Abstract
Despite intensive investigation, the mechanisms of T cell receptor (TCR)-mediated signal generation remain poorly understood. Here we review various dynamic processes at the cell membrane that might critically control this signaling. Firstly, we summarize recent reports providing new information on the sensitivity of TCR/ligand interaction to the membrane environment and particularly to applied forces. Secondly, we review recent evidence that forces and displacements are continuously generated at cell surfaces. Thirdly, we summarize recent experimental evidence demonstrating the capacity of forces to generate signals. Lastly, we provide a quantitative model to exemplify the capacity of dynamic processes to modulate TCR properties such as specificity that were previously difficult to explain with conventional models. It is concluded that the described dynamic processes must be integrated into current models of TCR signaling.
Keywords: T lymphocyte activation, T cell receptor, dynamics, forces, mechanotransduction, membrane curvature, rafts, antigen presenting cell
INTRODUCTION
Despite intensive investigation, the mechanisms of T cell receptor (TCR)-mediated signal generation remains poorly understood. It was suggested that the unique challenge set on TCR recognition might impose the simultaneous involvement of several triggering mechanisms including aggregation, conformational changes, and segregation (van der Merwe and Dushek, 2011). Here we review recent evidence showing that dynamic membrane phenomena influence signal generation at the TCR/antigen presenting cell (APC) interface. Indeed, these phenomena influence at the same time the species of molecules encountering each other and the physical conditions of molecular encounters, each of which are key components of signaling. Here, while numerous costimulatory molecules have known involvement in T cell activation, we focus exclusively on the TCR.
First, we present some quantitative parameters related to the TCR binding properties.
RECENT EVIDENCE PROVIDED NEW INSIGHT INTO THE EFFECT OF THE MEMBRANE ENVIRONMENT ON TCR/pMHC INTERACTION
Since TCR/pMHC interactions occur between surface-attached molecules, i.e., under two-dimensional (2D) conditions, it is difficult to relate the T cell response to the physical properties of the TCR/pMHC interaction measured in solution (Matsui et al., 1994; Aleksic et al., 2010), i.e., under 3D conditions. Indeed (Bongrand, 1999), 2D and 3D conditions differ for several reasons: First, the kinetics of bond formation between surface-attached molecules depends on the distance between surfaces, lateral motility of receptors, and size and flexibility of interacting molecules. Second, bond dissociation between surface-attached ligands and receptors depends on the relative motion of surfaces and applied forces. Third, the formation of multivalent attachments between surface-bound ligands and receptors depends on aforementioned parameters, surface roughness and ligand density, thus obscuring the link between affinity and avidity. Over the last 15 years, many investigators have devised new ways of monitoring bond formation and dissociation between surface-attached molecules. Devices have included laminar flow chambers (Kaplanski et al., 1993; Alon et al., 1995) and atomic force microscopes (Florin et al., 1994). Biomembrane force probes were introduced to dramatically improve the atomic force microscopes: the cantilever is replaced with a soft vesicle that acts as a tunable spring, thus increasing the dynamic range of measurements (Merkel et al., 1999). While these methods have yielded much information on interactions involving selectins, integrins, cadherins, and members of the immunoglobulin superfamily (Bongrand, 1999), it was only recently that the TCR/pMHC interaction was monitored at the single-molecule level (Huang et al., 2010; Huppa et al., 2010; Puech et al., 2011; Robert et al., 2012). However, some discrepancies remain, emphasizing the need for further studies. We shall briefly describe the available data.
Interactions between fluorescently labeled TCRs borne by T cells and pMHCs embedded in a lipid bilayer were visualized with fluorescence resonance energy transfer (Huppa et al., 2010). Molecular contacts were observed with an accuracy of a few nanometers. One major finding was that 2D TCR/pMHC dissociation was 4- to 12-fold more rapid than 3D separation, a difference that was abolished upon microfilament disruption with cytochalasin or latrunculin. This supported the hypothesis that forces are applied on membrane molecules in contact areas. Another unexpected finding was that 2D molecular associations between TCR and pMHC were about 1,000-fold more rapid than 3D bond formation, suggesting a strong influence of the conformation and orientation imposed on interacting molecules.
