrated that CTL from H596 and A2147 did not recognize the same optimal epitopes within peptides P6-9 and P6-10 (Table 4), confirming that the different alleles in these two horses presented different peptides.
Table 4.
Optimal epitopes in P6-9 and P6-10 peptides recognized by CTL from horses H596 and A2147
| Peptides | % Specific killing in 51Cr release assay
|
|
|---|---|---|
| H596 CTL | A2147 CTL | |
| PYPEFVDRLLSQIKSE (P6-9) | 25 | 9 |
| YPEFVDRLLS | NSa | 31 |
| FVDRLLSQI | 39 | NS |
| FVDRLLSQIKSEGHPQ (P6-10) | 29 | 9 |
| FVDRLLSQIK | 36 | NS |
| RLLSQIKSEG | NS | 30 |
NS indicates no significant % specific lysis.
When EC recognition by the eight horses with unique MHC class I alleles were considered, two recognized EC1, six recognized EC2, five recognized EC3, and four recognized EC4 (Table 5). Therefore, three of the four EC were broadly recognized by CTL from horses with diverse MHC class I alleles, with 50% or more of these horses having CTL to each of EC2–4. Based on these results, the hypothesis that CTL from EIAV-infected horses with diverse MHC class I alleles would recognize Gag EC was accepted.
Table 5.
Summary of EC recognition by CTL from eight EIAV-infected horses with unique MHC class I allelesa
| EC | Peptide and no. of aa | Horses with CTL to each EC (%) |
|---|---|---|
| 1 | MA 1–35, 35 aa | H593, H631 (25%) |
| 2 | MA 77–119, 43 aa | A2147, A2150, H596, H610, H614, H631 (75%) |
| 3 | CA156–187, 32 aa | A2147, H593, H596, H614, H631 (62.5%) |
| 4 | CA 192–232, 41 aa | A2147, H610, H614, H629 (50%) |
The eight horses with unique MHC class I alleles are the first eight horses listed in Table 1.
Analysis of aa sequence variability within EC1–4 and two epitope-lacking regions
To evaluate the aa sequence variability of EC1–4, all 79 complete EIAV Gag MA and CA sequences from GenBank containing Wyoming, Idaho, Texas, UK, PV, WSU5, and Japan strains, and their variants were analyzed after multiple alignment. Analysis of these available sequences was done to provide a minimum estimate of variability of the identified EC. EC1 had nine variant sequences among the 79 sequences, whereas EC2 had 15, EC3 had 11, and EC4 had 8 (Fig. 3). EC2 also had the largest number of changed aa with substitutions or deletions occurring in 20 of 43 residues (47%). EC4 had 15 residues with change (37%), EC1 had 9, (26%) and EC3 had 8 (25%). This was in comparison to epitope-lacking region1 and region2 with six (17%) residue changes each (Fig. 3). The overall percentage of aa residues with change was 30% for Gag MA and CA proteins. Therefore, EC2, which was recognized by CTL from the most horses with diverse MHC class I alleles, was the most variable EC. This region may have less functional importance, thereby allowing for more variation. Further, both epitope-lacking regions as well as EC1 and EC3 had fewer aa residues with changes than the overall sequence, suggesting more constraint on variation in these regions.
Fig. 3.
Amino acid sequence variation of EC1–4 in 79 EIAV proviral clones and plasma virus variants from GenBank. Only variable sequences are presented with the dots indicating aa identical to those in EIAVWSU5 and the dashes indicating amino acid deletions.
Comparison between EC recognition and control of disease
As a preliminary analysis to determine whether CTL responses to specific EC might be related with control of EIAV, days since the last clinical episode in the eight horses with unique MHC class I alleles were compared with the recognition of specific EC by CTL (Table 6). From this analysis, horses with CTL responses to EC4 were significantly more likely to have a fewer number of days since the last clinical episode (P = 0.03) than horses with CTL responses to other EC. In contrast, there was no significant relationship between days postinfection and EC recognition. The reason for a significant relationship between CTL responses to EC4 and fewer days since the last clinical episode is not known. It was surprising since the horses analyzed were infected with two different virus strains at different times and likely had different immune responses. However, the observation could be important and needs conformation with additional horses.
