Abstract
The induction of cytotoxic CD8+ T cell responses requires the presentation of antigenic peptides by MHC class I molecules (MHC I). MHC I usually present peptides derived from endogenous proteins. However, some subtypes of dendritic cells have developed the ability to efficiently present peptides derived from exogenous antigens on MHC I via a process called cross-presentation. Cross-presentation is intimately linked to the induction of anti-viral, -bacterial, and -tumor cytotoxic T cell (CTL) responses, as well as a wide variety of CTL-mediated diseases and transplant rejections. The molecular and cellular mechanisms underlying cross-presentation have been studied intensively since its original description, yet understanding of this process is incomplete and on the forefront of immunological research. Numerous pathways and models, some of them conflicting, have been described so far. Here, we review the various pathways reported as involved in cross-presentation, highlighting the complexity of this process. We also discuss in detail the different intracellular steps required, from antigen capture and routing, to processing, and finally peptide loading, emphasizing the need for a better understanding of the cell biology of this phenomenon.
Keywords: antigen, cross-presentation, dendritic cell, MHC class I, phagosome, vaccination, cross-priming, gap junctions
Introduction
The mammalian adaptive immune response is crucial in the clearance of many infections. Classically, immune cells present exogenous antigens on MHC Class II (MHC II) to “helper” CD4+ T cells, and endogenous antigens via MHC Class I (MHC I) to “cytotoxic” CD8+ T cells. In this review, we will focus on the CD8+ T cells that are key in the elimination of infected or cancer cells. CD8+ T cells are first activated when their unique T cell receptor (TCR) is triggered by fragments of microbial or tumor antigens in association with MHC I. All nucleated cells express MHC I, however dendritic cells (DCs) also express a range of co-stimulatory molecules, uptake receptors, and other key immune molecules to specifically initiate a cytotoxic CD8+ T cells response.
In cases where DCs are not infected directly, but a cytotoxic T cell (CTL) response is required, DCs have the capacity to sample antigens from their environment by a process called cross-presentation, as it differs from the normal pathway of MHC I antigen presentation where the antigens are merely cytosolic or nuclear. For cross-presentation, exogenous antigens (e.g., from an infected cell) are taken up by DCs and rerouted to the MHC I pathway for presentation to and activation of CD8+ T cells.
The cell biology of cross-presentation is clearly different from the classical MHC I antigen presentation. A myriad of studies have interrogated various cell biological pathways including the antigen uptake pathway, antigen translocation from endosome to cytosol, ER–phagosome fusion, the proteasome, the endosomal pH, the TAP transporter, and gap junctions (reviewed in Neefjes et al., 2011; Segura and Villadangos, 2011). Based on these findings, various pathways and mechanisms have been proposed that may all be correct or mutually exclusive. Here, we present a critical evaluation of the various models and observations reported on cross-presentation.
What is “True” in Cross-Presentation?
The classical pathway of MHC I antigen presentation is nowadays understood in detail (Neefjes et al., 2011). Cytosolic and nuclear antigens are degraded into peptide fragments by the proteasome, further trimmed by peptidases, and transported from the cytosol into the ER lumen by the peptide transporter TAP. Peptides are then loaded on newly synthesized MHC I and these complexes are released from the ER and transported to the cell surface via the Golgi.
As expected, the components of the classical MHC I antigen presentation have been tested for their involvement in the process of cross-presentation. In several studies, the proteasome has been implicated in cross-presentation in experiments where cells were treated with proteasome inhibitors for long periods of time (Brossart and Bevan, 1997; Rodriguez et al., 1999). Here an involvement of the proteasome in cross-presentation was implied due to its role in generation of peptides. However, proteasome inhibitors are known to alter the ubiquitin profile in cells by accumulating polyubiquitinated proteins and inducing alterations in transcription due to histone deubiquitination (Dantuma et al., 2006). Therefore, the inhibition of cross-presentation by proteasome inhibitors might be the direct consequence of a defect in peptide generation, or a subsequent event to transcriptome alterations or to any other process involving ubiquitin such as endosome formation.
Whether cross-presentation requires the peptide transporter TAP was also tested. If involved, this would imply that antigenic peptides originate from the cytosol, most likely after trimming by the proteasome. DCs isolated from TAP-deficient mice were found unable to cross-present (Brossart and Bevan, 1997). However, these observations have been contradicted by others (Dantuma et al., 2006). Moreover, a recent model proposed by Merzougui et al. implies a dissociation of TAP and proteasome dependence. In particular, a role for TAP has been implicated in the recycling of MHC I necessary for cross-presentation of particulate antigens (Merzougui et al., 2011).
Finally, the involvement of newly synthesized MHC I in cross-presentation was addressed using the chemical inhibitor Brefeldin A which blocks ER–golgi transport (Brossart and Bevan, 1997). Moreover, as it also affects the entire endosomal pathway, it is conceivable that the inhibition of cross-presentation does not reflect that peptide loading occurs on newly synthesized MHC I in the ER.