Huang et al. (2010) used micropipettes to make contacts between individual T cells and erythrocytes or artificial beads coated with pMHCs. Interactions were studied using a biomembrane force probe via a method of binding detection with improved sensitivity: the thermal fluctuation method in which bond formation was revealed by a significant decrease of the thermal fluctuations of pMHC-coated beads (Chen et al., 2008). The 2D dissociation was found to be up to 8,300-fold faster than the 3D dissociation. In addition, 2D and 3D dissociation rates measured on a same TCR with a series of pMHCs were negatively correlated.
Puech et al. (2011) used atomic force microscopy to study the interaction between a murine TCR and cognate or unrelated pMHCs. TCR/pMHC recognition was associated with higher association rate, but no difference could be found between the dissociation rates measured for each pMHC. These findings are in line with the serial triggering hypothesis (Valitutti et al., 1995) emphasizing the importance of multiple TCR engagement.
Finally, a laminar flow chamber was used to monitor bond formation and dissociation between planar surfaces coated with eight different pMHCs and TCR-coated microspheres (Robert et al., 2012): dissociation rates were well correlated to values previously obtained under 3D conditions with the same molecules (Aleksic et al., 2010). Thus aforementioned discrepancies between 2D and 3D interactions might be due to active cell phenomena, demonstrating the importance of assaying the TCR/pMHC interaction in cell-free models. In addition, the association kinetics and mechanical resistance of attachments was also studied by varying the shear rate: (i) 2D and 3D association kinetics were not correlated, in accordance with previous experiments performed to assess the specific properties of 2D biomolecular association (Robert et al., 2009). (ii) Bond lifetime was substantially altered by forces of several tens of piconewtons. (iii) Data suggested a negative correlation between force parameters and activation potencies, which was also consistent with the serial triggering hypothesis (Robert et al., 2012).
In conclusion, TCR/pMHC interaction is sensitive to applied forces and movement. Recent evidence showing that forces and displacements are generated in many cell–cell interfaces therefore warrants further review.
FORCES AND MOVEMENTS AT THE CELL SURFACE
FORCES RUNNING PARALLEL TO THE CELL MEMBRANE (LONGITUDINAL FORCES)
Adherent cells exert a continual traction on underlying surfaces (Harris et al., 1980). In addition, many cell types including leukocytes were found to generate lateral oscillations of several tens of nanometer amplitude and between 0.2 and 30 Hz frequency (Krol et al., 1990). Furthermore, many cells types, including lymphocytes, spread on adherent surfaces displayed sequential waves of membrane protrusion and retraction on their periphery with a timescale of several tens of seconds (Dobereiner et al., 2006). Finally, measuring of the forces generated by cultured neurons with optical tweezers revealed the presence of alternate pulling and pushing forces to the order of 50 pN with a total cycle duration of several seconds (Shahapure et al., 2010).
FORCES AND MOVEMENTS PERPENDICULAR TO THE CELL MEMBRANE (TRANSVERSE MOVEMENTS)
Lamellipodia or filopodia are cellular sensors located where active movements are generated and receptors are concentrated (Negulescu et al., 1996), however, force generation can occur anywhere on the cell membrane. Flexible surfaces display transverse undulations of nanometer-scale amplitude generated by thermal motion (Helfrich and Servuss, 1984). It had long been considered that the relatively high surface tension generated by myosin motors strongly reduced the existence of this kind of motion on the surface of nucleated cells. However, recent studies made with interference reflection microscopy on the initial interaction between monocytic (Zidovska and Sackmann, 2006; Pierres et al., 2008) or lymphocytic (Crétel et al., 2010) cells and planar surfaces revealed the existence of transverse undulations of nanometric amplitude and Hz frequency. These motions might be involved in the detection of activation cues by cells hovering on surfaces: When monocytic THP-1 cells were deposited on fibronectin-coated surfaces, membrane undulations of typically 5 nm amplitude and 5–10 s frequency were observed. The amplitude increased when the cell-to-surface separation fell below 50 nm. In addition, the undulation amplitude at close distances increased after a 1–2 min lag, while the cell-to-surface separation distance decreased (Pierres et al., 2008). When human T lymphocytes were deposited on non-activating (anti-HLA-coated) or activating (anti-CD3-coated) surfaces, the 1–2 min lag was followed by a rapid increase in contact area the rate of which was eightfold higher on activating than on non-activating surfaces (Crétel et al., 2010), as expected from previous reports (Bunnell et al., 2001). Thus, such movements might be involved in the analysis of surrounding surfaces by living cells (Pierres et al., 2009). While the mechanical strength of these undulations was not measured, recent observations made on fibroblasts with atomic force microscopy revealed the occurrence of pulsations of 20–200 nm amplitude and 2–35 s period (Pelling et al., 2007).