Table 6.
EC recognition by CTL from horses with unique MHC class I alleles and control of clinical disease episode
| Horse no. | Infection history
|
EC recognized | |
|---|---|---|---|
| Days postinfection | Days since the last clinical disease | ||
| A2147 | 1158 | 209 | 2, 3, 4 |
| A2150 | 1158 | 574 | 2 |
| H593 | 1442 | 780 | 1, 3 |
| H596 | 626 | 437 | 2, 3 |
| H610 | 490 | 120 | 2, 4 |
| H614 | 626 | 161 | 2, 3, 4 |
| H629 | 317 | 177 | 4 |
| H631 | 483 | 358 | 1, 2, 3 |
Discussion
Viral antigenic variation and host MHC class I polymorphism are the main barriers to inducing protective CTL responses against lentiviruses in humans and other outbred animals. One possible solution is to immunize with EC that are recognized by CTL from individuals with diverse MHC class I alleles (Buseyne et al., 1993; Goulder et al., 2000a; Yusim et al., 2002). This approach to vaccine design could include expressing EC in DNA plasmids or other vectors. There are several advantages to using smaller EC peptides instead of whole proteins to induce CTL. One is that there is a marked increase in peptide-MHC class I formation and killing of target cells expressing CTL epitopes from minigenes compared to expression of the whole protein suggesting that similar events occur in APC (Anton et al., 1997). Immunization with EC could also exclude regions of full-length proteins that induce dominant, but ineffective or adverse immune responses (Raabe et al., 1998; Wang et al., 1994; Woodberry et al., 1999). Additionally, the EC contain adjacent natural aa for epitope processing. Finally, several variant EC could be included in a vaccine construct.
CTL responses to Gag MA and CA could be a mechanism for controlling clinical disease episodes in naturally occurring lentiviral infections including EIAV. CTL from EIAV-infected horses with limited diversity of MHC class I haplotypes recognized Gag MA and CA proteins more frequently than other viral proteins (McGuire et al., 2000). Gag MA and CA in EIAV are the most abundant proteins, reaching 58% of total EIAV proteins, and are more conserved than other EIAV proteins including Env and Pol (Montelaro et al., 1993). Similarly, the CTL response to HIV-1 Gag proteins are more immunodominant than those responses to other viral proteins as there was significantly more IFN-gamma-secreting CD8+ T lymphocytes to HIV-1 Gag CA protein than to any other proteins (Novitsky et al., 2002). A study of HIV-1 Gag CTL responses in Caucasoid and African Americans infected with HIV-1 clade B as well as African adults and children in South Africa infected with clade C found three immunodominant regions in MA and CA proteins (Goulder et al., 2000a). Although the immunogenicity of these regions is partly explained by the clustering of similar or identical epitopes between clades, it generally reflects the predominance of HLA-A2- and -A3-restricted Gag MA-specific responses in Caucasoids and HLA-B42- and -B81-restricted CA-specific responses in Africans (Goulder et al., 2000a; Novitsky et al., 2002). Nevertheless, the data suggest that CTL EC occur in lentivirus Gag and indicate a need for more data from experimental groups with diverse MHC class I alleles. The goal of this study was to identify EC in EIAV Gag MA and CA proteins that could eventually be used to induce CTL in horses with diverse MHC class I alleles. Assuming that the CTL responses to MA and CA epitopes are important in the control of EIAV infection, CTL from horses that had controlled disease episodes for more than 120 days were used to identify EC in these proteins. Then, recognition of EC by CTL from horses with unique MHC class I alleles was evaluated.
