Abstract
The heart is a pump that is comprised of cardiac myocytes and other cell types and whose proper function is critical to quality of life. The ability to trigger regeneration of heart muscle following injury eludes adult mammals, a deficiency of great clinical impact. Major research efforts are attempting to change this through advances in cell therapy or activating endogenous regenerative mechanisms that exist only early in life. By contrast with mammals, lower vertebrates like zebrafish demonstrate an impressive natural capacity for cardiac regeneration throughout life. This review will cover recent progress in the field of heart regeneration with a focus on endogenous regenerative capacity and its potential manipulation.
1. Introduction
As more nations have become developed, heart failure has come to be a leading cause of death worldwide. This condition, in which the heart is unable to supply sufficient blood flow to other vital organs, is most often triggered by an ischemic event like myocardial infarction (MI) or non-ischemic events also causing myocardial loss. After cardiomyocyte death, there is no significant replacement of old muscle with new, and a collagen-rich scar is generated by fibroblasts concomitant with removal of cell debris. While scarring is essential as a quick fix, a non-contractile scar cannot restore cardiac output and can cause arrhythmia. In many cases, heart failure signs can be medically managed, while other cases require cardiac transplantation. Undoubtedly, regenerative strategies that improve cardiac function by restoring electrically coupled cardiomyocytes to injured or diseased hearts would be manna from heaven to cardiologists and their patients. In this review, we will focus on the efforts that have been made to uncover, understand, and enhance regenerative capacity after cardiomyocyte loss.
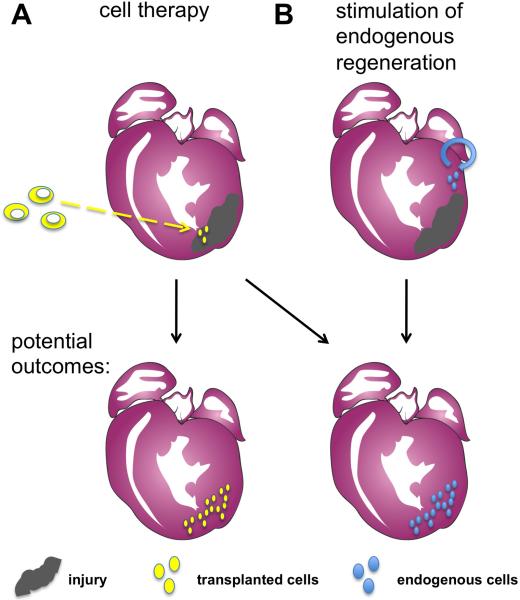
One option for replenishing a depleted cardiomyocyte population is transplantation of exogenous cells (Figure 1A). Recent studies of embryogenesis and stem cell biology have improved our understanding of cardiomyocyte differentiation from progenitor cells. Drawing from these advances, various cell types from embryonic stem (ES) cells to differentiated cardiomyocytes have been injected into injured mammalian hearts and examined for their potential to engraft and generate new cardiomyocytes. While progress has been made, there remain technical and biological hurdles that limit current success of these approaches. Most notably, while functional improvements have been reported, there exists no consensus agreement on a cell type and delivery strategy that provides faithful incorporation and electrical coupling of the transplanted cells and/or their progeny.
Figure 1.
Approaches to stimulating heart regeneration. (A) One strategy for heart regeneration involves the injection of exogenous cells into the infarcted area of the heart. These transplanted cells could proliferate and repopulate the injured area with myocardium, or may signal to endogenous cardiac cells and promote their proliferation. (B) A second fundamental approach relies on the regenerative capacity of uninjured progenitor cells or cardiomyocytes adjacent to the infarcted area. These cells proliferate and/or differentiate in situ to build new cardiac muscle.
Complementary regenerative strategies to cell transplantation are simultaneously being pursued by the field. A seemingly less invasive option to cell therapy is to stimulate growth of new cardiomyocytes from within the milieu of the cardiac muscle spared by injury, regenerating new heart tissue from endogenous players (Figure 1B). While there are currently few clues on how to unlock the regenerative capacity of the mammalian heart, this notion is not far-fetched. Regeneration is an ancestral trait of animals that is selectively distributed among species and their organ systems (Poss, 2010). While certain organisms are able to re-grow missing body parts like heads and appendages, the regenerative capabilities of mammals are relatively modest. In humans, skin, blood, skeletal muscle, and liver regenerate efficiently following injury. In fact, evidence from studies in zebrafish and neonatal mammalians have stoked the idea that the regenerative capacity can be provoked after an injury to the adult mammalian heart.
2. Cell Therapy and Heart Regeneration
Cardiomyocyte proliferation is the dominant mechanism for cardiac growth throughout the first several days of life in rodent model systems, while cardiomyocyte hypertrophy and limited cell division is the hallmark of juvenile and adult hearts (discussed later) (Adler and Costabel, 1975; Li et al., 1996). Given the relative scarcity of natural cardiomyocyte hyperplasia in mature animals, significant effort has been invested in identifying cells that might differentiate into cardiomyocytes upon transplantation into an injured heart. The burgeoning field of stem cell biology has driven this quest, providing opportunity to investigate and direct cardiomyocyte differentiation from undifferentiated cells isolated from the heart or other tissues. One goal of these studies has been to identify markers to allow prospective isolation of cardiac stem and progenitor cell populations from embryonic and adult tissue. There are multiple sources, techniques, and markers used by different groups for isolating progenitor cells, as is the case with any tissue stem cell(s). In addition, there are new paradigms for generating cardiomyocytes in vitro from pluripotent stem cells, advances that are relevant to potential cellular therapies. Here, we will provide an overview with descriptions of a few of these promising cell populations, and we refer readers to other reviews with more extensive coverage (Hansson et al., 2009; Laflamme and Murry, 2011; Laflamme et al., 2007b; Seidel et al., 2009; Wu et al., 2008).
2.1. Adult cardiac progenitor cells
Studies of heart development in embryos have yielded a wealth of information about embryonic progenitors of cardiomyocytes, as described earlier in this issue. Logically, there is value in determining the presence and nature of analogous cardiac progenitor cells that might exist in the adult heart, as they would represent a natural source of regenerating cardiomyocytes and perhaps other important cell types. These investigations have revealed multiple distinct cell populations that can be isolated from the adult murine heart; e.g. defined by surface expression of the markers c-kit, Sca1, or the ATP-binding cassette transporter (Beltrami et al., 2003; Martin et al., 2004; Oh et al., 2003). Such cells comprise less than 1% of the total cell population in the mouse heart, and when cultured in vitro are able to differentiate into cells expressing contractile genes.
While these adult cell populations may be related to each other, their origins and potential relationship to embryonic cardiac progenitor cells remain unclear. Also, the success of these cardiac progenitor cells, particularly the population expressing the surface marker c-kit, in generating mature cardiomyocytes in vivo has varied in the literature. A population of cells expressing c-kit was isolated from the human heart and showed similar abilities to give rise to cardiomyocytes in vivo (Bearzi et al., 2007), prompting cell therapy studies. While some work indicated significant formation of new myocardium and blood vessels after addition of c-kit+ cells to a rodent MI (Bearzi et al., 2007), other results suggested modest gains (Tang et al., 2010). Some reports have been unable to detect adult myogenesis from transplanted c-kit+ cells or derivation of cardiomyocytes from adult c-kit+ cells in culture (Tallini et al., 2009; Zaruba et al., 2010). Further characterization of these cardiac cell populations is necessary both to understand their endogenous roles in healthy and injured hearts and to optimize their use in cell therapy.