In conclusion, cell membranes continuously generate longitudinal and transverse oscillations that involve forces of several tens of piconewtons and periods of a few seconds. We shall now review recent evidence that TCRs subjected to such forces may generate signaling cascades.
TCRs CAN GENERATE SIGNALING CASCADES IN RESPONSE TO FORCES
Intracellular calcium rise is a hallmark of T cell activation. It was recently reported that a tangential force but not a normal force of 50 pN applied on T cells through anti-CD3-coated magnetic beads could induce a calcium rise within a few minutes, and this effect was ascribed to a quaternary conformational change induced by forces (Kim et al., 2009). In other experiments, when T cells were bound to APCs through an engineered CD3 ligand, no calcium rise was observed in static conditions, whereas mild hydrodynamic forces triggered a calcium rise within minutes (Li et al., 2010). However, hydrodynamic forces did not induce any calcium rise in T cells tethered through CD28 receptors rather than CD3.
Thus, TCR can act as a mechanotransducer, i.e., it can generate signaling cascades in response to forces. While this role of TCRs is clear, the precise molecular mechanisms involved remain poorly understood. An in-depth review of the mechanotransduction mechanisms that were recently demonstrated in a number of cell models other than immune cells is therefore warranted to assess the possible relevance of such mechanisms to T cells.
GENERAL MECHANISMS OF MECHANOTRANSDUCTION
For the sake of clarity, we shall discuss separately (i) some mechanisms potentially involved in TCR mechanotransduction, and (ii) the expected dependence of these mechanisms on the dynamics of lipid organization in the cell membrane.
GENERAL MECHANISMS FOR MECHANOTRANSDUCTION
Membrane ionic channels may act as force transducers (Chalfie, 2009)
However, while TCR-mediated T lymphocyte activation involves an early calcium rise, most evidence suggests that the opening of calcium channels is a secondary event in the signaling cascade (Smith-Garvin et al., 2009).
Force-induced conformational changes
A force-induced conformational change of membrane-associated proteins might generate new docking sites. As previously emphasized (Pierres et al., 2009), a force F acting on a protein can induce a conformational change if this would generate a displacement d such that F × d is substantially higher than kBT, where kB is Boltzmann’s constant and T is the absolute temperature. Since kBT is about 4 pN × nanometer, a force of 40 pN could induce a significant conformational shift provided it involved a deformation of at least 0.1 nm. Interestingly, integrins may undergo deformations that are in the order of 1 nm (Salas et al., 2004). Also, a force of a few piconewtons applied on talin exposed docking sites allowing vinculin binding (Del Rio et al., 2009). Also (Ehrlicher et al., 2011), forces were found to influence the connection of actin to integrins through the actin binding protein filamin.
Force-induced displacement of a receptor relative to the membrane
A force might move membrane molecules relative to the lipid bilayer, thus masking or unmasking docking sites for cytoplasmic molecules. Indeed, a force of 20 pN might suffice to uproot a membrane-embedded molecule (Bell, 1978). It was recently suggested that the interaction of the CD3/TCR complex with membrane bilayers could result in the sequestration of some key tyrosine residues that might be exposed after ligand binding (Ma et al., 2008; Xu et al., 2008).