Seventeen horses with EIAV infection were used for initial identification of EC. Gag MA, CA proteins, or both were recognized by CTL from all but one horse and the peptides recognized were in four clusters (EC1–4). One or more of these EC were recognized by CTL from 15 of the horses, whereas one other horse had CTL recognizing Gag peptides outside the EC. However, it was not possible to identify a universal EC recognized by CTL from all the responding horses. The two horses (A2153 and H540) that did not have CTL to EC were evaluated for CTL to Gag epitopes early in infection in previous studies (Mealey et al., 2003; Zhang et al., 1998). No CTL to Gag epitopes were detected in A2153 PBMC soon after infection with EIAVWSU5 (Mealey et al., 2003) and none were found later in infection in the current study. H540 responded to one CA peptide outside the defined EC early after infection EIAVWSU5 (Zhang et al., 1998), but had no response to EC peptides in either that study or the current study. Failure of these two horses to make CTL to EC peptides either early or late after infection suggests that the lack of response was because of haplotype or other factors affecting immunodominance rather than sequence variation. The identification of EC1–4 and the availability of eight EIAV-infected horses with unique MHC class I alleles allowed us to test the hypothesis that CTL from EIAV-infected horses with diverse MHC class I alleles recognize EC in Gag MA and CA proteins. Three of the four EC were recognized by CTL from 50% or more of eight horses with diverse MHC class I alleles and the hypothesis was accepted. This broad CTL recognition of EIAV EC exceeds that of the major EC in HIV-1 Gag MA and CA proteins, which was recognized by CTL from 39% of a test group that included two predominant HLA-A types (Goulder et al., 2000a). Whether the broad EC recognition by CTL from these horses will extend to larger groups of horses will have to be tested.
Gag has been generally considered a highly conserved lentiviral protein; however, three of five immunodominant regions identified in HIV-1C Gag CA protein were less variable than overall Gag MA suggesting different functional and immunological pressures in different Gag regions (Novitsky et al., 2002). Likewise, EIAV Gag sequences vary among strains as shown in EIAVID which had 12% aa differences from some other EIAV strains including EIAVWSU5 (Zhang et al., 1999). The minimum variability of the identified EC and non-EC regions was analyzed in the 79 available sequences from GenBank which included sequences from at least seven different EIAV strains and their variants. The overall percentage of EIAV Gag MA and CA aa positions with changes was 30%, whereas EC1 (26%) and EC3 (25%) had fewer aa residue changes than the overall proteins and EC2 (47%) and EC4 (37%) had more changes. The relative conservation of EIAV Gag EC1 and EC3 sequences could be related to their involvement in the function of MA and CA, respectively, as sequences essential for virus function have less variation and therefore lower probability of immune escape (Goulder et al., 1997; Kelleher et al., 2001; Wagner et al., 1999; Walker and Korber, 2001). For instance, EIAV MA aa 11–19 in EC1 contains a leucine-rich nuclear export signal (Hatanaka et al., 2002), which is similar to the location of a nuclear export signal in HIV-1 MA (Dupont et al., 1999), and this may constrain the amount of variation. Interestingly, CTL from each of seven horses responding to EC1 recognized only one EC1 epitope, but most horses with CTL recognizing the more variable EC4 recognized multiple EC4 epitopes (Fig. 2). Further, a significant relationship between fewer days since the last clinical episode and horses with CTL to EC4 could be due to CTL escape variants in this region. Finding this relationship was surprising since the horses analyzed were infected with two different virus strains at different times and likely had different immune responses.
Conclusively, four EC in MA and CA proteins were identified using CTL from EIAV-infected horses. Three of these (EC2–4) were recognized by CTL from more than 50% of eight horses with unique MHC class I alleles. These EC, which include the more conserved EC3, will be further evaluated to determine which have more high avidity CTL epitopes because immunization with such epitopes may be a way to overcome both MHC class I diversity and viral variation.
Materials and methods
EIAV infection of horses
A total of 17 EIAV-infected horses were used in this study (Table 1). Seven horses were infected with 107 50% tissue culture infective doses (TCID50) of EIAVWSU5 injected intravenously. These included horses H593 and H631, and previously infected horses H540 (Zhang et al., 1998), A2140, A2147, A2150, and A2153 (Mealey et al., 2001). Another five horses including H596, H601, H610, H614 and H629 were infected with 300 TCID50 of EIAVPV. The origin of EIAVPV has been described (Rwambo et al., 1990b). EIAVWSU5 (GenBank accession no. NP_056901) and EIAVPV (GenBank accession no. AAC03760) have identical Gag MA and CA protein sequences allowing the use of horses infected with both strains for EC identification. Finally, published epitope mapping data (Zhang et al., 1998) using another five horses infected with EIAVWSU5 (H507, H513, H521, H529, H532) were used to identify EC. Autologous EK cells for CTL targets were obtained from kidney biopsies before infection of each horse (McGuire et al., 1994) and maintained as frozen cell lines. Before use in CTL studies, all the horses were free of clinical disease for at least 3 months determined by measuring body temperature daily and counting platelets weekly.