2.2. Pluripotent stem cells
While cardiac progenitor cells have relatively limited lineage potential, embryonic stem (ES) cells and induced pluripotent stem (iPS) cells have the capacity to develop into almost any cell type. The key to harnessing these potentially unlimited sources of cardiomyocytes lies in the ability to consistently initiate differentiation into functional, mature cells. Using traditional differentiation protocols, embryonic stem cells give rise to approximately 1% cardiomyocytes in embryoid body cultures (Kehat et al., 2001). Guided by principles of developmental biology, multiple groups have optimized these protocols for cardiogenic purposes with reported yields of ~30–60% human embryonic stem cell-derived cardiomyocytes (Laflamme et al., 2007a; Zhu et al., 2010). These derived cells express cardiac transcription factors such as Nkx2.5 and Isllet1 and structural proteins characteristic of developing cardiomyocytes. However, these cells are still distinct from adult cardiomyocytes in morphology, function, and expression profiles. There is also interest in differentiating pluripotent cells to a mesodermal or cardiovascular progenitor cell state as they may be able to self-renew and proliferate upon transplantation (Bu et al., 2009; Kattman et al., 2011).
Progress has been made in cardiomyocyte derivation protocols, but challenges remain before ES cell- or iPS cell-derived cardiomyocytes can be used for therapeutic purposes. Human ES cell use remains controversial and any cells derived from these lineages will require immunosuppression upon transplantation. iPS cells eliminate the need for immunosuppression, but the methods used to deliver reprogramming factors have occasionally been tumorigenic, raising questions about their subsequent use in cell therapy. While these issues are being addressed, murine or human ES cell-derived cardiomyocytes have been injected into various infarcted animal hearts. These cardiomyocytes have been shown to engraft and proliferate within the scar zone with reported short-term improvements in overall heart function (Caspi et al., 2007; Fernandes et al., 2010; Laflamme et al., 2005). However, cardiomyocytes of different species have important differences (e.g. they contract at different rates), that in combination with non-contractile scar tissue obstructs successful coupling of transplanted and host cardiomyocytes.
2.3. Non-cardiac tissue stem cells
Several cell types of non-cardiac lineages have also been tested for therapeutic effects, some based on reports that they might transdifferentiate into cardiomyocytes in the heart. For instance, both haematopoietic stem cells and stromal cells derived from the bone marrow were initially reported to transdifferentiate into cardiomyocytes (Orlic et al., 2001; Toma et al., 2002), although subsequent studies have argued against these interpretations (Balsam et al., 2004; Murry et al., 2004). Interestingly, infarcted hearts that were injected with cells of either lineage have shown functional improvement in multiple studies, suggesting that they provide beneficial paracrine effects (Hatzistergos et al., 2010; Mirotsou et al., 2007). In marrow stromal cells, the release of antagonists of the Wnt pathway, among other secreted proteins, are likely contributors to these effects (Mirotsou et al., 2007). Such paracrine effects continue past the death of the injected marrow stromal cells, which only survive in the infarcted hearts for a few days or weeks (Murry et al., 2004). Skeletal muscle cells have also been examined as a potential source of cardiomyocytes for the injured heart. Similarly to bone marrow-derived cells, skeletal myoblasts engraft at least temporarily and can lead to improvements in heart function. Close examination of the transplanted cells in these studies have confirmed that transdifferentiation does not occur, as the myoblasts do not express typical cardiac markers or the cell junction proteins required for coupling to endogenous cardiomyocytes (Reinecke et al., 2002). As with bone marrow-derived cells, the majority of engrafted myoblasts that survive transplantation die within weeks, and their mechanism of action remains unclear (Seidel et al., 2009).
3. Natural Cardiac Regeneration
While the various approaches towards cell therapy provide hope for cardiac repair in mammalian models, significant steps are required to optimize their survival and functional incorporation into an injured heart. Continued fate-mapping experiments are necessary to definitively determine the outcome of the transplanted cells, as is explanation and extension of short-term beneficial effects toward possible long-term healing. Notably, any improvement observed from paracrine effects indicates that there might be value in examining and enhancing the regenerative potential of endogenous heart cells, a line of investigation relevant to the study of naturally occurring heart regeneration (Figure 1B).
In fact, natural cardiac regeneration is now well-documented in multiple non-mammalian species. Initial experiments performed in the 1960's and 1970's probed amphibians like frogs and salamanders for the ability to heal mechanical injuries to the cardiac ventricle (Rumyantsev, 1973; Rumyantsev, 1977). These studies were inspired by the discoveries that salamanders like newts regenerate many tissues that adult mammals cannot, include major appendages, tail, spinal cord, lens, jaws, portions of intestine, and retina (Tsonis, 1996). The extent of regeneration after cardiac injury varied among the reports on the topic, but a synthesis of these and more recent findings highlights a sharp contrast with responses by mammals. Markers of cell proliferation such as 3H-thymidine and BrdU incorporation were detected in cardiomyocytes, as were presence of mitotic figures (Flink, 2002; Oberpriller and Oberpriller, 1974). Removal of ~25% of the ventricle at the apex typically resulted in cardiac scarring, although minced ventricular muscle transplanted to the injury site displayed a more vigorous response of cardiomyocyte proliferation and reorganization (Bader and Oberpriller, 1979; Bader and Oberpriller, 1978; Oberpriller and Oberpriller, 1974). A recent report indicated that newts show more robust regeneration when the injury is inflicted at the base of the ventricle (Witman et al., 2011). Bolstering these in vivo results is demonstrable proliferation by adult newt cardiomyocytes cultured in vitro (Oberpriller et al., 1995), events that are rare in cultured adult mammalian cardiomyocytes (Engel et al., 2005). Thus, early studies in leopard frogs, newts, and axolotls laid the foundation for developing and understanding additional models of natural heart regeneration.
4. Heart Regeneration in Zebrafish
4.1. Injury models
The examination of heart regeneration in lower vertebrates has continued in zebrafish, a popular vertebrate model organism with embryologists for the past 20 years. Zebrafish have been a particularly tractable system with low maintenance costs, relatively short generation times, easy breeding and large clutch sizes, and transparency during early development. A large collection of mutant and transgenic strains exists, as does a growing body molecular genetic tools. An increasing number of groups have become interested in the regenerative potential of adult zebrafish, which compare to salamanders in that transected spinal cords, damaged retinae, and amputated fins regenerate (Becker et al., 1997; Otteson and Hitchcock, 2003; Poss et al., 2003; Vihtelic and Hyde, 2000). Moreover, almost 10 years ago, it was discovered that zebrafish have a robust capacity for cardiac muscle regeneration.