Signal generation following a change of membrane curvature
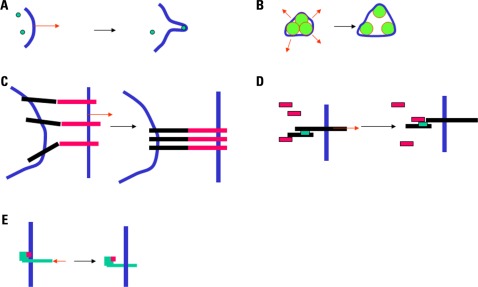
Sucking lymphocytes into a micropipette with an inner diameter in the order of a few micrometers and a pressure of a few tens of Pascals thereby generating a force of several hundreds of piconewtons (Foa et al., 1988; Tözeren et al., 1989) might generate a protrusion with a radius of curvature in the order of a micrometer. Also, the application of a pulling force of about 25 pN to a fibroblast could generate the formation of a membrane tube of about 200 nm radius (Dai and Sheetz, 1995; Raucher and Sheetz, 1999). Thus, the transverse forces described in Section “Forces and Movements Perpendicular to the Cell Membrane (Transverse Movements)” might change the membrane curvature and generate nanometer-scale protrusions. This might trigger a signal through two non-exclusive mechanisms: (i) Curvature-sensitive proteins might be recruited into a localized area and nucleate a signaling scaffold (Peter et al., 2004; Suetsugu et al., 2006). (ii) If traction forces are exerted through cell membrane adhesion receptors, these receptors may be gathered into the contact area, resulting in a local change in protein composition. All these mechanisms are depicted in Figure 1.
FIGURE 1.
FIGURE 1. Mechanisms for mechanotransduction. Five potential mechanisms for mechanotransduction are depicted. (A) A local force (red arrow) may generate a protrusion or a tether, resulting in the recruitment of curvature-sensitive molecules (green disks; Suetsugu et al., 2006). (B) Increased membrane tension (red arrows) may result in the opening of mechanosensitive channels (Chalfie, 2009). (C) A separating force (red arrow) exerted on attached cells may result in the concentration of adhesion receptors into a smaller area (Tözeren et al., 1989). (D) A force applied to a protein may result in a conformational change and exposure of docking sites shown as green rectangles (Del Rio et al., 2009). (E) A pulling or pushing force (red arrow) may alter the position of a protein with respect to the plasma membrane, resulting in the exposure of docking sites shown as pink rectangles (Xu et al., 2008).
Importantly, all these mechanisms can be strongly influenced by the nanometer-scale organization of membrane lipids. We shall now briefly discuss this point.
MECHANOTRANSDUCTION IS MODULATED BY THE HIGHLY DYNAMIC PLASMA MEMBRANE
T cell receptor/pMHC interaction is likely strongly influenced by the membrane organization and dynamics. It is thus legitimate to ask whether membrane domains such as lipid rafts could play a role in the TCR triggering mechanism. After more than one decade of intensive investigation and debate, current views converge on the notion that lipid rafts exist in the cell membrane as fluctuating molecular assemblies/domains with typical sizes of less than 100 nm (Lingwood and Simons, 2010; He and Marguet, 2011). It has been suggested that in the resting state TCRs might partition into raft nanodomains (He and Marguet, 2008; Simons and Gerl, 2010). In such a scenario, raft nanodomains could be involved in TCR triggering in several ways.
Docking sites generated by forces or enzyme processing will preferentially interact with molecules localized within the same nanodomains as the TCR (Lingwood et al., 2008)
In a recent study using single-molecule near-field scanning optical microscopy it was shown that raftophilic proteins such as CD55 or LFA-1, but not the non-raft protein CD71, were recruited to regions proximal (<150 nm) to CTxB-GM1 raft nanodomains without physical intermixing (van Zanten et al., 2010). Therefore, TCR partitioning into rafts could facilitate its encounter with Lck (Zhang et al., 2011), since the latter is most likely also a raftophilic protein.