Evaluation of MHC class I haplotypes
Nine of the horses listed in Table 1 were evaluated by sequencing RT-PCR products of MHC class I alleles amplified with classical MHC class I-specific primers (Chung et al., 2003). The MHC class I ELA-A haplotypes of all 17 horses were determined by lymphocyte micro-cytotoxicity (Table 1) using antisera A1-A10 and W11 (Bailey, 1980). The ELA-A haplotypes are associated with a putative ELA-A locus (Bailey et al., 2000; Bernoco et al., 1987; Lazary et al., 1988).
PBMC isolation and stimulation of memory CTL
PBMC were isolated from the EIAV-infected horses by centrifugation on Histopaque (specific gravity 1.077), washed four times, and viability determined (Wyatt et al., 1988). Monocytes in PBMC were infected with EIAVWSU5 at a multiplicity of infection of one or incubated with peptides (103 nM) in 5 ml of RPMI 1640 with 20% FBS at 37 °C for 2 h with gentle mixing every 15 min (McGuire et al., 2000; Zhang et al., 1998). Stimulations were done in a 175 cm2 tissue culture flask containing RPMI 1640 with 10% FCS, 5 × 10−5 M 2-mercaptoethanol, and 20 mM Hepes. After incubation in a humidified chamber with 5% CO2 at 37 °C for 7 days, viable lymphocytes were counted using trypan blue dye exclusion in a hemocytometer for use in the 51Cr release assay (McGuire et al., 2000).
Synthetic peptides of EIAV Gag MA and CA proteins
Fifty-two peptides of 15 aa overlapping by 11 aa and 21 peptides of 16 aa overlapping by 12 aa with C-terminal OH and N-terminal H groups were synthesized to cover the entire EIAVWSU5 Gag MA and CA proteins (Sigma-Genosys, The Woodlands, TX) (Tables 1 and 2). The peptides were dissolved in 100% dimethyl sulfoxide and stored in −20 °C before dilution for use in CTL mapping to identify EC.
CTL assay
The 51Cr-release assay was used to identify EC recognized by CTL from EIAV-infected horses. EK target cells (3 × 104/well) were incubated in collagen-coated wells of 96-well plates at 37 °C with 5% CO2 for 24 h before 51Cr labeling and peptide pulsing. These cells were pulsed with 104 nM synthetic peptides in 50 μl/well of DMEM containing 5% FBS and 1.25 μCi of 51Cr at 37 °C for 2 h and then washed three times with DMEM. Effectors (effector-to-target cell ratio of 50:1) were incubated with target cells in 200 μl/well RPMI containing 10% FBS at 37 °C with 5% CO2 for 17 h (McGuire et al., 1994), and then 100 μl of supernatant removed from each well to determine 51Cr release. Percent specific lysis was calculated as [(E − S)/(M − S)] × 100, where E was the mean of three test wells, S was the mean spontaneous release from three wells without effector cells, and M was the mean maximal release from three wells with 2% Triton X-100 (Siliciano et al., 1985). The standard error (SE) of % specific lysis was calculated taking into consideration the variability of E, S, and M as described previously (Siliciano et al., 1985). Only assays with a spontaneous lysis of <30% were used. Specific lysis of peptide-pulsed target cells that was >5% and exceeded lysis of nonpulsed target cells by >2.5 SE was considered significant.
Peptide mapping to identify EC
Initial peptide mapping used seven peptide pools, each containing 10–14 synthetic peptides each with a final concentration of 104 nM, to pulse target cells. A second mapping used pools of two peptides and a final mapping used individual peptides. Regions with the most peptides identified by CTL from the most horses were designated as EC. Epitope-lacking regions were defined as MA or CA regions without recognition by CTL from EIAV-infected horses in this and a previous study (Zhang et al., 1998).