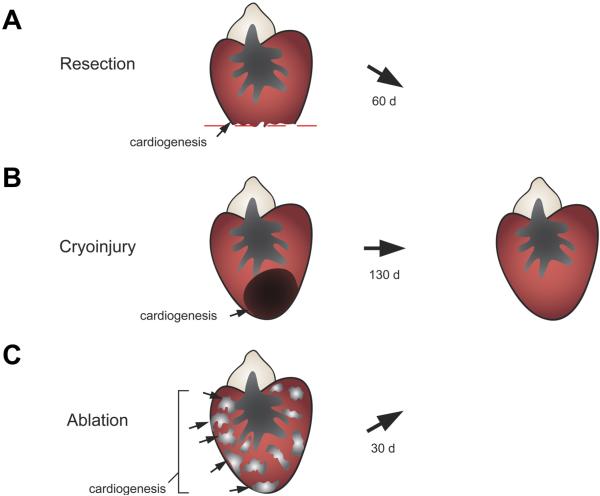
Initial studies of heart regeneration in zebrafish employed a resection model as had been performed in amphibians (Poss et al., 2002b). In these studies, approximately 20% of their single ventricle was removed with iridectomy scissors – a large injury that penetrates the lumen and causes massive bleeding. Wounds clotted quickly, become filled with mature fibrin within one week, and then with cardiac muscle over the next several days to weeks. Adult zebrafish cardiomyocytes showed indices of elevated proliferation, beginning at the end of the first week after injury. Cardiomyocyte proliferation was highest within the first 2 weeks of resection, but could be observed for weeks beyond this. By 60 days post-amputation (dpa), the ventricular wall had recovered approximately the amount of tissue that was removed (Figure 2A). The zebrafish ventricle is comprised of two main types of cardiac muscle: 1) inner myofibers organized into long, elaborate trabeculae separated from the lumen by the endocardial layer; and 2) a thin wall of compact muscle enveloped by the epicardial layer. The regenerated muscle returned largely in the form of a thickened compact myocardial wall (Poss et al., 2002b).
Figure 2.
Injury and regeneration in zebrafish. (A) In the resection model, 20% of the ventricular apex is surgically removed. Within two months, the resected tissue is replaced through local cardiomyocyte proliferation. (B) In the cryoablation model, a cooled probe is applied directly to the heart to induce localized cell death affecting up to 25% of the ventricle. The lesions can remain detectable past 60 days post-injury, but the majority of collagen is eventually cleared coincident with local cardiomyocyte proliferation. (C) Using a conditional genetic approach, over 60% of the ventricular (and atrial) myocardium is destroyed throughout the heart. Regeneration is particularly rapid and robust, with full recovery within one month after ablation.
Resection provides a major, complex injury, and stimulates a dramatic regenerative response akin to that which regenerates an amputated limb. Because an MI is characterized by local death of cardiac muscle rather than physical removal, there was considerable interest in probing whether high regenerative capacity as in the case of zebrafish can function in the setting of an infarct. In mouse and rat, this is achieved by occluding a coronary artery. However, it is unclear the extent to which one could create an ischemic MI in zebrafish, due to the small ventricular size (~ one microliter) and the fact that the coronary vasculature perfuses only a relatively small proportion of the ventricular muscle. Therefore, multiple groups recently used a cryoinjury model, creating a large region of necrotic myocardium through application of dry ice or a liquid nitrogen-cooled probe directly to the ventricle (Chablais et al., 2011; Gonzalez-Rosa et al., 2011; Schnabel et al., 2011). Like the resection model, cryoinjury killed a target region of ~25% of the ventricular muscle without impacting animal survival (Figure 2B). Moreover, while taking slightly longer to complete, the dead tissue was cleared and replaced by a contiguous myocardial wall. Vigorous cardiomyocyte proliferation accompanied these events, clear evidence for heart regeneration.
Finally, another recent study employed an inducible, genetic ablation technique for producing major cardiac injuries in zebrafish (Wang et al., 2011). Here, a double transgenic system was generated consisting of a tamoxifen-inducible CreER recombinase driven specifically in cardiac muscle cells via the cardiac myosin light chain 2 (cmlc2) promoter, combined with a transgene enabling release of expression of cytotoxic diptheria toxin A chain (DTA) by Cre-mediated recombination. After tamoxifen application, cardiomyocyte cell death was triggered diffusely throughout both cardiac chambers over the course of the next 5 to 7 days. The amount of ablation was tunable by tamoxifen administration, with histological analysis indicating that more than 60% of the ventricular myocardium can be ablated without effects on animal survival (Figure 2C). Robust cardiomyocyte proliferation and muscle recovery followed, with almost half of the ventricular cardiomyocytes replaced within a week after initial assessment (14 days post-tamoxifen treatment), and full muscular recovery by 30 days post-treatment. This injury model presents certain advantages. First, because of the extreme severity of the injuries versus other models, zebrafish showed clear, outward signs of heart failure after injury, including lethargy, reduced exercise tolerance, and stress sensitivity, that were ameliorated during regeneration. This reversal of heart failure is a striking example of the impact of high cardiac regenerative capacity. Second, as a non-surgical model, the procedure for injury is simple and offers the potential for more consistent cardiomyocyte loss and recovery among different animals. Third, genetic ablation directly injures only one of the major cardiac cell types, providing specificity that could also be employed with similar tools to ablate other cardiac cell types, and suggesting that cardiomyocyte cell death is sufficient to stimulate regeneration.
Overall, the similarities between the regenerative events after the three cardiac injury models are greater than their differences, and several themes of heart regeneration can be highlighted from this work.
4.2. Cellular hallmarks of heart regeneration
Lack of scarring
Scarring is a major obstacle to functional recovery in the infarcted mammalian heart. By contrast, scarring is a minimal component of repair in the injured zebrafish heart. The initial fibrin clot that fills a resection injury accumulates low levels of collagen, but there is typically no significant accumulation after an injury (Poss et al., 2002b). Cardiomyocyte ablation injuries also show very little collagen deposition (Wang et al., 2011). Importantly, this is not to say that zebrafish hearts cannot repair through scarring. This was first deduced from experiments using mutants in the mitotic checkpoint gene, mps1, which fail to regenerate heart muscle after ventricular resection. Instead of filling the wound with cardiac muscle, injured mutants healed injuries with large, collagen-rich scars (Poss et al., 2002b). Similar results were obtained when cardiac regeneration was inhibited by transgenic blockade of Fgf receptors (see below) (Lepilina et al., 2006). Thus, zebrafish have the capacity to repair cardiac injuries by scarring, but this mechanism is typically overridden by a robust regenerative process. Recovery from cryoablation injuries includes collagen deposition to greater extents, yet the majority of this collagen is cleared during the regenerative process (Gonzalez-Rosa et al., 2011). Therefore, while some collagen deposition occurs after resection injury and moreso after cryoinjury, a mature scar forms ostensibly only when muscle regenerative mechanisms are disabled.
Cardiac regenerative capacity would potentially have its greatest impact in humans if it were effective in MI survivors with existing ventricular scars. To test whether the zebrafish heart mounts a regenerative response in the presence of mature scar tissue, Kikuchi et al. performed resection surgery and arrested regeneration by Fgfr inhibition to create a scar (Kikuchi et al., 2010). After restoring Fgf signaling, many animals built a new, contiguous wall of muscle around the scar. These findings indicated that even in the presence of established scars, zebrafish cardiomyocytes retain regenerative capacity.
A cardiomyocyte source
Arguably the most important initial goal in investigating a regenerative process is to identify the cellular source(s) of new tissue (Tanaka and Reddien, 2011); in this case, new cardiomyocytes. In zebrafish, as in amphibians, it is clear that adult cardiomyocytes proliferate to replace new muscle, but there are several mechanisms by which these cardiomyocytes might initially arise. These include: 1) a stem or progenitor cell as during embryonic development; 2) direct proliferation by cardiomyocytes; and/or 3) reduction of the contractile program in mature myocytes, or dedifferentiation, to facilitate cell proliferation. Early studies suggested that undifferentiated progenitor cells might give rise to the regenerating cardiomyocytes, based on the induction of embryonic cardiogenic markers and the indication of cells activating the contractile gene cmlc2 de novo (Lepilina et al., 2006).