Conformational changes induced by forces and/or ligand binding may be influenced by the local composition of the membrane
Indeed, through promoting selective lipid–protein interactions, raft nanodomains could enhance certain conformations/spatial orientations of TCR/CD3 complexes that might either positively or negatively regulate receptor activity as was demonstrated on other models: Coskun et al. (2011) recently reported that cholesterol-rich ld/lo, but not cholesterol-poor ld membrane environments enhanced the GM3 ganglioside-dependent inactivation of EGFR autophosphorylation. In the case of TCR, acidic phospholipids including several phosphoinositides were shown to bind to the cytoplasmic domain of CD3ε and CD3ζ, and were proposed to regulate the access of ITAMs by Lck (Xu et al., 2008; Deford-Watts et al., 2009,2011). Raft nanodomains could be involved in such a binding mechanism since it has been shown that they strongly contributed to the membrane recruitment of pleckstrin homology (PH) domain-containing proteins by the phosphatidylinositol-3,4,5-trisphosphate (Lasserre et al., 2008).
The recruitment of curvature-sensitive molecules should be influenced by local nanodomains
Finally, experimental evidence suggests that membrane curvature is controlled by its constituent molecules, and conversely, curvature could participate in organizing membrane proteins and lipids (Groves, 2007).
In conclusion, available evidence suggests that dynamic phenomena do influence TCR signaling. This may strongly influence the performance (i.e., sensitivity and specificity) of TCR recognition.
A SIMPLE MODEL ILLUSTRATES THE NEED TO INCORPORATE DYNAMIC PROCESSES TO BE ABLE TO EXPLAIN THE PERFORMANCE OF TCR-MEDIATED SIGNALING
The kinetic proofreading mechanism was suggested to account for the extraordinary specificity of TCR-mediated ligand recognition: how can a TCR robustly discriminate between ligands that bind with fairly comparable kinetics and affinity (McKeithan, 1995)? We shall address this question by comparing the information provided by TCR engagement under static and dynamic conditions.
The T cell decision to become activated after encountering a pMHC is at least partly linked to the lifetime of the TCR/pMHC interaction (Matsui et al., 1994). How could a T cell discriminate between two pMHCs (1) and (2) that its TCR binds with dissociation rates of respectively, say, k1 = 0.5 and k2 = 2 s-1? Suppose the criterion used is whether a bond survives for at least time t. The probability that pMHCs (1) and (2) will meet this criterion are respectively P1 = exp(-k1t) and P2 = exp(-k2t). If we require that P1/P2 be higher than, say, 100 to ensure specificity, we obtain:
This emphasizes two limitations of the discrimination procedure: firstly, the decision takes at least 3 s, in line with the kinetic proofreading mechanism, secondly, the probability that the pMHC with the lowest dissociation rate k1 be bound for at least 3 s is exp(-k1 × 3) = 0.223. Thus, the detection sensitivity is only 22%.
Sensitivity might be improved without decreasing the specificity by performing two determinations and starting activation if at least one of the binding events lasts for more than time t. This gives:
The minimum time required for a specificity ratio P1/P2 of 100 is now 3.14 s, and measurement time is at least 2 × 3.14 = 6.28 s. The detection sensitivity is [2 exp(-k1t) - exp(-2k1t)] = 0.37. Thus, specificity and sensitivity can both be increased by increasing the number of measurements. However, this also increases the measurement time, which impairs recognition speed. This might be avoided by pulling at bonds, thus increasing the dissociation rate in accordance with Bell’s law (Robert et al., 2012). This thus demonstrates how forces could help increase detection efficiency.
CONCLUSION
Recent results support the conclusion that (i) the region of initial contact between T cells and APCs is highly dynamic and thus generates forces; (ii) these forces influence TCR/pMHC binding and dissociation as well as signal generation; and (iii) these forces may strongly influence the performance of TCR recognition.
Since these mechanisms are not exclusive of previously suggested triggering mechanisms (van der Merwe and Dushek, 2011), TCR triggering might involve a combination of these and previously suggested mechanisms. Predicting the outcome of a T cell/APC encounter will thus require quantitative modeling to account for a combination of multiple mechanisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by institutional grants from INSERM and CNRS, and by specific grants from the Fondation pour la Recherche Médicale and the Agence Nationale de la Recherche (ANR-08-PCVI-0034; ANR-10-BLAN-1509). We thank Lee Leserman and AngloScribe for English language editing.