Analysis of aa sequence variability in EC
Query sequence EIAVWSU5 Gag MA and CA had 79 blast hits in GenBank. Fifteen sequences were Wyoming strain and its variants, 16 were Idaho strain and its variants, 30 were WSU5 strain and its variants, and seven were Texas strain and its variants. The others were two Japan strains, UK, PV, their variants, and seven unknown strains. Those sequences were compared using a multiple alignment program. The variabilities of EC1–4 and in epitope-lacking regions1 and region2 were calculated as the total number of aa residues with changes.
Statistical analysis
The relationship between CTL to specific EC and days postinfection or days since the last clinical episode was evaluated by analysis of contingency tables based on the Fisher’s exact test using GraphPad InStat version 3.05 (GraphPad Software, Inc., San Diego, CA). Significance was determined by P < 0.05.
Acknowledgments
This research was supported in part by U.S. Public Health Service, National Institutes of Health grants AI24291, AI47660, and AI01575, and Morris Animal Foundation grant D01EQ-09. The authors acknowledge the important technical assistance of Emma Karel and valuable discussions with Dr. Darrilyn G. Fraser and Dr. Jeffrey R. Abbott.
References
- Anton LC, Yewdell JW, Bennink JR. MHC class I-associated peptides produced from endogenous gene products with vastly different efficiencies. J Immunol. 1997;158:2535–2542. [PubMed] [Google Scholar]
- Bailey E. Identification and genetics of horse lymphocyte alloantigens. Immunogenetics. 1980;11:499–506. doi: 10.1007/BF01567818. [DOI] [PubMed] [Google Scholar]
- Bailey E, Marti E, Fraser DG, Antczak DF, Lazary S. Immunogenetics of the horse. In: Bowling AT, Ruvinsky A, editors. The Genetics of the Horse. CABI Publishing Inc; UK: 2000. pp. 123–155. [Google Scholar]
- Bernoco D, Byrns G, Bailey E, Lew AM. Evidence of a second polymorphic ELA class I (ELA-B) locus and gene order for three loci of the equine major histocompatibility complex. Anim Genet. 1987;18:103–118. doi: 10.1111/j.1365-2052.1987.tb00749.x. [DOI] [PubMed] [Google Scholar]
- Buseyne F, McChesney M, Porrot F, Kovarik S, Guy B, Riviere Y. Gag-specific cytotoxic T lymphocytes from human immunodeficiency virus type 1-infected individuals: gag epitopes are clustered in three regions of the p24gag protein. J Virol. 1993;67:694–702. doi: 10.1128/jvi.67.2.694-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S, Evans LH, Sevoian M, Chesebro B. Role of the host immune response in selection of equine infectious anemia virus variants. J Virol. 1987;61:3783–3789. doi: 10.1128/jvi.61.12.3783-3789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C, Leib SR, Fraser DG, Ellis SA, McGuire TC. Novel classical MHC class I alleles identified in horses by sequencing clones of reverse transcription-PCR products. Eur J Immunogenet. 2003;30:387–396. doi: 10.1111/j.1365-2370.2003.00420.x. [DOI] [PubMed] [Google Scholar]
- Coggins L. Carriers of equine infectious anemia virus. J Am Vet Med Assoc. 1984;184:279–281. [PubMed] [Google Scholar]
- Dupont S, Sharova N, DeHoratius C, Virbasius CM, Zhu X, Bukrinskaya AG, Stevenson M, Green MR. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature. 1999;402:681–685. doi: 10.1038/45272. [DOI] [PubMed] [Google Scholar]
- Ellis SA, Martin AJ, Holmes EC, Morrison WI. At least four MHC class I genes are transcribed in the horse: phylogenetic analysis suggests an unusual evolutionary history for the MHC in this species. Eur J Immunogenet. 1995;22:249–260. doi: 10.1111/j.1744-313x.1995.tb00239.x. [DOI] [PubMed] [Google Scholar]