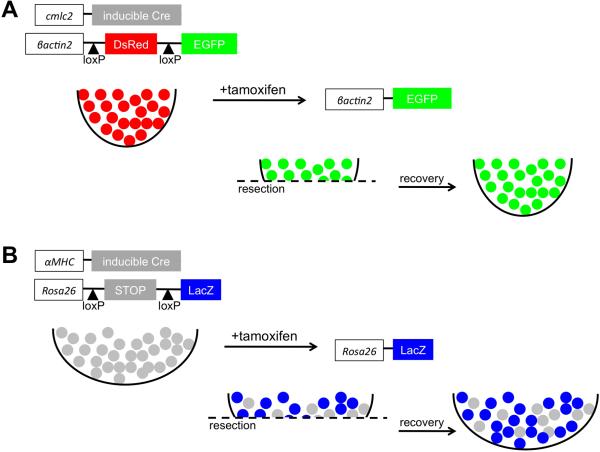
More recently, genetic fate-mapping was applied in attempts to conclusively assess existing cardiomyocytes as a source tissue (Jopling et al., 2010; Kikuchi et al., 2010). Using inducible fate-mapping techniques that employ Cre recombinase (Buckingham and Meilhac, 2011), two groups independently labeled cmlc2-expressing cardiomyocytes prior to injury. Both found that most, if not all, newly regenerated cardiomyocytes were derived from existing cardiomyocytes (Figure 3A). Similar results were found when cardiac muscle regeneration after genetic ablation injury was assessed by this method (Wang et al., 2011), suggesting a common source mechanism that is likely to also apply to the cryoinjury model.
Figure 3.
Cellular source of regenerating cardiomyocytes. (A) Genetic fate-mapping to determine the source of new cardiomyocytes during zebrafish heart regeneration. Treatment with tamoxifen leads to fluorescent EGFP marking of nearly all cardiomyocytes. Following resection, new cardiomyocytes expressed EGFP, indicating derivation from existing cardiomyocytes rather than a different unmarked cell source. (B) A similar fate-mapping experiment was performed in the neonatal mouse heart, in this case with an inducible LacZ reporter. Although the labeling of cardiomyocytes with this system was ~60%, the ratio of labeled cells was maintained between the regenerate and uninjured myocardium. This result indicates that the new cardiomyocytes are derived from existing cardiomyocytes rather than a progenitor cell.
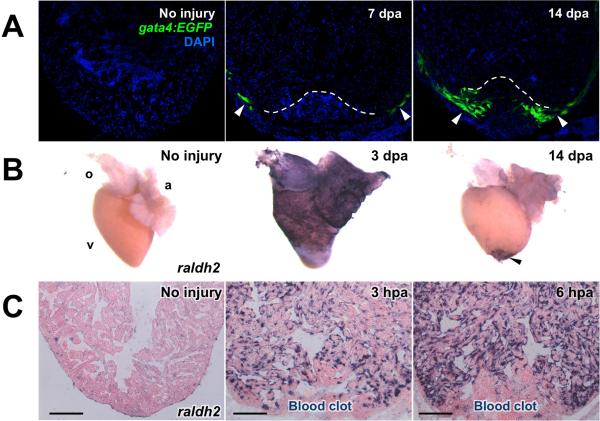
Kikuchi et al. additionally found using a transgenic reporter line that regulatory sequences of the cardiogenic transcription factor gata4 are activated throughout the peripheral compact muscle by 7 dpa (Figure 4A), and appears to label most of the regenerated ventricular wall by 14 and 30 dpa (Kikuchi et al., 2010). At 7 and 14 dpa, many of these cells co-label with BrdU, suggesting that they have recently undergone DNA synthesis. While it is known that gata4 is necessary for embryonic heart development and vascularization (Heicklen-Klein and Evans, 2004; Holtzinger and Evans, 2005), its role in heart regeneration is not yet determined. That gata4 induction marks regenerating cardiac muscle suggests that the tissue has activated an embryonic program. This is consistent with results of transmission electron microscopy and sarcomere stains, which indicated that regenerating cardiomyocytes acquire a less organized sarcomeric structure during regeneration (Jopling et al., 2010; Kikuchi et al., 2010; Wang et al., 2011). Thus, the current thinking in the field is that dedifferentiation of existing cardiomyocytes is the dominant source mechanism for heart regeneration in zebrafish.
Figure 4.
Three major cardiac cell types activate developmental gene expression in a chamber-wide manner after resection injury. (A) Myocardial activation, represented by the activation of a gata4:EGFP reporter, is first detected throughout the compact layer of the myocardium by 7 days post-amputation (dpa) and becomes localized to the regenerating myocardium by 14–30 dpa (arrowheads). Dotted lines indicate amputation planes. (B) Ostensibly the entire epicardium activates developmental genes such as raldh2 (violet) within several days following injury. Enhanced gene expression localizes to the wound area by two weeks post-injury (arrowheads). (C) Endocardial cells undergo structural remodeling following injury and upregulate raldh2 (violet) within hours of amputation. Modified from Lepilina et al., 2006, Kikuchi et al., 2010, and Kikuchi et al., 2011b.
The re-entry of resident cardiomyocytes into the cell cycle is a crucial aspect of replacing the injured cardiomyocyte population. Equally important is that these new cardiomyocytes incorporate functionally with existing cardiac muscle and do not generate arrhythmias. This was investigated using optical voltage mapping of surface myocardium at various stages of regeneration following resection surgery (Kikuchi et al., 2010). At 7 dpa, when cardiomyocytes begin to proliferate, muscle at the regenerating apex was uncoupled. A week later, coupling was evident, and by 30 dpa, electrical conduction through the apex occurred at normal velocities. Loss, and recovery, of conduction velocities was also evident after diffuse genetic ablation, and then regeneration, of cardiomyocytes (Wang et al., 2011). Thus, in addition to replacing lost tissue, the newly regenerated cardiomyocytes show evidence of functional integration.
Epicardial and endocardial activation
Another characteristic of the regenerative process in zebrafish is the activation and participation of two other major cardiac cell types, the epicardium and the endocardium, mentioned briefly earlier. These cell layers that line the cardiac muscle appear to play important structural and signaling roles during heart regeneration.
The epicardium covers the periphery of the chambers the heart and interacts closely with the myocardium during development, both as a source of fibroblasts and vascular support cells as well as cardiomyocyte mitogens. In all three injury models, trauma stimulates the epicardium to proliferate and upregulate several genes that mark the embryonic epicardium. These include wt1b and tbx18 genes that are markers of epicardium in the developing embryo (Chablais et al., 2011; Gonzalez-Rosa et al., 2011; Kikuchi et al., 2011a; Lepilina et al., 2006; Schnabel et al., 2011; Wang et al., 2011). Additionally, expression of raldh2, an enzyme involved in the synthesis of retinoic acid (RA), is strongly induced in epicardial cells throughout the chambers within the first 1–3 days of injury (Lepilina et al., 2006) (Figure 4B). These markers localize to cells within the injury site within the first week, where they subsequently incorporate into the regenerate. Various reports have suggested that epicardial cells might have the capacity to transdifferentiate and give rise to cardiomyocytes in mice (Cai et al., 2008; Smart et al., 2011; Zhou et al., 2008a). However, lineage-tracing experiments in zebrafish using a tamoxifen-inducible Cre driven by the epicardial specific tcf21 promoter indicated that this is not the case during heart development or regeneration in zebrafish. Rather, epicardial cells play a supporting role for neovascularization that occurs during the process (Kikuchi et al., 2011a). Additionally, these cells appear to promote cardiomyocyte proliferation, at least partially through the upregulation of the RA signaling pathway (described further below) (Kikuchi et al., 2011b).