REFERENCES
- Aleksic M., Dushek O., Zhang H., Shenderov E., Chen J.-L., Cerundolo V., Coombs D., van der Merwe P. A. (2010). Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity 32 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon R., Hammer D. A., Springer T. A. (1995). Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature 374 539–542 (Correction 1995: Nature 377, 86) [DOI] [PubMed] [Google Scholar]
- Bell G. I. (1978). Models for the specific adhesion of cells to cells. Science 200 618–627 [DOI] [PubMed] [Google Scholar]
- Bongrand P. (1999). Ligand–receptor interactions. Rep. Prog. Phys. 62 921–968 [Google Scholar]
- Bunnell S. C., Kapoor V., Trible R. P., Zhang W., Samelson L. E. (2001). Dynamic actin polymerization drives T cell receptor-induced spreading: a role for the signal transduction adaptor LAT. Immunity 14 315–329 [DOI] [PubMed] [Google Scholar]
- Chalfie M. (2009). Neurosensory mechanotransduction. Nat. Rev. Mol. Cell Biol. 10 44–52 [DOI] [PubMed] [Google Scholar]
- Chen W., Zarnitzyna V. I., Sarangapani K. K., Huang J., Zhu C. (2008). Measuring receptor–ligand binding kinetics on cell surfaces: from adhesion frequency to thermal fluctuation methods. Cell. Mol. Bioeng. 1 276–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun üU., Grzybek M., Drechsel D., Simons K. (2011). Regulation of human EGF receptor by lipids. Proc. Natl. Acad. Sci. U.S.A. 108 9044–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crétel E., Touchard D., Benoliel A.-M., Bongrand P., Pierres A. (2010). Early contacts between T lymphocytes and activating surfaces. J. Phys. Condens. Matter 22 194107 [DOI] [PubMed] [Google Scholar]
- Dai J., Sheetz M. P. (1995). Mechanical properties of neuronal growth cone membranes studies by tether formation with laser optical tweezers. Biophys. J. 68 988–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deford-Watts L. M., Dougall D. S., Belkaya S., Johnson B. A., Eitson J. L., Roybal K. T., Barylko B., Albanesi J. P., Wülfing C., van Oers N. S. (2011). The CD3 zeta subunit contains a phosphoinositide-binding motif that is required for the stable accumulation of TCR–CD3 complex at the immunological synapse. J. Immunol. 186 6839–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deford-Watts L. M., Tassin T. C., Becker A. M., Medeiros J. J., Albanesi J. P., Love P. E., Wülfing C., van Oers N. S. (2009). The cytoplasmic tail of the T cell receptor CD3 epsilon subunit contains a phospholipid-binding motif that regulates T cell functions. J. Immunol. 183 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009). Stretching single talin rod molecules activates vinculin binding. Science 323 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobereiner H. G., Dubin-Thaler B. J., Hofman J. M., Xenias H. S., Sims T. N., Giannone G., Dustin M., Wiggins C. H., Sheetz M. P. (2006). Lateral membrane waves constitute a universal dynamic pattern of motile cells. Phys. Rev. Let. 97 038102 [DOI] [PubMed] [Google Scholar]
- Ehrlicher A. J., Nakamura F., Hartwig J. H., Weitz D. A., Stossel T. P. (2011). Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478 260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin E. L., Moy V. T., Gaub H. E. (1994). Adhesion forces between individual ligand–receptor pairs. Science 264 415–417 [DOI] [PubMed] [Google Scholar]
- Foa C., Mège J. L., Capo C., Benoliel A. M., Galindo J. R., Bongrand P. (1988). T-cell-mediated cytolysis: analysis of killer and target cell deformability and deformation during conjugate formation. J. Cell Sci. 89 561–573 [DOI] [PubMed] [Google Scholar]
- Groves J. T. (2007). Bending mechanics and molecular organization in biological membranes. Annu. Rev. Phys. Chem. 58 697–717 [DOI] [PubMed] [Google Scholar]
- Harris A. K., Wild P., Stopak D. (1980). Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208 177–179 [DOI] [PubMed] [Google Scholar]