To conclude, some well accepted concepts concerning cross-presentation can be contested, and it is important to reconsider and take into account the different options.
Cross-Presentation, Who is Allocated to Do It?
Cross-presentation is considered an exclusive feature to DCs. Amongst the many subsets of DCs, the CD8α+ DCs in mouse and their human equivalent, the BDCA3+ DCs, the monocytes-derived DCs (Mo-DCs), and the migratory CD103+ DCs are presumed to be unique in their capacity to cross-present the antigen (in vitro and in vivo; den Haan et al., 2000; del Rio et al., 2007; Hildner et al., 2008; Bedoui et al., 2009; Bachem et al., 2010; Jongbloed et al., 2010; Poulin et al., 2010). As our main focus in this review is the cell biological aspect of cross-presentation, and as there has been none performed with the CD103+ DCs, we will focus mainly on the CD8α+ DCs and the Mo-DCs.
The obvious question is what distinguishes the cross-presenting DC subsets from the others. Although the DC types have been profiled by a series of techniques including microarray and mass spectrometry, a clear “cross-presentation signature” has not been defined (Robbins et al., 2008; Luber et al., 2010; Segura et al., 2010). One option is that all cells (including non-immune cells) are able to cross-present but the cross-presenting DCs are just remarkable because they display the perfect arsenal of the required characteristics for this process, from the surface expression of specific uptake receptors, a low endosomal protease activity, an extremely high expression of MHC I and finally a series of co-stimulatory molecules to allow cross-priming. These DCs may therefore be better equipped, without being unique. This would explain why cross-presentation has also been observed, but to a lesser extent, in other immune cells like CD8α− DCs (den Haan and Bevan, 2002), Langerhans cells (Oh et al., 2011), B cells (de Wit et al., 2010), macrophages (Asano et al., 2011), as well as in non-immune cells (Gromme et al., 1999; Neijssen et al., 2005; Pang et al., 2009). Alternatively, the cross-presenting DCs may have a cell biological secret that is yet to be revealed.
Receptor-Mediated Antigen Uptake and Routing to the MHC I Pathway
One issue that is not contested is that exogenous antigens have to be captured by DCs for cross-presentation (Figure 1). Antigens can be taken up by fluid phase or by receptor-mediated endocytosis. A number of endocytosis/phagocytosis receptors have long been thought to be critical for antigen uptake as they concentrate antigens in the endocytic pathway. However, many of them have a more important function in rerouting and targeting the antigen to defined endosomal compartments: efficient uptake in combination with cargo delivery to the appropriate compartments is a decisive event for antigen cross-presentation (Burgdorf et al., 2007; Caminschi et al., 2008; Sancho et al., 2009; Tacken et al., 2011).
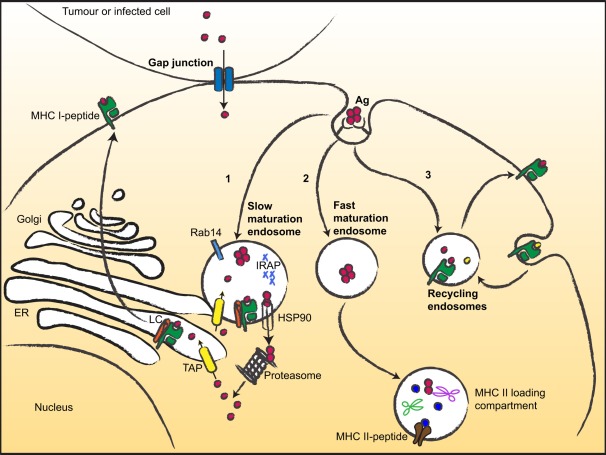
Figure 1.
Multiple ways for MHC I cross-presentation. After uptake, antigens can follow different routes and be targeted to (1) slow maturating endosomes (or phagosome) displaying low proteolytic activity. Then, antigens can escape to the cytosol (via HSP90) and are degraded by the proteasome into peptides that are translocated to the ER or back to the endocytic compartment for loading; (2) Fast maturating endosomes, addressing the antigens to low pH- and high proteolytic- compartments suited for MHC II antigen presentation; (3) Recycling endosomes, where loading occurs on recycling MHC I molecules. Gap junctions mediate transfer of small peptides between neighboring cells. LC, loading complex.
A series of receptors have been implicated including Fcγ receptors (den Haan and Bevan, 2002; Schuurhuis et al., 2002) and lectin receptors such as the mannose receptor that mediates the uptake of soluble but not cell-associated antigens (Burgdorf et al., 2006), Dectin-1 that is involved in uptake and cross-presentation of cellular antigen (Weck et al., 2008), Clec9A that mediates the capture and cross-presentation of antigens derived from necrotic cells (Caminschi et al., 2008; Sancho et al., 2009), and others.
