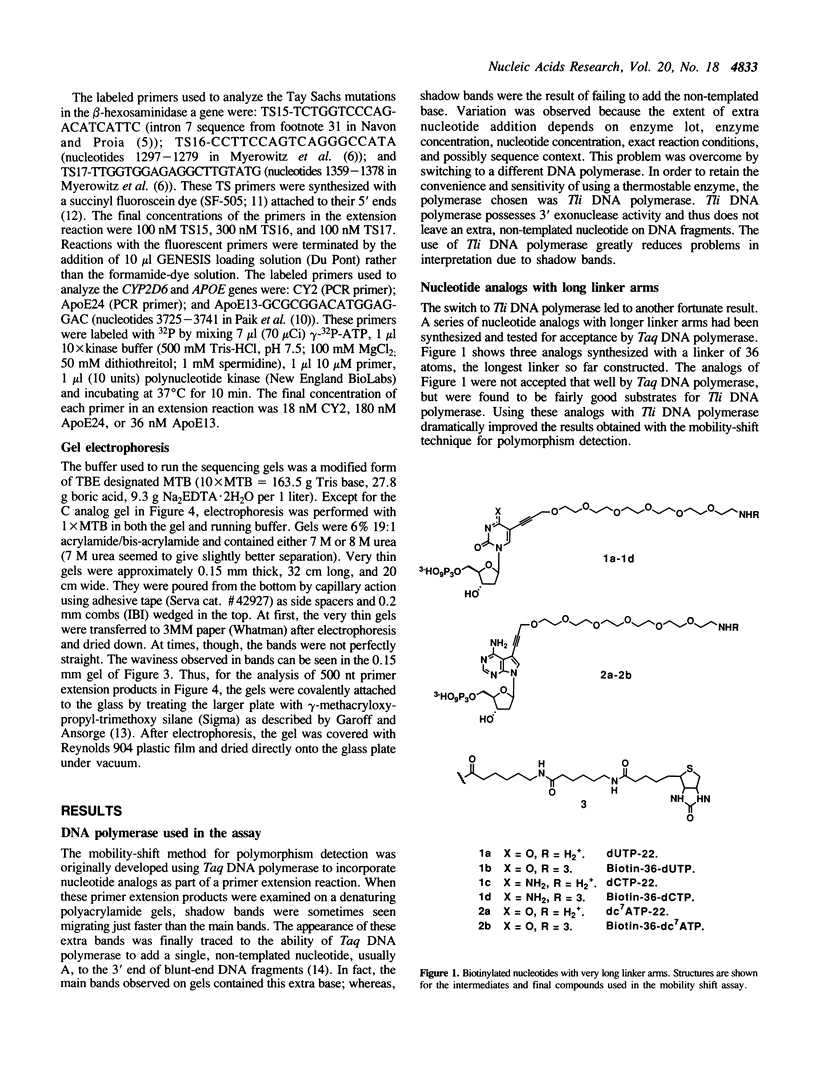
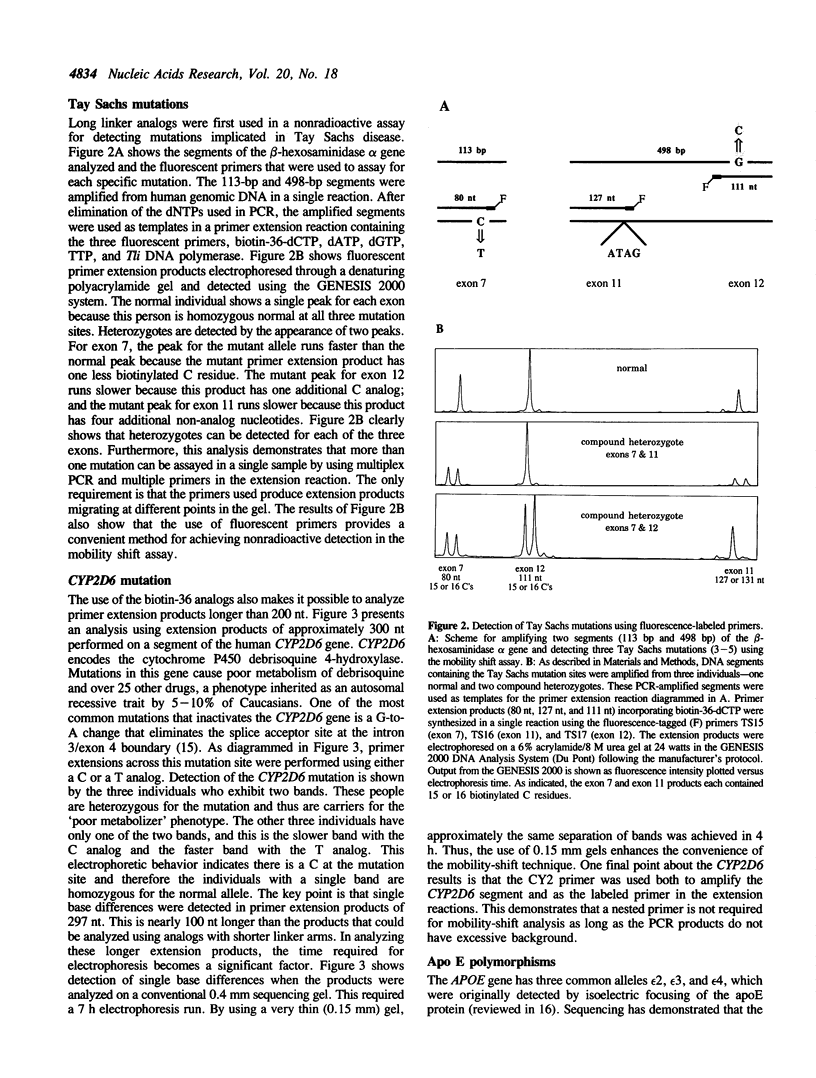
Abstract
A simple primer extension method for detecting nucleotide differences is based on the substitution of mobility-shifting analogs for natural nucleotides (1). This technique can detect any single-base difference that might occur including previously unknown mutations or polymorphisms. Two technical limitations of the original procedure have now been addressed. First, switching to Thermococcus litoralis DNA polymerase has eliminated variability believed to be due to the addition of an extra, non-templated base to the 3' end of DNA by Taq DNA polymerase. Second, with the analogs used in the original study, the mobility shift induced by a single base change can usually be resolved only in DNA segments 200 nt or smaller. This size limitation has been overcome by synthesizing biotinylated nucleotides with extraordinarily long linker arms (36 atom backbone). Using these new analogs and conventional sequencing gels (0.4 mm thick), mutations in the human beta-hexosaminidase alpha and CYP2D6 genes have been detected in DNA segments up to 300 nt in length. By using very thin (0.15 mm) gels, single-base polymorphisms in the human APOE gene have been detected in 500-nt segments.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brumley R. L., Jr, Smith L. M. Rapid DNA sequencing by horizontal ultrathin gel electrophoresis. Nucleic Acids Res. 1991 Aug 11;19(15):4121–4126. doi: 10.1093/nar/19.15.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. M. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988 Oct 25;16(20):9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton R. G., Rodrigues N. R., Campbell R. D. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davignon J., Gregg R. E., Sing C. F. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988 Jan-Feb;8(1):1–21. doi: 10.1161/01.atv.8.1.1. [DOI] [PubMed] [Google Scholar]
- Emi M., Wu L. L., Robertson M. A., Myers R. L., Hegele R. A., Williams R. R., White R., Lalouel J. M. Genotyping and sequence analysis of apolipoprotein E isoforms. Genomics. 1988 Nov;3(4):373–379. doi: 10.1016/0888-7543(88)90130-9. [DOI] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Heim M., Meyer U. A. Genotyping of poor metabolisers of debrisoquine by allele-specific PCR amplification. Lancet. 1990 Sep 1;336(8714):529–532. doi: 10.1016/0140-6736(90)92086-w. [DOI] [PubMed] [Google Scholar]
- Kagimoto M., Heim M., Kagimoto K., Zeugin T., Meyer U. A. Multiple mutations of the human cytochrome P450IID6 gene (CYP2D6) in poor metabolizers of debrisoquine. Study of the functional significance of individual mutations by expression of chimeric genes. J Biol Chem. 1990 Oct 5;265(28):17209–17214. [PubMed] [Google Scholar]