- Evans DT, Knapp LA, Jing P, Mitchen JL, Dykhuizen M, Montefiori DC, Pauza CD, Watkins DI. Rapid and slow progressors differ by a single MHC class I haplotype in a family of MHC-defined rhesus macaques infected with SIV. Immunol Lett. 1999;66:53–59. doi: 10.1016/s0165-2478(98)00151-5. [DOI] [PubMed] [Google Scholar]
- Goulder P, Price D, Nowak M, Rowland-Jones S, Phillips R, McMichael A. Co-evolution of human immunodeficiency virus and cytotoxic T-lymphocyte responses. Immunol Rev. 1997;159:17–29. doi: 10.1111/j.1600-065x.1997.tb01004.x. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Brander C, Annamalai K, Mngqundaniso N, Govender U, Tang Y, He S, Hartman KE, O’Callaghan CA, Ogg GS, Altfeld MA, Rosenberg ES, Cao H, Kalams SA, Hammond M, Bunce M, Pelton SI, Burchett SA, McIntosh K, Coovadia HM, Walker BD. Differential narrow focusing of immunodominant human immunodeficiency virus gag-specific cytotoxic T-lymphocyte responses in infected African and Caucasoid adults and children. J Virol. 2000a;74:5679–5690. doi: 10.1128/jvi.74.12.5679-5690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder PJ, Tang Y, Pelton SI, Walker BD. HLA-B57-restricted cytotoxic T-lymphocyte activity in a single infected subject toward two optimal epitopes, one of which is entirely contained within the other. J Virol. 2000b;74:5291–5299. doi: 10.1128/jvi.74.11.5291-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder PJ, Pasquier C, Holmes EC, Liang B, Tang Y, Izopet J, Saune K, Rosenberg ES, Burchett SK, McIntosh K, Barnardo M, Bunce M, Walker BD, Brander C, Phillips RE. Mother-to-child transmission of HIV infection and CTL escape through HLA-A2-SLYNTVATL epitope sequence variation. Immunol Lett. 2001;79:109–116. doi: 10.1016/s0165-2478(01)00272-3. [DOI] [PubMed] [Google Scholar]
- Hammond SA, Cook SJ, Lichtenstein DL, Issel CJ, Montelaro RC. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SA, Li F, McKeon BM, Cook SJ, Issel CJ, Montelaro RC. Immune responses and viral replication in long-term inapparent carrier ponies inoculated with equine infectious anemia virus. J Virol. 2000;74:5968–5981. doi: 10.1128/jvi.74.13.5968-5981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka H, Iourin O, Rao Z, Fry E, Kingsman A, Stuart DI. Structure of equine infectious anemia virus matrix protein. J Virol. 2002;76:1876–1883. doi: 10.1128/JVI.76.4.1876-1883.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issel CJ, Horohov DW, Lea DF, Adams WV, Hagius SD, McManus JM, Allison AC, Montelaro RC. Efficacy of inactivated whole-virus and subunit vaccines in preventing infection and disease caused by equine infectious anemia virus. J Virol. 1992;66:3398–3408. doi: 10.1128/jvi.66.6.3398-3408.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A, Alexander L, Staprans SI, Denekamp L, Hale CL, McClure HM, Feinberg MB, Desrosiers RC, Johnson RP. Emergence of cytotoxic T lymphocyte escape mutations in nonpathogenic simian immunodeficiency virus infection. Eur J Immunol. 2001;31:3207–3217. doi: 10.1002/1521-4141(200111)31:11<3207::aid-immu3207>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono Y. Viremia and immunological responses in horses infected with equine infectious anemia virus. Natl Inst Anim Health Q (Tokyo) 1969;9:1–9. [PubMed] [Google Scholar]
- Kono Y, Kobayashi K, Fukunaga Y. Immunization of horses against equine infectious anemia (EIA) with an attenuated EIA virus. Natl Inst Anim Health Q (Tokyo) 1970;10:113–122. [PubMed] [Google Scholar]
- Kono Y, Kobayashi K, Fukunaga Y. Antigenic drift of equine infectious anemia virus in chronically infected horses. Arch Gesamte Virusforsch. 1973;41:1–10. doi: 10.1007/BF01249923. [DOI] [PubMed] [Google Scholar]