Collectively, these studies illustrate that different antigens can use multiple uptake mechanisms and pathways leading to cross-presentation of antigenic peptides on MHC I.
Are all Endosomes Equal for Cross-Presentation?
As written in classical textbooks, captured antigens enter the endocytic pathway. Antigens first enter early endosomes, then late endosomes, and finally lysosomes. In the case of cell-associated antigens or bacteria, the endosomes are enlarged and called phagosomes. Each of these compartments has specific markers and pH. Early endosomes are positive for Rab5 and are mildly acidic without many proteases while late endosomes and lysosomes are positive for Rab7 and more acidic (around pH 5.0) with a substantial proteolytic activity. However, this concept of a simple endosomal pathway, where material moves from early to late endosomes then to lysosomes as their inescapable fate, has been challenged for many years especially through the description of lysosome-related organelles such as cytotoxic granules, melanosomes, and MHC II loading compartments (MIIC). The endosomal pathway can therefore undergo specialization, and this may be applied in antigen cross-presentation (Lakadamyali et al., 2006; Burgdorf et al., 2007; Saveanu et al., 2009; Tacken et al., 2011).
In this regard, Lakadamyali et al. showed that internalized proteins can be sorted into two different categories of early endosomes, “dynamic” or “static.” The dynamic population matured rapidly toward late endosomes and subsequently fused with lysosomes yielding material for MHC II antigen presentation. The more “static” population displayed a slower maturation rate (Lakadamyali et al., 2006), that would favor cross-presentation, as it displayed a low proteolytic activity, which is believed to protect antigens from excessive destruction (Savina et al., 2006; Jancic et al., 2007). Interestingly, the mannose receptor (Burgdorf et al., 2007) and DC-SIGN (Tacken et al., 2011) are surface receptors that target antigens to these low maturating compartments and that mediate cross-presentation.
But what specifies these endosomes involved in cross-presentation? They are marked by the GTPase Rab14, and contain MHC I and the trimming peptidase IRAP (Saveanu et al., 2009). Of note, Salmonella-containing phagosomes are also marked by Rab14, a GTPase that controls phagosomal fusion with lysosomes (Kuijl et al., 2007).
Some studies have provided functional evidence for the presence of endosomal compartments in DCs dedicated for long-term storage of antigen, therefore providing material for cross-presentation by MHC I over long periods of time after uptake (Faure et al., 2009; van Montfoort et al., 2009). Whether the “static Rab14 endosome” corresponds to the long-term antigen storage organelle is, however, unclear.
Antigen Processing for Cross-Presentation
As MHC I classically presents fragments of antigens in the form of 8–10 amino acid long peptides, antigens have to be processed for cross-presentation (Neefjes et al., 2011). A low proteolytic endosomal activity, characteristic of the CD8+ DCs, is believed to be decisive to avoid excessive antigen destruction and allow its processing for cross-presentation (Delamarre et al., 2005; Savina et al., 2006; Jancic et al., 2007). Endocytosed antigens can be processed for cross-presentation by different pathways most likely displaying distinct proteolytic specificities: (1) by endosomal proteases such as cathepsin S and D (Fonteneau et al., 2003; Shen et al., 2004) and (2) by the cytosolic proteasomal machinery used in the classical MHC I antigen presentation pathway (Fonteneau et al., 2003; Shen et al., 2004; Neefjes et al., 2011) which implies that exogenous antigens find a way to enter the cytosol for degradation by the proteasome. Various reports present different solutions to this topological problem.
Antigen translocation from the endosome to the cytosol has been proposed to be specific to internalized antigen and dependent on the size (Rodriguez et al., 1999). As most proteins do not spontaneously pass lipid bilayers, this would require a dedicated – unknown – transporter. Most models imply the ER retrotranslocation machinery (ERAD) involved in the translocation of misfolded ER proteins into the cytosol for degradation by the proteasome (Haug et al., 2003; Taylor et al., 2010; Imai et al., 2011; Oura et al., 2011). Two recent studies demonstrate a role for the chaperone HSP90 in this process (Imai et al., 2011; Oura et al., 2011). An independent study of the group of Cresswell showed that some antigens might be transported from endosomes back to the ER and delivered to the cytosol by the ERAD machinery followed by degradation by the proteasome (Ackerman et al., 2005). This mechanism is used by some toxins to enter the cytosol, but is poorly – if at all – used by most proteins (Neefjes et al., 1988). It is therefore unlikely to be the major route. A role for the mannose receptor in antigen translocation to the cytosol has also been suggested (Burgdorf et al., 2008; Zehner et al., 2011). The underlying mechanisms remain unclear but it is unlikely that the receptor itself performs the translocation step.