The endocardial cell layer lining the inside of the cardiac chambers also plays an important role in development of the myocardium (Smith and Bader, 2007), suggesting that it may also facilitate adult heart regeneration. Following ventricular resection injury, endocardial cells not only adjacent to the injury site, but throughout the ventricle, undergo rapid morphological changes (Kikuchi et al., 2011b). Additionally, within hours, endocardial cells throughout the ventricle upregulate developmental genes including raldh2 and heart of glass (Figure 4C). This developmental activation of the endocardium completes a trifecta of chamber-wide cellular responses: the compact myocardium (gata4), the epicardium (raldh2 and others), and the endocardium each respond to a local injury in a rapid, global manner (Figure 4). How the appropriate signals are transmitted throughout the chamber, and just how much the response underlies regenerative capacity, are key issues being pursued.
This early endocardial response is transient, as by 1 dpa, endocardial cells away from the wound recover their uninjured appearance. By contrast, the morphological changes and raldh2 expression persist near the injury site at least through 7 dpa. This suggests that paracrine RA signaling from the endocardium, and presumably also from the epicardium, play a role in localizing cardiomyocyte proliferation to the injury site. Indeed, the use of transgenic lines that inhibited RA signaling had very strong inhibitory effects on proliferation during regeneration (Kikuchi et al., 2011b). Thus, both the epicardium and endocardium appear to be important cell layers for regenerating cardiac muscle, as they are for building embryonic myocardium.
4.3. Molecular factors controlling heart regeneration
The primary goal of most heart regeneration research is to identify molecules and manipulations that block or enhance the process. These discoveries in turn can be considered in the context of the poorly regenerative mammalian heart. RA signaling mentioned above is an intriguing pathway whose manipulation impacts heart regeneration. While expression analyses have identified many possible candidates (Lien et al., 2006; Raya et al., 2003), only a small list of additional developmental signaling pathways have also been functionally implicated in zebrafish heart regeneration. For example, members of the Fgf signaling pathway are upregulated following resection injury (Lepilina et al., 2006). Expression of the ligand fgf17b is activated in injured myocardium, corresponding with upregulation of the receptors fgfr2 and fgfr4 in epicardial cells within the regenerate. Pathway inhibition using a dominant-negative Fgfr transgene prevented completion of regeneration and led to the formation of a scar, as mentioned earlier. During Fgfr inhibition, epicardial cell integration and neovascularization of regenerating myocardium was decreased, suggesting that Fgfs recruit epicardial cells into the regenerate to enable its vascularization during cardiomyocyte proliferation. Lien and colleagues performed a microarray to compare gene expression at 3, 7, and 17 dpa and identified two members of the Platelet derived growth factor (Pdgf) family as exhibiting increased expression in the regenerating heart. Pdgf was demonstrated to promote DNA synthesis in cultured cardiomyocytes, suggesting a possible role in activating proliferation (Lien et al., 2006). Follow-up studies determined that Pdgf signaling involved the epicardium, and revealed that the likely primary role of Pdgf signaling during heart regeneration involves reactivating vascular development within the injury site (Kim et al., 2010).
As heart regeneration is driven by cardiomyocyte proliferation, manipulation of genes associated with the cell cycle can impact regeneration. In mps1 mutants, mentioned earlier, the injured heart retains collagen and fibrin in the place of a new, regenerated myocardial wall. On an interesting note, the mps1 mutation was identified through a forward genetic screen (in this case designed towards identifying genes involved in caudal fin regeneration), highlighting one of the advantages of studying regeneration in a genetic model organism where screens are common practice (Poss et al., 2002a). Pharmacologic inhibition of Polo-like kinase 1, a regulator of cell-cycle progression, reduced cardiomyocyte proliferation and slowed regeneration (Jopling et al., 2010). With a growing number of highly specific pharmacologic agents, this approach is quick and might translate more easily for comparing responses and requirements in mammalian tissues.
5. Endogenous Regenerative Capacity of Mice
The existence of high regenerative capacity in lower vertebrates is a unique vantage point, but key differences in cardiac and cardiomyocyte biology from mammalians model system must be taken into consideration. The four-chambered human heart is several thousand times larger and functions at much higher pressure than the two-chambered zebrafish heart. Notwithstanding, the heart rate of adult zebrafish (~120 beats/minute) is much closer than mouse (300–600) to the human resting heart rate (60–90). The majority of fish cardiomyocytes remain diploid and mononucleated into adulthood and lack the T-tubule system characteristic of mammalian cardiomyocytes (Poss, 2007). By contrast, a high percentage of mammalian cardiomyocytes increase their DNA in a stage-dependent manner. In mouse, this occurs as most cardiomyocytes withdraw from the cell cycle during the first week after birth, with the majority of these cells undergoing a final round of DNA replication followed by nuclear division to generate binucleated cardiomyocytes (Li et al., 1996). In humans, cardiomyocyte proliferation continues for several months after birth before withdrawal from the cell cycle, when cardiomyocytes undergo DNA replication without nuclear or cell division resulting in mononucleated cells with tetraploid (or higher) DNA content (Adler and Costabel, 1975; Bergmann et al., 2009). Because of available genetic tools, mice are especially attractive mammalian models for interpreting and potentially influencing human cardiac regenerative capacity.
5.1. Fetal and neonatal capacity
The paucity of cardiomyocyte proliferation in adult mammalian hearts has been well documented (detailed below), but, as mentioned earlier, cardiomyocytes in developing embryos proliferate to build muscle. To investigate whether this proliferation might be able to compensate for injury in the fetal heart, Drenckhahn et al. used a conditional X-linked knockout mouse to disrupt half of the cardiomyocytes in a fetal female mouse heart (Drenckhahn et al., 2008). The deleted gene, Holocytochrome c synthase (Hccs), is required for energy generation in cardiomyocytes, and cells lacking the gene can no longer function appropriately. Interestingly, in spite of the loss of roughly half the cardiomyocytes at 12.5 dpc, the heterozygous knockout females survived until birth. At this stage, lineage-tracing studies demonstrated that the hearts are composed of only ~10% Hccs-deficient cells, with the change in proportion being driven by an increase in the number of wild-type cells. While the precise source of the new cells was not determined by this study, these results suggested that the fetal mouse heart possesses high regenerative capacity following cardiomyocyte loss.
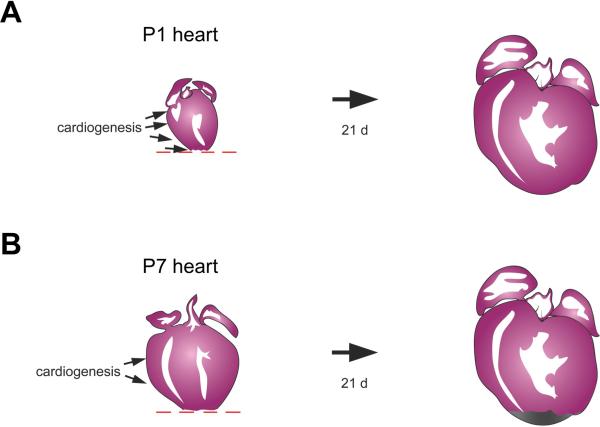
Intriguingly, it was recently reported that mice into the neonatal period are able to regenerate cardiac muscle following injury (Porrello et al., 2011). When the ventricular apex of a 1-day-old mouse was resected, an initial blood clot was replaced by regenerating cardiomyocytes over the following weeks. Most new cardiomyocytes were derived from existing cardiomyocytes, as determined by a Myh6-Cre lineage tracing experiment (Figure 3B), and proliferation was accompanied by sarcomere disassembly. This regenerative capacity was lost by 7 days after birth, when resection surgery led to the formation of a fibrotic scar (Figure 5). It will be critical to explore the differences in gene expression between cardiac tissues at these two developmental stages to perhaps uncover cues that enable cardiomyocyte regeneration. Neonatal mouse heart regeneration appears similar to adult zebrafish heart regeneration in many mechanistic ways, like muscle source and epicardial activation (Porrello et al., 2011). Different from the phenomenon in zebrafish is that the mouse heart is rapidly increasing its size during its window of regenerative potential, and that baseline cardiomyocyte is quite high both near to and away from the resection injury. Thus, the regenerated myocardium is likely to be a product of an interesting compensatory proliferation response away from the injury in addition to local proliferation. It is exciting to have a new model for natural cardiac regeneration in mammals, and its commonalities and differences with respect to events in zebrafish can inform the biology of required and flexible mechanisms for regenerative success.