- Kuroda MJ, Schmitz JE, Charini WA, Nickerson CE, Lifton MA, Lord CI, Forman MA, Letvin NL. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J Immunol. 1999;162:5127–5133. [PubMed] [Google Scholar]
- Lazary S, Antczak DF, Bailey E, Bell TK, Bernoco D, Byrns G, McClure JJ. Joint report of the Fifth International Workshop on Lymphocyte Alloantigens of the Horse, Baton Rouge, Louisiana, 31 October–1 November 1987. Anim Genet. 1988;19:447–456. doi: 10.1111/j.1365-2052.1988.tb00836.x. [DOI] [PubMed] [Google Scholar]
- McGuire TC, Tumas DB, Byrne KM, Hines MT, Leib SR, Brassfield AL, O’Rourke KI, Perryman LE. Major histocompatibility complex-restricted CD8+ cytotoxic T lymphocytes from horses with equine infectious anemia virus recognize Env and Gag/PR proteins. J Virol. 1994;68:1459–1467. doi: 10.1128/jvi.68.3.1459-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire TC, Leib SR, Lonning SM, Zhang W, Byrne KM, Mealey RH. Equine infectious anaemia virus proteins with epitopes most frequently recognized by cytotoxic T lymphocytes from infected horses. J Gen Virol. 2000;81:2735–2739. doi: 10.1099/0022-1317-81-11-2735. [DOI] [PubMed] [Google Scholar]
- Mealey RH, Fraser DG, Oaks JL, Cantor GH, McGuire TC. Immune reconstitution prevents continuous equine infectious anemia virus replication in an Arabian foal with severe combined immunodeficiency: lessons for control of lentiviruses. Clin Immunol. 2001;101:237–247. doi: 10.1006/clim.2001.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealey RH, Zhang B, Leib SR, Littke MH, McGuire TC. Epitope specificity is critical for high and moderate avidity cytotoxic T lymphocytes associated with control of viral load and clinical disease in horses with equine infectious anemia virus. Virology. 2003;313:537–552. doi: 10.1016/s0042-6822(03)00344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelaro RC, Parekh B, Orrego A, Issel CJ. Antigenic variation during persistent infection by equine infectious anemia virus, a retrovirus. J Biol Chem. 1984;259:10539–10544. [PubMed] [Google Scholar]
- Montelaro RC, Ball JM, Rushlow KE. Equine retroviruses. In: Levy JA, editor. The Retroviridae. Plenum Press; New York, N.Y: 1993. pp. 257–360. [Google Scholar]
- Novitsky V, Cao H, Rybak N, Gilbert P, McLane MF, Gaolekwe S, Peter T, Thior I, Ndung’u T, Marlink R, Lee TH, Essex M. Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J Virol. 2002;76:10155–10168. doi: 10.1128/JVI.76.20.10155-10168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Cerundolo V, Hurley A, Markowitz M, Ho DD, Nixon DF, McMichael AJ. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- O’Rourke K, Perryman LE, McGuire TC. Antiviral, anti-glycoprotein and neutralizing antibodies in foals with equine infectious anaemia virus. J Gen Virol. 1988;69:667–674. doi: 10.1099/0022-1317-69-3-667. [DOI] [PubMed] [Google Scholar]
- Perryman LE, O’Rourke KI, McGuire TC. Immune responses are required to terminate viremia in equine infectious anemia lentivirus infection. J Virol. 1988;62:3073–3076. doi: 10.1128/jvi.62.8.3073-3076.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe ML, Issel CJ, Cook SJ, Cook RF, Woodson B, Montelaro RC. Immunization with a recombinant envelope protein (rgp90) of EIAV produces a spectrum of vaccine efficacy ranging from lack of clinical disease to severe enhancement. Virology. 1998;245:151–162. doi: 10.1006/viro.1998.9142. [DOI] [PubMed] [Google Scholar]