An alternative model implicating a fusion of the ER with phagosomes has been suggested by several groups (Gagnon et al., 2002; Guermonprez et al., 2003; Houde et al., 2003). This process may involve a Sec22b-dependent fusion of ER–golgi intermediate compartments with the phagosome (Cebrian et al., 2011), an event that would allow a recruitment of the ERAD machinery to the phagosome, and therefore antigen translocation to the cytosol where proteasomal degradation finally occurs. Peptides can then either be transported by TAP into the newly formed ER–phagosome hybrid organelle or to the ER for loading (Gagnon et al., 2002; Guermonprez et al., 2003; Houde et al., 2003). These models are fairly unique in cell biology and make assumptions that (1) the energy for ERAD is available in the ER–phagosome hybrid organelle, and (2) the chaperones involved in directing antigens to ERAD recognize and consider the antigens sufficiently misfolded for degradation. These considerations complicate the involvement of the ERAD system in cross-presentation and reinforces the fact that the antigen export machinery and especially the actual retrotranslocon pore remains to be defined.
Peptide Loading
Although there may be various pathways for cross-presentation and different locations for antigen processing, peptides and MHC I finally have to meet in the same compartment or organelle. These meeting point options suggested so far are:
– In the ER. This pathway is the most evident from the MHC I point of view as it represents the classical pathway for efficient peptide loading in the presence of the complete MHC I machinery. This pathway has been extensively studied and is nowadays understood in detail (Neefjes et al., 2011). The major issue is how exogenous antigens enter this pathway in an efficient manner to compete for loading with the myriad of peptides that are in the ER.
– In endosomes. This pathway would be simpler as it does not require (unknown) mechanisms for delivering antigens to the cytosol. Antigens would simply be degraded by endosomal proteases and peptides loaded on recycling MHC I. Peptide release and exchange can occur efficiently under mild acidic conditions, without the support of chaperones (Gromme et al., 1999; Burgdorf et al., 2006; Di Pucchio et al., 2008; Zou et al., 2009; Win et al., 2011).
Alternative Mechanisms for Cross-Presentation
Cross-presentation may also result from the transfer of MHC I-peptide complexes from infected cells or tumor to DCs via exosomes or trogocytosis (Wolfers et al., 2001; Herrera et al., 2004; Wakim and Bevan, 2011). Whether these processes are relevant in vivo is unclear.
An alternative that would solve the topological problem of exogenous antigen delivery to the cytosol of DCs is peptide transfer via gap junctions (Neijssen et al., 2005). Gap junctions are small channels directly connecting the cytosol of two adjacent cells (Neijssen et al., 2007). The proteins constituting the gap junctions are called connexins and these are upregulated on monocytes and DCs upon exposure to danger signals (Pang and Neefjes, 2010; Saccheri et al., 2010). Gap junctions are able to mediate the transfer of small peptides from apoptotic and tumor cells to DCs (Pang et al., 2009; Saccheri et al., 2010). The transferred peptides have been shown to be efficiently presented by MHC I at the cell surface to trigger activation of specific CD8+ T cells (Neijssen et al., 2005; Pang and Neefjes, 2010; Saccheri et al., 2010).
The Final Step: Cell Surface Delivery for Cross-Presentation
Ultimately, MHC I molecules have to present antigenic information at the cell surface. Depending on the site of peptide loading, the route of the MHC I-peptides complexes to the cell surface differs. If peptide loading occurs in the ER, complexes simply follow the standard secretory pathway (Neefjes et al., 2011). If MHC I molecules acquire antigenic peptides in endosomal compartments, they may use the pathway also used by MHC II molecules after peptide loading in the MIIC (Gromme et al., 1999).
Concluding Remarks and Beyond
The last few decades have uncovered profound insights into the biology of antigen presentation, but many components still have to be defined to unequivocally understand the complex cross-presentation logistics from antigen uptake and routing, processing, and peptide loading. The many conflicting opinions in the field might be partially explained by the fact that the process has different requirements depending on the cell type (e.g., CD8α+ vs CD8α− DC or Mo-DCs), on the antigen form (soluble, antibody-, or cell- associated), and source (necrotic, apoptotic, infected, or tumor cell), and on the uptake route (gap junctions, receptor-mediated endocytosis). Also, we cannot exclude the option that multiple pathways may be active in the cross-presentation of a specific antigen. Moreover, a study on the cell biology of cross-presentation usually focuses on one step of the process, without relating it to the upstream and downstream events. For the above reasons, drawing a general consensual scheme of cross-presentation based on independent studies is akin to putting together a picture puzzle using a collection of pieces originating from different sources.