- He H. T., Marguet D. (2008). T-cell antigen receptor triggering and lipid rafts: a matter of space and time scales. Talking Point on the involvement of lipid rafts in T-cell activation. EMBO Rep. 9 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H. T., Marguet D. (2011). Detecting nanodomains in living cell membrane by fluorescence correlation spectroscopy. Annu. Rev. Phys. Chem. 62 417–436 [DOI] [PubMed] [Google Scholar]
- Helfrich W., Servuss R. M. (1984). Undulations, steric interaction and cohesion of fluid membranes. Nuovo Cimento D 3 137–151 [Google Scholar]
- Huang J., Zarnitsyna V., Liu B., Edwards L. J., Jiang N., Evavold B. D., Zhu C. (2010). The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 464 932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa J. B., Axmann M., Mortelmaier M. A., Lillemeier B. F., Newell E. W., Brameshuber M., Klein L. O, Schutz, G. J., Davis M. M. (2010). TCR–peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463 963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplanski G., Farnarier C., Tissot O., Pierres A., Benoliel A.-M., Alessi M.-C., Kaplanski S., Bongrand P. (1993). Granulocyte-endothelium initial adhesion. Analysis of transient binding events mediated by E-selectin in a laminar shear flow. Biophys. J. 64 1922–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. T., Takeuchi K., Sun Z.-Y. J., Touma M., Castro C. E., Fahmy A., Lang M. J. (2009). The T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 284 31028–31037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A. Y., Grinfeldt M. G., Levin S. V., Smilgavichus A. D. (1990). Local mechanical oscillations of the cell surface within the range 0.2–30 Hz. Eur. Biophys. J. 19 93–99 [DOI] [PubMed] [Google Scholar]
- Lasserre R., Guo X. J., Conchonaud F., Hamon Y., Hawchar O., Bernard A. M., Soudja S. M., Lenne P. F., Rigneault H., Olive D., Bismuth G., Nunès J. A., Payrastre B., Marguet D., He H. T. (2008). Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat. Chem. Biol. 4 538–547 [DOI] [PubMed] [Google Scholar]
- Li Y.-C., Chen B.-M., Wu P.-C., Cheng T.-L., Kao L.-S., Tao M.-H., Lieber A., Roffler S. R. (2010). Cutting edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J. Immunol. 184 5959–5963 [DOI] [PubMed] [Google Scholar]
- Lingwood D., Ries J., Schwille P., Simons K. (2008). Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc. Natl. Acad. Sci. U.S.A. 105 10005–10010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D., Simons K. (2010). Lipid rafts as a membrane-organizing principle. Science 327 46–50 [DOI] [PubMed] [Google Scholar]
- Ma Z., Janmey P. A., Finkel T. H. (2008). The receptor deformation model of TCR triggering. FASEB J. 22 1002–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Boniface J. J., Steffner P., Reay P. A., Davis M. M. (1994). Kinetics of T-cell receptor binding to peptide/I-Ek complexes: correlation of the dissociation rate with T-cell responsiveness. Proc. Natl. Acad. Sci. U.S.A. 91 12862–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeithan T. W. (1995). Kinetic proofreading in T-cell receptor signal transduction. Proc. Natl. Acad. Sci. U.S.A. 92 5042–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel R., Nassoy P., Leung A., Ritchie K., Evans E. (1999). Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 397 50–53 [DOI] [PubMed] [Google Scholar]
- Negulescu P. A., Krasieva T. B., Khan A., Kerschbaum H. H., Cahalan M. D. (1996). Polarity of T cell shape, motility and sensitivity to antigen. Immunity 4 421–430 [DOI] [PubMed] [Google Scholar]
- Pelling A. E., Veraitc F. S., Chu C. P. K., Nicholls B. M., Hemsley A. L., Mason C., Horton M. A. (2007). Mapping correlated membrane pulsations and fluctuations in human cells. J. Mol. Recognit. 20 467–475 [DOI] [PubMed] [Google Scholar]