- Rwambo PM, Issel CJ, Adams WV, Jr, Hussain KA, Miller M, Montelaro RC. Equine infectious anemia virus (EIAV) humoral responses of recipient ponies and antigenic variation during persistent infection. Arch Virol. 1990a;111:199–212. doi: 10.1007/BF01311054. [DOI] [PubMed] [Google Scholar]
- Rwambo PM, Issel CJ, Hussain KA, Montelaro RC. In vitro isolation of a neutralization escape mutant of equine infectious anemia virus (EIAV) Arch Virol. 1990b;111:275–280. doi: 10.1007/BF01311062. [DOI] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Sellon DC, Fuller FJ, McGuire TC. The immunopathogenesis of equine infectious anemia virus. Virus Res. 1994;32:111–138. doi: 10.1016/0168-1702(94)90038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano RF, Keegan AD, Dintzis RZ, Dintzis HM, Shin HS. The interaction of nominal antigen with T cell antigen receptors: I. Specific binding of multivalent nominal antigen to cytolytic T cell clones. J Immunol. 1985;135:906–914. [PubMed] [Google Scholar]
- Wagner R, Leschonsky B, Harrer E, Paulus C, Weber C, Walker BD, Buchbinder S, Wolf H, Kalden JR, Harrer T. Molecular and functional analysis of a conserved CTL epitope in HIV-1 p24 recognized from a long-term nonprogressor: constraints on immune escape associated with targeting a sequence essential for viral replication. J Immunol. 1999;162:3727–3734. [PubMed] [Google Scholar]
- Walker BD, Korber BT. Immune control of HIV: the obstacles of HLA and viral diversity. Nat Immunol. 2001;2:473–475. doi: 10.1038/88656. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Rushlow KE, Issel CJ, Cook RF, Cook SJ, Raabe ML, Chong YH, Costa L, Montelaro RC. Enhancement of EIAV replication and disease by immunization with a baculovirus-expressed recombinant envelope surface glycoprotein. Virology. 1994;199:247–251. doi: 10.1006/viro.1994.1120. [DOI] [PubMed] [Google Scholar]
- Wilson CC, Kalams SA, Wilkes BM, Ruhl DJ, Gao F, Hahn BH, Hanson IC, Luzuriaga K, Wolinsky S, Koup R, Buchbinder SP, Johnson RP, Walker BD. Overlapping epitopes in human immunodeficiency virus type 1 gp120 presented by HLA A, B, and C molecules: effects of viral variation on cytotoxic T-lymphocyte recognition. J Virol. 1997;71:1256–1264. doi: 10.1128/jvi.71.2.1256-1264.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodberry T, Gardner J, Mateo L, Eisen D, Medveczky J, Ramshaw IA, Thomson SA, Ffrench RA, Elliott SL, Firat H, Lemonnier FA, Suhrbier A. Immunogenicity of a human immunodeficiency virus (HIV) polytope vaccine containing multiple HLA A2 HIV CD8(+) cytotoxic T-cell epitopes. J Virol. 1999;73:5320–5325. doi: 10.1128/jvi.73.7.5320-5325.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt CR, Davis WC, McGuire TC, Perryman LE. T lymphocyte development in horses: I. Characterization of monoclonal antibodies identifying three stages of T lymphocyte differentiation. Vet Immunol Immunopathol. 1988;18:3–18. doi: 10.1016/0165-2427(88)90032-3. [DOI] [PubMed] [Google Scholar]
- Yusim K, Kesmir C, Gaschen B, Addo MM, Altfeld M, Brunak S, Chigaev A, Detours V, Korber BT. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J Virol. 2002;76:8757–8768. doi: 10.1128/JVI.76.17.8757-8768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Lonning SM, McGuire TC. Gag protein epitopes recognized by ELA-A-restricted cytotoxic T lymphocytes from horses with long-term equine infectious anemia virus infection. J Virol. 1998;72:9612–9620. doi: 10.1128/jvi.72.12.9612-9620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Auyong DB, Oaks JL, McGuire TC. Natural variation of equine infectious anemia virus Gag protein cytotoxic T lymphocyte epitopes. Virology. 1999;261:242–252. doi: 10.1006/viro.1999.9862. [DOI] [PubMed] [Google Scholar]