Another consideration is the difference in kinetics between the different mechanisms. The endosomal cross-presentation pathway may be faster that the cytosolic one, allowing rapid expression of significant levels of MHC I with cross-presented peptides at the cell surface (Burgdorf et al., 2008; Di Pucchio et al., 2008). Moreover, the pathways engaged for cross-presentation appear to be cell type dependent (Segura et al., 2009). While inflammatory Mo-DCs may be relying mainly on the fast endosomal pathway, the ability of “steady state” CD8α+ DCs to cross-present seems to depend more on the cytosolic pathway. All these variations should have major implications when the cell biology of cross-presentation is translated into the design of new therapies aiming to target cross-presentation by DCs to stimulate specific CTL to control infections and cancer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by European Research Council (ERC) and Netherlands Organization for Scientific Research (NWO) grants to Jacques Neefjes. Robbert Spaapen, Sander van Kasteren, and Shalin Naik are warmly acknowledged for their critical reading of the review.
References
- Ackerman A. L., Kyritsis C., Tampe R., Cresswell P. (2005). Access of soluble antigens to the endoplasmic reticulum can explain cross-presentation by dendritic cells. Nat. Immunol. 6, 107–113 10.1038/ni1147 [DOI] [PubMed] [Google Scholar]
- Asano K., Nabeyama A., Miyake Y., Qiu C. H., Kurita A., Tomura M., Kanagawa O., Fujii S., Tanaka M. (2011). CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity 34, 85–95 10.1016/j.immuni.2010.12.011 [DOI] [PubMed] [Google Scholar]
- Bachem A., Guttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H. W., Henn V., Kloetze P. M., Gurka S., Kroczek R. A. (2010). Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207, 1273–1281 10.1084/jem.20100348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoui S., Whitney P. G., Waithman J., Eidsmo L., Wakim L., Caminschi I., Allan R. S., Wojtasiak M., Shortman K., Carbone F. R., Brooks A. G., Heath W. R. (2009). Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat. Immunol. 10, 488–495 10.1038/ni.1724 [DOI] [PubMed] [Google Scholar]
- Brossart P., Bevan M. J. (1997). Presentation of exogenous protein antigens on major histocompatibility complex class I molecules by dendritic cells: pathway of presentation and regulation by cytokines. Blood 90, 1594–1599 [PMC free article] [PubMed] [Google Scholar]
- Burgdorf S., Kautz A., Bohnert V., Knolle P. A., Kurts C. (2007). Distinct pathways of antigen uptake and intracellular routing in CD4 and CD8 T cell activation. Science 316, 612–616 10.1126/science.1137971 [DOI] [PubMed] [Google Scholar]
- Burgdorf S., Lukacs-Kornek V., Kurts C. (2006). The mannose receptor mediates uptake of soluble but not of cell-associated antigen for cross-presentation. J. Immunol. 176, 6770–6776 [DOI] [PubMed] [Google Scholar]
- Burgdorf S., Scholz C., Kautz A., Tampe R., Kurts C. (2008). Spatial and mechanistic separation of cross-presentation and endogenous antigen presentation. Nat. Immunol. 9, 558–566 10.1038/ni.1601 [DOI] [PubMed] [Google Scholar]
- Caminschi I., Proietto A. I., Ahmet F., Kitsoulis S., Shin Teh J., Lo J. C., Rizzitelli A., Wu L., Vremec D., van Dommelen S. L., Campbell I. K., Maraskovsky E., Braley H., Davey G. M., Mottram P., van de Velde N., Jensen K., Lew A. M., Wright M. D., Heath W. R., Shortman K., Lahoud M. H. (2008). The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood 112, 3264–3273 10.1182/blood-2008-05-155176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian I., Visentin G., Blanchard N., Jouve M., Bobard A., Moita C., Enninga J., Moita L. F., Amigorena S., Savina A. (2011). Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell 147, 1355–1368 10.1016/j.cell.2011.11.021 [DOI] [PubMed] [Google Scholar]
- Dantuma N. P., Groothuis T. A., Salomons F. A., Neefjes J. (2006). A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol. 173, 19–26 10.1083/jcb.200510071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J., Souwer Y., Jorritsma T., Klaasse Bos H., ten Brinke A., Neefjes J., van Ham S. M. (2010). Antigen-specific B cells reactivate an effective cytotoxic T cell response against phagocytosed Salmonella through cross-presentation. PLoS ONE 5, e13016. 10.1371/journal.pone.0013016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio M. L., Rodriguez-Barbosa J. I., Kremmer E., Forster R. (2007). CD103- and CD103+ bronchial lymph node dendritic cells are specialized in presenting and cross-presenting innocuous antigen to CD4+ and CD8+ T cells. J. Immunol. 178, 6861–6866 [DOI] [PubMed] [Google Scholar]