- Peter B. J., Kent H. M., Mill I. G., Vallis Y., Butle P. J. G., Evans P. R., McMahon H. T. (2004). BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303 495–499 [DOI] [PubMed] [Google Scholar]
- Pierres A., Benoliel A. M., Touchard D., Bongrand P. (2008). How cells tiptoe on adhesive surfaces before sticking. Biophys. J. 94 4114–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierres A., Monnet-Corti V., Benoliel A.-M., Bongrand P. (2009). Do membrane undulations help cells probe the world? Trends Cell Biol. 19 428–433 [DOI] [PubMed] [Google Scholar]
- Puech P.-H., Nevoltris D., Robert P., Limozin L., Boyer C., Bongrand P. (2011). Force measurements of TCR/pMHC recognition at T cell surface. PLoS ONE 6 e22344 10.1371/journal.pone.0022344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucher D., Sheetz M. P. (1999). Characteristics of a membrane reservoir buffering membrane tension. Biophys. J. 77 1992–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert P., Aleksic M., Dushek O., Cerundolo V., Bongrand P., van der Merwe A. (2012). Kinetics and mechanics of 2D interactions between T cell receptors and different activating ligands. Biophys. J. 102 248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert P., Limozin L., Pierres A., Bongrand P. (2009). Biomolecule association rates do not provide a complete description of bond formation. Biophys. J. 96 4642–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A., Shimaoka M., Kogan A. N, Harwood, C., von Andrian U., Springer T. A. (2004). Rolling adhesion through an extended conformation of integrin αLβ2 and relation to α I and β I-like domain interaction. Immunity 20 393–406 [DOI] [PubMed] [Google Scholar]
- Shahapure R., Difato F., Laio A., Bisson G., Ercolini E., Amin L., Ferrari E., Torre V. (2010). Force generation in lamellipodia is a probabilistic process with fast growth and retraction events. Biophys. J. 98 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Gerl M. J. (2010). Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11 688–699 [DOI] [PubMed] [Google Scholar]
- Smith-Garvin J. E., Koretzky G. A., Jordan M. S. (2009). T cell activation. Annu. Rev. Immunol. 27 591–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S., Murayama K., Sakamoto A., Hanawa-Suetsugu K., Seto A., Oikawa T. Mishima, C., Shirouzu M., Takenawa T., Yokoyama S. (2006). The RAC binding domain/IRSP53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J. Biol. Chem. 281 35347–35358 [DOI] [PubMed] [Google Scholar]
- Tüozeren A., Sung K. L. P., Chien S. (1989). Theoretical and experimental study on cross-bridge migration during cell disaggregation. Biophys. J. 55 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valitutti S., Müuller S., Cella M., Padovan E., Lanzavecchia A. (1995). Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature 375 148–151 [DOI] [PubMed] [Google Scholar]
- van der Merwe P. A., Dushek O. (2011). Mechanisms for T cell receptor triggering. Nat. Rev. Immunol. 11 47–55 [DOI] [PubMed] [Google Scholar]
- van Zanten T. S., Gómez J., Manzo C., Cambi A., Buceta J., Reigada R., Garcia-Parajo M. F. (2010). Direct mapping of nanoscale compositional connectivity on intact cell membranes. Proc. Natl. Acad. Sci. U.S.A. 107 15437–15442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Gagnon E., Call M. E., Schnell J. R., Schwieters C. D., Carman C. V., Chou J. J., Wucherpfennig K. W. (2008). Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell 135 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Cordoba S. P., Dushek O., van der Merwe P. A. (2011). Basic residues in the T-cell receptor ζ cytoplasmic domain mediate membrane association and modulate signaling. Proc. Natl. Acad. Sci. U.S.A. 108 19323–19328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidovska A., Sackmann E. (2006). Brownian motion of nucleated cell envelopes impedes adhesion. Phys. Rev. Let. 96 048103 [DOI] [PubMed] [Google Scholar]