Figure 5.
Transient regenerative capacity of the neonatal mouse heart after ventricular resection. (A) Rather than inducing fibrosis, resection injuries to the 1-day neonate stimulate replacement with new cardiomyocytes, as assessed 21 days later. Cardiomyocyte proliferation stimulated away from the injury is also likely to be a key component of this regenerative potential. (B) By contrast, fibrosis is the dominant response after resection of ventricular muscle in the 7-day neonate.
5.2. Adult capacity
Cardiomyocyte cell cycle entry is extremely rare in adult mammals, with one study identifying just one murine ventricular cardiomyocyte in 180,000 undergoing DNA synthesis (0.0006%) over the course of 36 hours (Soonpaa and Field, 1997). Cardiomyocyte DNA synthesis was measured after focal cauterization of the myocardium in the same study, and a slightly higher proliferation index of 0.0083% was observed. Even after cardiomyocyte cell cycle entry, an additional event of cytokinesis would be necessary to generate new contractile cells. This is a hurdle indeed, given that unlike zebrafish cardiomyocytes, a high percentage (~75%) of adult mouse cardiomyocytes are binucleate (Li et al., 1996). However, even that rare cell cycle entry events occur suggests that there is opportunity for enhancing a naturally occurring process.
Many studies have attempted to override the roadblocks to adult mammalian cardiomyocyte proliferation in vitro or in vivo by experimental introduction of potential cell-autonomous or paracrine effectors. A recent study examined the effects of continuous transgenic expression of the cell cycle activator cyclin D2 specifically in cardiomyocytes (Hassink et al., 2008). This manipulation contributed to improved cardiac function after MI, an effect attributed to more myocardium and less scar tissue in the transgenic hearts. The result suggests, as in zebrafish, that cardiomyocyte proliferation may be sufficient outcompete the scarring process in mammals. In a second study, Bersell and colleagues found that neuregulin1 (NRG1) promotes proliferation of differentiated mononucleated cardiomyocytes in cell culture (Bersell et al., 2009). Following MI injury in mouse, injection of NRG1 for 12 weeks improved cardiac function as measured by ejection fraction, and decreased scar size by 46%. This improved function was correlated with a 4.4-fold increase in cardiomyocyte proliferation indices, expected to result in approximately 700,000 new cardiomyocytes after 15 weeks of treatment. This group also visualized proliferating cardiomyocytes and found evidence that they disassemble their sarcomeres to facilitate cytokinesis. That a secreted factor could be added systemically to bolster a cardiac regenerative response is an attractive idea. It will be important to understand which cardiomyocytes respond favorably to NRG1, and to reconcile its apparent hyperplastic effects with its reported hypertrophic effects (Baliga et al., 1999).
5.3. Possible non-myocyte sources
There is great interest in determining the cell types with potential to give rise to new cardiomyocytes in adult mice. To this end, a fate-mapping experiment similar to those described above was performed in injured and uninjured animals (Hsieh et al., 2007). In this study, transgenic mice carrying a tamoxifen-inducible Cre under the control of the cardiomyocyte-specific α-myosin heavy chain (Myh6) promoter were used along with a lineage labeling strain. The researchers found that during normal aging, the percentage of labeled cardiomyocytes did not change, suggesting that any new cardiomyocytes that arise during homeostasis are not contributed by non-myocytes. Interestingly, following injury, the percentage of labeled cardiomyocytes decreased, a result that could be interpreted as dilution with cardiomyocytes derived from unlabeled cardiac progenitors. Because the Myh6-Cre initially labeled only 80% of the cardiomyocyte population, it could not be ruled out that the new cardiomyocytes might also arise from existing, unlabeled cardiomyocytes.
In a follow-up study, this system was used to examine the effects of injecting bone marrow-derived c-kit+ cells into the infarcted heart (Loffredo et al., 2011). Lineage tracing revealed that upon cell injection, the percentage of labeled cardiomyocytes was further decreased compared to sham or injections with bone marrow-derived mesenchymal stem cells. These results suggest that the addition of c-kit+ cells to the injured heart stimulates greater amounts of cardiomyocyte regeneration from an endogenous progenitor or unlabeled cardiomyocyte source than in hearts without added c-kit+ cells. Additionally, by reversing the system and transplanting c-kit+ cells of the lineage-tracing line into sex-mismatched wild-type recipients, Loffredo and colleagues could exclude transdifferentiation with a high degree of certainty. The sex-mismatched aspect of the experiment also allowed the group to examine and argue against cell fusion of the c-kit+ cells with host cells as a possible source of new cardiomyocytes. Taken together, the results point to paracrine effects from a yet-uncharacterized effector signal(s) from the transplanted cells that stimulate de novo cardiomyocyte creation.
Because of their abundance, epicardial cells and fibroblasts represent potential sources of cardiomyocytes for the injured mammalian heart. As mentioned earlier, fate-mapping experiments performed a few years ago with regulatory sequences of the epicardial markers Wt1 and Tbx18 suggested that embryonic cells of this lineage contribute significantly to ventricular myocardium (Cai et al., 2008; Zhou et al., 2008a). Conclusive interpretation of these data requires that Wt1 and Tbx18 regulatory sequences are inactive in embryonic cardiomyocytes, yet subsequent findings revealed Tbx18 expression in these cells (Christoffels et al., 2009). Although zebrafish wt1 sequences drive expression in embryonic zebrafish cardiomyocytes in addition to epicardium (Kikuchi et al., 2011a), the parallel in mice has not yet been reported. Interestingly, Wt1 is upregulated in epicardial cells in the adult mouse heart following MI, where lineage-tracing evidence indicates Wt1-expressing cells normally contribute to the canonical epicardial lineage (epicardium, fibroblasts, perivascular cells) (Zhou et al., 2011). Yet, a recent study indicates that these cells might at low frequencies differentiate into mature cardiomyocytes in the presence of injury and exogenously added Thymosin beta-4 (Smart et al., 2011). These are intriguing results with major implications, and advancing these observations with additional markers and mechanistic exploration is anticipated. This finding might explain in part the reported effects of Thymosin beta 4 on improving cardiac function in rodents after MI (Bock-Marquette et al., 2004).
While the extent to which transdifferentiation between cardiac lineages occurs naturally is debated, recent studies indicate that overexpression of certain cardiogenic genes can facilitate this process. This line of research is based on successes in fibroblast reprogramming to iPS cells (Takahashi and Yamanaka, 2006) or neurons (Vierbuchen et al., 2010), and direct reprogramming of pancreatic exocrine cells to beta cells (Zhou et al., 2008b). An initial study along these lines uncovered that a minimal combination of factors (Gata4, Tbx5, and BAF60C) are necessary to differentiate mouse mesoderm into beating cardiomyocytes (Takeuchi and Bruneau, 2009). Building on this research, a cocktail of three overexpressed transcription factors (Mef2C, Gata4, and Tbx5) was found to transform cardiac or skin fibroblasts into beating cardiomyocytes at low frequency (Ieda et al., 2010). A similar study reprogrammed mouse embryonic fibroblasts to cardiomyocytes, but in this case the cells were initially directed towards an induced pluripotent stem cell state with the transcription factors Oct4, Sox2, Klf4, and c-Myc before provocation to a cardiogenic fate by Bmp4 addition (Efe et al., 2011). Thus far, these studies have been performed in vitro, but the idea of reprogramming scar-forming fibroblasts into cardiomyocytes within the infarcted heart is undeniably attractive.