- Delamarre L., Pack M., Chang H., Mellman I., Trombetta E. S. (2005). Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307, 1630–1634 10.1126/science.1108003 [DOI] [PubMed] [Google Scholar]
- den Haan J. M., Bevan M. J. (2002). Constitutive versus activation-dependent cross-presentation of immune complexes by CD8(+) and CD8(−) dendritic cells in vivo. J. Exp. Med. 196, 817–827 10.1084/jem.20020295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan J. M., Lehar S. M., Bevan M. J. (2000). CD8(+) but not CD8(−) dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192, 1685–1696 10.1084/jem.192.12.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pucchio T., Chatterjee B., Smed-Sorensen A., Clayton S., Palazzo A., Montes M., Xue Y., Mellman I., Banchereau J., Connolly J. E. (2008). Direct proteasome-independent cross-presentation of viral antigen by plasmacytoid dendritic cells on major histocompatibility complex class I. Nat. Immunol. 9, 551–557 10.1038/ni.1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure F., Mantegazza A., Sadaka C., Sedlik C., Jotereau F., Amigorena S. (2009). Long-lasting cross-presentation of tumor antigen in human DC. Eur. J. Immunol. 39, 380–390 10.1002/eji.200838669 [DOI] [PubMed] [Google Scholar]
- Fonteneau J. F., Kavanagh D. G., Lirvall M., Sanders C., Cover T. L., Bhardwaj N., Larsson M. (2003). Characterization of the MHC class I cross-presentation pathway for cell-associated antigens by human dendritic cells. Blood 102, 4448–4455 10.1182/blood-2003-06-1801 [DOI] [PubMed] [Google Scholar]
- Gagnon E., Duclos S., Rondeau C., Chevet E., Cameron P. H., Steele-Mortimer O., Paiement J., Bergeron J. J., Desjardins M. (2002). Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110, 119–131 10.1016/S0092-8674(02)00797-3 [DOI] [PubMed] [Google Scholar]
- Gromme M., Uytdehaag F. G., Janssen H., Calafat J., van Binnendijk R. S., Kenter M. J., Tulp A., Verwoerd D., Neefjes J. (1999). Recycling MHC class I molecules and endosomal peptide loading. Proc. Natl. Acad. Sci. U.S.A. 96, 10326–10331 10.1073/pnas.96.18.10326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermonprez P., Saveanu L., Kleijmeer M., Davoust J., Van Endert P., Amigorena S. (2003). ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature 425, 397–402 10.1038/nature01911 [DOI] [PubMed] [Google Scholar]
- Haug G., Leemhuis J., Tiemann D., Meyer D. K., Aktories K., Barth H. (2003). The host cell chaperone Hsp90 is essential for translocation of the binary Clostridium botulinum C2 toxin into the cytosol. J. Biol. Chem. 278, 32266–32274 10.1074/jbc.M303980200 [DOI] [PubMed] [Google Scholar]
- Herrera O. B., Golshayan D., Tibbott R., Salcido Ochoa F., James M. J., Marelli-Berg F. M., Lechler R. I. (2004). A novel pathway of alloantigen presentation by dendritic cells. J. Immunol. 173, 4828–4837 [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B. T., Purtha W. E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B. U., Unanue E. R., Diamond M. S., Schreiber R. D., Murphy T. L., Murphy K. M. (2008). Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M., Bertholet S., Gagnon E., Brunet S., Goyette G., Laplante A., Princiotta M. F., Thibault P., Sacks D., Desjardins M. (2003). Phagosomes are competent organelles for antigen cross-presentation. Nature 425, 402–406 10.1038/nature01912 [DOI] [PubMed] [Google Scholar]
- Imai T., Kato Y., Kajiwara C., Mizukami S., Ishige I., Ichiyanagi T., Hikida M., Wang J. Y., Udono H. (2011). Heat shock protein 90 (HSP90) contributes to cytosolic translocation of extracellular antigen for cross-presentation by dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 108, 16363–16368 10.1073/pnas.1108372108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancic C., Savina A., Wasmeier C., Tolmachova T., El-Benna J., Dang P. M., Pascolo S., Gougerot-Pocidalo M. A., Raposo G., Seabra M. C., Amigorena S. (2007). Rab27a regulates phagosomal pH and NADPH oxidase recruitment to dendritic cell phagosomes. Nat. Cell Biol. 9, 367–378 10.1038/ncb1552 [DOI] [PubMed] [Google Scholar]