- Kimura S., Umeno M., Skoda R. C., Meyer U. A., Gonzalez F. J. The human debrisoquine 4-hydroxylase (CYP2D) locus: sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am J Hum Genet. 1989 Dec;45(6):889–904. [PMC free article] [PubMed] [Google Scholar]
- Kornher J. S., Livak K. J. Mutation detection using nucleotide analogs that alter electrophoretic mobility. Nucleic Acids Res. 1989 Oct 11;17(19):7779–7784. doi: 10.1093/nar/17.19.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer P. R., Waldrop A. A., Ward D. C. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myerowitz R., Costigan F. C. The major defect in Ashkenazi Jews with Tay-Sachs disease is an insertion in the gene for the alpha-chain of beta-hexosaminidase. J Biol Chem. 1988 Dec 15;263(35):18587–18589. [PubMed] [Google Scholar]
- Myerowitz R., Piekarz R., Neufeld E. F., Shows T. B., Suzuki K. Human beta-hexosaminidase alpha chain: coding sequence and homology with the beta chain. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7830–7834. doi: 10.1073/pnas.82.23.7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myerowitz R. Splice junction mutation in some Ashkenazi Jews with Tay-Sachs disease: evidence against a single defect within this ethnic group. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3955–3959. doi: 10.1073/pnas.85.11.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Lumelsky N., Lerman L. S., Maniatis T. Detection of single base substitutions in total genomic DNA. Nature. 1985 Feb 7;313(6002):495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- Navon R., Proia R. L. The mutations in Ashkenazi Jews with adult GM2 gangliosidosis, the adult form of Tay-Sachs disease. Science. 1989 Mar 17;243(4897):1471–1474. doi: 10.1126/science.2522679. [DOI] [PubMed] [Google Scholar]
- Orita M., Suzuki Y., Sekiya T., Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989 Nov;5(4):874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- Paik Y. K., Chang D. J., Reardon C. A., Davies G. E., Mahley R. W., Taylor J. M. Nucleotide sequence and structure of the human apolipoprotein E gene. Proc Natl Acad Sci U S A. 1985 May;82(10):3445–3449. doi: 10.1073/pnas.82.10.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober J. M., Trainor G. L., Dam R. J., Hobbs F. W., Robertson C. W., Zagursky R. J., Cocuzza A. J., Jensen M. A., Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987 Oct 16;238(4825):336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- Schmitz G. G., Walter T., Seibl R., Kessler C. Nonradioactive labeling of oligonucleotides in vitro with the hapten digoxigenin by tailing with terminal transferase. Anal Biochem. 1991 Jan;192(1):222–231. doi: 10.1016/0003-2697(91)90212-c. [DOI] [PubMed] [Google Scholar]
- Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. B., Carvalho M., Derse D., O'Brien S. J., Dean M. Detecting single base substitutions as heteroduplex polymorphisms. Genomics. 1992 Feb;12(2):301–306. doi: 10.1016/0888-7543(92)90377-5. [DOI] [PubMed] [Google Scholar]