6. The regenerative capacity of human hearts
The consensus of the majority in the field is that adult human hearts have little or no significant regenerative capacity after injury. The detection of cardiomyocyte proliferation itself has historically been challenging, given reliance on pathology samples. Several groups have attempted to count cardiomyocyte numbers at different life stages with varying results (Adler, 1975; Adler and Costabel, 1975; Herget et al., 1997). To address this problem in a lineage-tracing paradigm, a recent study took advantage of the residual 14C left in the atmosphere from the testing of nuclear weapons in the 1950s (Bergmann et al., 2009). Following the agreement of the Limited Nuclear Test Ban Treaty in 1963, the levels of atmospheric 14C dropped sharply. These conditions enabled an elegant pulse-chase experiment where people alive during that time could be evaluated posthumously for the age of their cardiomyocytes versus actual birthdate. This was achieved by sorting cardiomyocyte nuclei from non-cardiomyocytes (using a specific marker, cardiac troponin T) and dating them according to 14C concentration. These experiments indicated that cardiomyocyte DNA was synthesized many years after birth in this population, an indication of cell turnover. Interestingly, the rate of turnover appeared to decrease with age, as ~1% of cardiomyocytes are estimated to turn over annually at age 25, and only 0.45% at age 75. Thus, over the course of an average lifetime, almost half of the cardiomyocyte population is replaced, suggesting existence of an endogenous mechanism of low-frequency, homeostatic regeneration. Importantly, it is unclear whether the cellular origin of new muscle is a progenitor cell or differentiated cardiomyocyte, or both. As there are large communities attempting to define mechanisms of regeneration and identify how to stimulate regeneration from both types of source, there is reason to be optimistic that either source mechanism could be accommodated and activated.
7. Perspectives
Cardiac regeneration is a fascinating biological phenomenon that is distributed unequally among species. As a potential panacea for common human heart diseases, there is urgency to secure the blueprint. While various strategies may appeal differently to developmental biologists, stem cell biologists, cardiologists, or pathologists in the field, stimulating human heart regeneration is the shared goal. This will ultimately occur via success in cell therapy and/or stimulating endogenous regenerative mechanisms. For the former, continued searches for sources, and systematic tracking of transplanted cells, is needed to improve our understanding of beneficial effects and guide the development of consistently successful therapies. For the latter, it is key to interpret examples of high regenerative capacity and to use these systems to identify the cells and molecules that drive successful heart regeneration.
From zebrafish and early studies in neonatal mice, we know that regenerative capacity is a product of at least two major components. These are: 1) a cardiomyocyte source that can be activated for regeneration; and 2) a dynamic cardiac environment comprised of non-myocardial tissue that facilitates regeneration from this source. Future studies anticipate procurement of molecular strategies for stimulating regeneration, and may inform cell therapy approaches by suggesting optimal combinations of cardiac cell types and/or cofactors, or inhibitory responses to be suppressed after cell delivery. The heart regeneration research community has gained momentum and become multi-faceted in the past 10–15 years, and it is exciting to consider what interventions will arise in the near future from a synthesis of findings.
Acknowledgments
We thank Amy Dickson for artwork. The authors were funded by postdoctoral fellowships from National Heart, Lung, and Blood Institute and American Heart Association to W.Y.C., and grants from National Heart, Lung, and Blood Institute, American Heart Association, and American Federation for Aging Research to K.D.P. K.D.P. is an Early Career Scientist of the Howard Hughes Medical Institute.
References
- Adler CP. Recent Adv Stud Cardiac Struct Metab. 1975;8:373–86. [PubMed] [Google Scholar]
- Adler CP, Costabel U. Recent Adv Stud Cardiac Struct Metab. 1975;6:343–55. [PubMed] [Google Scholar]
- Bader D, Oberpriller J. J Exp Zool. 1979;208:177–93. doi: 10.1002/jez.1402080206. [DOI] [PubMed] [Google Scholar]
- Bader D, Oberpriller JO. J Morphol. 1978;155:349–57. doi: 10.1002/jmor.1051550307. [DOI] [PubMed] [Google Scholar]
- Baliga RR, Pimental DR, Zhao YY, Simmons WW, Marchionni MA, Sawyer DB, Kelly RA. Am J Physiol. 1999;277:H2026–37. doi: 10.1152/ajpheart.1999.277.5.H2026. [DOI] [PubMed] [Google Scholar]
- Balsam LB, Wagers AJ, Christensen JL, Kofidis T, Weissman IL, Robbins RC. Nature. 2004;428:668–73. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- Bearzi C, Rota M, Hosoda T, Tillmanns J, Nascimbene A, De Angelis A, Yasuzawa-Amano S, Trofimova I, Siggins RW, Lecapitaine N, Cascapera S, Beltrami AP, D'Alessandro DA, Zias E, Quaini F, Urbanek K, Michler RE, Bolli R, Kajstura J, Leri A, Anversa P. Proc Natl Acad Sci U S A. 2007;104:14068–73. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. J Comp Neurol. 1997;377:577–95. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Cell. 2003;114:763–76. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kuhn B. Cell. 2009;138:257–70. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D. Nature. 2004;432:466–72. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR. Nature. 2009;460:113–7. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Buckingham ME, Meilhac SM. Dev Cell. 2011;21:394–409. doi: 10.1016/j.devcel.2011.07.019. [DOI] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. Nature. 2008;454:104–8. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi O, Huber I, Kehat I, Habib M, Arbel G, Gepstein A, Yankelson L, Aronson D, Beyar R, Gepstein L. J Am Coll Cardiol. 2007;50:1884–93. doi: 10.1016/j.jacc.2007.07.054. [DOI] [PubMed] [Google Scholar]
- Chablais F, Veit J, Rainer G, Jazwinska A. BMC Dev Biol. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, Kispert A. Nature. 2009;458:E8–9. doi: 10.1038/nature07916. discussion E9–10. [DOI] [PubMed] [Google Scholar]
- Drenckhahn JD, Schwarz QP, Gray S, Laskowski A, Kiriazis H, Ming Z, Harvey RP, Du XJ, Thorburn DR, Cox TC. Dev Cell. 2008;15:521–33. doi: 10.1016/j.devcel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Nat Cell Biol. 2011;13:215–22. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- Engel FB, Schebesta M, Duong MT, Lu G, Ren S, Madwed JB, Jiang H, Wang Y, Keating MT. Genes Dev. 2005;19:1175–87. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes S, Naumova AV, Zhu WZ, Laflamme MA, Gold J, Murry CE. J Mol Cell Cardiol. 2010;49:941–9. doi: 10.1016/j.yjmcc.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flink IL. Anat Embryol (Berl) 2002;205:235–44. doi: 10.1007/s00429-002-0249-6. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Development. 2011 doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- Hansson EM, Lindsay ME, Chien KR. Cell Stem Cell. 2009;5:364–77. doi: 10.1016/j.stem.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Hassink RJ, Pasumarthi KB, Nakajima H, Rubart M, Soonpaa MH, de la Riviere AB, Doevendans PA, Field LJ. Cardiovasc Res. 2008;78:18–25. doi: 10.1093/cvr/cvm101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzistergos KE, Quevedo H, Oskouei BN, Hu Q, Feigenbaum GS, Margitich IS, Mazhari R, Boyle AJ, Zambrano JP, Rodriguez JE, Dulce R, Pattany PM, Valdes D, Revilla C, Heldman AW, McNiece I, Hare JM. Circ Res. 2010;107:913–22. doi: 10.1161/CIRCRESAHA.110.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heicklen-Klein A, Evans T. Dev Biol. 2004;267:490–504. doi: 10.1016/j.ydbio.2003.09.042. [DOI] [PubMed] [Google Scholar]