- Jongbloed S. L., Kassianos A. J., McDonald K. J., Clark G. J., Ju X., Angel C. E., Chen C. J., Dunbar P. R., Wadley R. B., Jeet V., Vulink A. J., Hart D. N., Radford K. J. (2010). Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 207, 1247–1260 10.1084/jem.20092140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijl C., Savage N. D., Marsman M., Tuin A. W., Janssen L., Egan D. A., Ketema M., van den Nieuwendijk R., van den Eeden S. J., Geluk A., Poot A., van der Marel G., Beijersbergen R. L., Overkleeft H., Ottenhoff T. H., Neefjes J. (2007). Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature 450, 725–730 10.1038/nature06345 [DOI] [PubMed] [Google Scholar]
- Lakadamyali M., Rust M. J., Zhuang X. (2006). Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell 124, 997–1009 10.1016/j.cell.2005.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber C. A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O’Keeffe M., Mann M. (2010). Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289 10.1016/j.immuni.2010.01.013 [DOI] [PubMed] [Google Scholar]
- Merzougui N., Kratzer R., Saveanu L., van Endert P. (2011). A proteasome-dependent, TAP-independent pathway for cross-presentation of phagocytosed antigen. EMBO Rep. 12, 1257–1264 10.1038/embor.2011.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefjes J., Jongsma M. L., Paul P., Bakke O. (2011). Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 [DOI] [PubMed] [Google Scholar]
- Neefjes J. J., Verkerk J. M., Broxterman H. J., van der Marel G. A., van Boom J. H., Ploegh H. L. (1988). Recycling glycoproteins do not return to the cis-Golgi. J. Cell Biol. 107, 79–87 10.1083/jcb.107.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijssen J., Herberts C., Drijfhout J. W., Reits E., Janssen L., Neefjes J. (2005). Cross-presentation by intercellular peptide transfer through gap junctions. Nature 434, 83–88 10.1038/nature03290 [DOI] [PubMed] [Google Scholar]
- Neijssen J., Pang B., Neefjes J. (2007). Gap junction-mediated intercellular communication in the immune system. Prog. Biophys. Mol. Biol. 94, 207–218 10.1016/j.pbiomolbio.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Oh J. Z., Kurche J. S., Burchill M. A., Kedl R. M. (2011). TLR7 enables cross-presentation by multiple dendritic cell subsets through a type I IFN-dependent pathway. Blood 118, 3028–3038 10.1182/blood-2011-04-348839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura J., Tamura Y., Kamiguchi K., Kutomi G., Sahara H., Torigoe T., Himi T., Sato N. (2011). Extracellular heat shock protein 90 plays a role in translocating chaperoned antigen from endosome to proteasome for generating antigenic peptide to be cross-presented by dendritic cells. Int. Immunol. 23, 223–237 10.1093/intimm/dxq475 [DOI] [PubMed] [Google Scholar]
- Pang B., Neefjes J. (2010). Coupled for cross-presentation in tumor immunotherapy. Sci. Transl. Med. 2, 44ps40. 10.1126/scitranslmed.3001245 [DOI] [PubMed] [Google Scholar]
- Pang B., Neijssen J., Qiao X., Janssen L., Janssen H., Lippuner C., Neefjes J. (2009). Direct antigen presentation and gap junction mediated cross-presentation during apoptosis. J. Immunol. 183, 1083–1090 10.4049/jimmunol.0900861 [DOI] [PubMed] [Google Scholar]
- Poulin L. F., Salio M., Griessinger E., Anjos-Afonso F., Craciun L., Chen J. L., Keller A. M., Joffre O., Zelenay S., Nye E., Le Moine A., Faure F., Donckier V., Sancho D., Cerundolo V., Bonnet D., Sousa C. (2010). Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 207, 1261–1271 10.1084/jem.20092618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins S. H., Walzer T., Dembele D., Thibault C., Defays A., Bessou G., Xu H., Vivier E., Sellars M., Pierre P., Sharp F. R., Chan S., Kastner P., Dalod M. (2008). Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9, R17. 10.1186/gb-2008-9-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Regnault A., Kleijmeer M., Ricciardi-Castagnoli P., Amigorena S. (1999). Selective transport of internalized antigens to the cytosol for MHC class I presentation in dendritic cells. Nat. Cell Biol. 1, 362–368 10.1038/14058 [DOI] [PubMed] [Google Scholar]
- Saccheri F., Pozzi C., Avogadri F., Barozzi S., Faretta M., Fusi P., Rescigno M. (2010). Bacteria-induced gap junctions in tumors favor antigen cross-presentation and antitumor immunity. Sci. Transl. Med. 2, 44ra57. 10.1126/scitranslmed.3000739 [DOI] [PubMed] [Google Scholar]
- Sancho D., Joffre O. P., Keller A. M., Rogers N. C., Martinez D., Hernanz-Falcon P., Rosewell I., Reis e Sousa C. (2009). Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature 458, 899–903 10.1038/nature07750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu L., Carroll O., Weimershaus M., Guermonprez P., Firat E., Lindo V., Greer F., Davoust J., Kratzer R., Keller S. R., Niedermann G., van Endert P. (2009). IRAP identifies an endosomal compartment required for MHC class I cross-presentation. Science 325, 213–217 10.1126/science.1172845 [DOI] [PubMed] [Google Scholar]
- Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I. C., Lennon-Dumenil A. M., Seabra M. C., Raposo G., Amigorena S. (2006). NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell 126, 205–218 10.1016/j.cell.2006.05.035 [DOI] [PubMed] [Google Scholar]
- Schuurhuis D. H., Ioan-Facsinay A., Nagelkerken B., van Schip J. J., Sedlik C., Melief C. J., Verbeek J. S., Ossendorp F. (2002). Antigen-antibody immune complexes empower dendritic cells to efficiently prime specific CD8+ CTL responses in vivo. J. Immunol. 168, 2240–2246 [DOI] [PubMed] [Google Scholar]
- Segura E., Albiston A. L., Wicks I. P., Chai S. Y., Villadangos J. A. (2009). Different cross-presentation pathways in steady-state and inflammatory dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 106, 20377–20381 10.1073/pnas.0808263106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Kapp E., Gupta N., Wong J., Lim J., Ji H., Heath W. R., Simpson R., Villadangos J. A. (2010). Differential expression of pathogen-recognition molecules between dendritic cell subsets revealed by plasma membrane proteomic analysis. Mol. Immunol. 47, 1765–1773 10.1016/j.molimm.2010.02.028 [DOI] [PubMed] [Google Scholar]
- Segura E., Villadangos J. A. (2011). A modular and combinatorial view of the antigen cross-presentation pathway in dendritic cells. Traffic 12, 1677–1685 10.1111/j.1600-0854.2011.01254.x [DOI] [PubMed] [Google Scholar]
- Shen L., Sigal L. J., Boes M., Rock K. L. (2004). Important role of cathepsin S in generating peptides for TAP-independent MHC class I crosspresentation in vivo. Immunity 21, 155–165 10.1016/j.immuni.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Tacken P. J., Ginter W., Berod L., Cruz L. J., Joosten B., Sparwasser T., Figdor C. G., Cambi A. (2011). Targeting DC-SIGN via its neck region leads to prolonged antigen residence in early endosomes, delayed lysosomal degradation, and cross-presentation. Blood 118, 4111–4119 10.1182/blood-2011-07-367615 [DOI] [PubMed] [Google Scholar]
- Taylor M., Navarro-Garcia F., Huerta J., Burress H., Massey S., Ireton K., Teter K. (2010). Hsp90 is required for transfer of the cholera toxin A1 subunit from the endoplasmic reticulum to the cytosol. J. Biol. Chem. 285, 31261–31267 10.1074/jbc.M109.088559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfoort N., Camps M. G., Khan S., Filippov D. V., Weterings J. J., Griffith J. M., Geuze H. J., van Hall T., Verbeek J. S., Melief C. J., Ossendorp F. (2009). Antigen storage compartments in mature dendritic cells facilitate prolonged cytotoxic T lymphocyte cross-priming capacity. Proc. Natl. Acad. Sci. U.S.A. 106, 6730–6735 10.1073/pnas.0900969106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim L. M., Bevan M. J. (2011). Cross-dressed dendritic cells drive memory CD8+ T-cell activation after viral infection. Nature 471, 629–632 10.1038/nature09863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck M. M., Appel S., Werth D., Sinzger C., Bringmann A., Grunebach F., Brossart P. (2008). hDectin-1 is involved in uptake and cross-presentation of cellular antigens. Blood 111, 4264–4272 10.1182/blood-2006-10-051375 [DOI] [PubMed] [Google Scholar]
- Win S. J., Ward V. K., Dunbar P. R., Young S. L., Baird M. A. (2011). Cross-presentation of epitopes on virus-like particles via the MHC I receptor recycling pathway. Immunol. Cell Biol. 89, 681–688 10.1038/icb.2010.161 [DOI] [PubMed] [Google Scholar]
- Wolfers J., Lozier A., Raposo G., Regnault A., Thery C., Masurier C., Flament C., Pouzieux S., Faure F., Tursz T., Angevin E., Amigorena S., Zitvogel L. (2001). Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 7, 297–303 10.1038/85438 [DOI] [PubMed] [Google Scholar]
- Zehner M., Chasan A. I., Schuette V., Embgenbroich M., Quast T., Kolanus W., Burgdorf S. (2011). Mannose receptor polyubiquitination regulates endosomal recruitment of p97 and cytosolic antigen translocation for cross-presentation. Proc. Natl. Acad. Sci. U.S.A. 108, 9933–9938 10.1073/pnas.1102397108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Zhou J., Zhang J., Li J., Liu N., Chai L., Li N., Liu T., Li L., Xie Z., Liu H., Wan Y., Wu Y. (2009). The GTPase Rab3b/3c-positive recycling vesicles are involved in cross-presentation in dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 106, 15801–15806 10.1073/pnas.0911963106 [DOI] [PMC free article] [PubMed] [Google Scholar]