- Herget GW, Neuburger M, Plagwitz R, Adler CP. Cardiovasc Res. 1997;36:45–51. doi: 10.1016/s0008-6363(97)00140-5. [DOI] [PubMed] [Google Scholar]
- Holtzinger A, Evans T. Development. 2005;132:4005–14. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- Hsieh PC, Segers VF, Davis ME, Macgillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT. Nat Med. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Nature. 2010;464:606–9. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Cell Stem Cell. 2011;8:228–40. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. J Clin Invest. 2001;108:407–14. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, Poss KD. Development. 2011a;138:2895–902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Dev Cell. 2011b;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Nature. 2010;464:601–5. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wu Q, Zhang Y, Wiens KM, Huang Y, Rubin N, Shimada H, Handin RI, Chao MY, Tuan TL, Starnes VA, Lien CL. Proc Natl Acad Sci U S A. 2010;107:17206–10. doi: 10.1073/pnas.0915016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Nat Biotechnol. 2007a;25:1015–24. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Laflamme MA, Gold J, Xu C, Hassanipour M, Rosler E, Police S, Muskheli V, Murry CE. Am J Pathol. 2005;167:663–71. doi: 10.1016/S0002-9440(10)62041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE. Nature. 2011;473:326–35. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Zbinden S, Epstein SE, Murry CE. Annu Rev Pathol. 2007b;2:307–39. doi: 10.1146/annurev.pathol.2.010506.092038. [DOI] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. Cell. 2006;127:607–19. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Li F, Wang X, Capasso JM, Gerdes AM. J Mol Cell Cardiol. 1996;28:1737–46. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. PLoS Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo FS, Steinhauser ML, Gannon J, Lee RT. Cell Stem Cell. 2011;8:389–98. doi: 10.1016/j.stem.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CM, Meeson AP, Robertson SM, Hawke TJ, Richardson JA, Bates S, Goetsch SC, Gallardo TD, Garry DJ. Dev Biol. 2004;265:262–75. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Mirotsou M, Zhang Z, Deb A, Zhang L, Gnecchi M, Noiseux N, Mu H, Pachori A, Dzau V. Proc Natl Acad Sci U S A. 2007;104:1643–8. doi: 10.1073/pnas.0610024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Soonpaa MH, Reinecke H, Nakajima H, Nakajima HO, Rubart M, Pasumarthi KB, Virag JI, Bartelmez SH, Poppa V, Bradford G, Dowell JD, Williams DA, Field LJ. Nature. 2004;428:664–8. doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]
- Oberpriller JO, Oberpriller JC. J Exp Zool. 1974;187:249–53. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- Oberpriller JO, Oberpriller JC, Matz DG, Soonpaa MH. Ann N Y Acad Sci. 1995;752:30–46. doi: 10.1111/j.1749-6632.1995.tb17404.x. [DOI] [PubMed] [Google Scholar]
- Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Entman ML, Schneider MD. Proc Natl Acad Sci U S A. 2003;100:12313–8. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine DM, Leri A, Anversa P. Nature. 2001;410:701–5. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- Otteson DC, Hitchcock PF. Vision Res. 2003;43:927–36. doi: 10.1016/s0042-6989(02)00400-5. [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Science. 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD. Semin Cell Dev Biol. 2007;18:36–45. doi: 10.1016/j.semcdb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Poss KD. Nature reviews. Genetics. 2010;11:710–22. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Keating MT, Nechiporuk A. Dev Dyn. 2003;226:202–10. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Development. 2002a;129:5141–9. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Science. 2002b;298:2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Raya A, Koth CM, Buscher D, Kawakami Y, Itoh T, Raya RM, Sternik G, Tsai HJ, Rodriguez-Esteban C, Izpisua-Belmonte JC. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11889–95. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke H, Poppa V, Murry CE. J Mol Cell Cardiol. 2002;34:241–9. doi: 10.1006/jmcc.2001.1507. [DOI] [PubMed] [Google Scholar]
- Rumyantsev PP. Z Zellforsch Mikrosk Anat. 1973;139:431–50. doi: 10.1007/BF00306596. [DOI] [PubMed] [Google Scholar]
- Rumyantsev PP. Int Rev Cytol. 1977;51:187–273. [PubMed] [Google Scholar]
- Schnabel K, Wu CC, Kurth T, Weidinger G. PLoS One. 2011;6:e18503. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel M, Borczynska A, Rozwadowska N, Kurpisz M. Cell Transplant. 2009;18:695–707. doi: 10.3727/096368909X470810. [DOI] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. Nature. 2011;474:640–4. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TK, Bader DM. Semin Cell Dev Biol. 2007;18:84–9. doi: 10.1016/j.semcdb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonpaa MH, Field LJ. Am J Physiol. 1997;272:H220–6. doi: 10.1152/ajpheart.1997.272.1.H220. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Nature. 2009 doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini YN, Greene KS, Craven M, Spealman A, Breitbach M, Smith J, Fisher PJ, Steffey M, Hesse M, Doran RM, Woods A, Singh B, Yen A, Fleischmann BK, Kotlikoff MI. Proc Natl Acad Sci U S A. 2009;106:1808–13. doi: 10.1073/pnas.0808920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM, Reddien PW. Dev Cell. 2011;21:172–85. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XL, Rokosh G, Sanganalmath SK, Yuan F, Sato H, Mu J, Dai S, Li C, Chen N, Peng Y, Dawn B, Hunt G, Leri A, Kajstura J, Tiwari S, Shirk G, Anversa P, Bolli R. Circulation. 2010;121:293–305. doi: 10.1161/CIRCULATIONAHA.109.871905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma C, Pittenger MF, Cahill KS, Byrne BJ, Kessler PD. Circulation. 2002;105:93–8. doi: 10.1161/hc0102.101442. [DOI] [PubMed] [Google Scholar]
- Tsonis PA. Limb Regeneration. Cambridge University Press; Cambridge: 1996. [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Nature. 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. J Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, Werdich AA, Yelon D, Macrae CA, Poss KD. Development. 2011;138:3421–30. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman N, Murtuza B, Davis B, Arner A, Morrison JI. Dev Biol. 2011;354:67–76. doi: 10.1016/j.ydbio.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Wu SM, Chien KR, Mummery C. Cell. 2008;132:537–43. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaruba MM, Soonpaa M, Reuter S, Field LJ. Circulation. 2010;121:1992–2000. doi: 10.1161/CIRCULATIONAHA.109.909093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, Gise A, Zhou P, Hu YW, Wang G, Zhang B, Wang L, Hall JL, Moses MA, McGowan FX, Pu WT. J Clin Invest. 2011;121:1894–904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Nature. 2008a;454:109–13. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. Nature. 2008b;455:627–32. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA. Circ Res. 2010;107:776–86. doi: 10.1161/CIRCRESAHA.110.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]