Abstract
A number of clever recombinant methodologies have been developed that recapitulate the valencies of IgG’s (bivalent) and IgA’s (tetravalent). Although, higher synthetic valencies have been achieved by conjugation of either monoclonal antibodies or single-chain antibodies to nanoparticles and liposomes, a method for the preparation of recombinant antibodies with valencies similar to IgM’s (decavalent) but considerably less than what is generally found after antibody particle conjugation has yet to be devised. Recently, we have developed a methodology for the design of bivalent Chemically Self-Assembled Antibody Nanorings (CSANs). We now report the crystal structure of the nanoring subunit composed of the E. coli DHFR dimer and a methotrexate dimerizer (MTX2-C9) containing visible nine methylene linker and a protocol for the preparation of CSANs from this subunit with valencies similar to IgMs, ranging from 8–10 single chain antibodies (scFvs). The multivalent CSANs were reversibly assembled from a fusion protein dihydrofolate reductase (DHFR)-DHFR-antiCD3 scFv containing a single glycine linker between the two DHFR scaffolding proteins. We also demonstrate that, similar to the parental bivalent anti-CD3 monoclonal antibody (MAB), anti-CD3 CSANs selectively bind to CD3+ leukemia cells, and undergo rapid internalization through a caveolin-independent pathway that requires cholesterol, actin polymerization and protein tyrosine kinase activation. While treatment with the monoclonal antibody leads to T-cell activation and nearly complete loss (i.e. 90%) of surface displayed T-cell receptor (TCR), only 25–30% of the TCR down regulate and no significant T-cell proliferation is observed after treatment of peripheral blood mononuclear cells (PBMCs) with anti-CD3 CSANs. Consistent with the proliferation findings, 15–25 % less CD25 (IL-2 receptor) was found on the surface of PBMCs treated with either the polyvalent or bivalent anti-CD3 CSANs, respectively, than on PBMCs treated with the parental MAB. Comparative experiments with F(ab')2 derived from the MAB confirm that the activation of the T-cells by the MAB is dependent on the Fc domain, and thus interactions of the PBMC T-cells with accessory cells, such as macrophages. Taken together, our results demonstrate that anti-CD3 CSANs with valencies ranging from 2 to 8 could be employed for radionuclide, drug or potentially oligonucleotide delivery to T-cells without, as has been observed for other antibody conjugated nanoparticles, the deleterious affects of activation observed for MAB. Further the CSAN construct may be adapted for the preparation of other multivalent scFvs.
Antibodies and recombinant antibodies are a rapidly growing area for the development of therapeutics and imaging agents.1–4 Typically, antibodies are found in nature as either bivalent (IgG), tetravalent (IgA) or decavalent (IgM) species. Numerous clever and imaginative strategies for the design of recombinant antibodies have been developed.5–15 In general, these approaches have relied on the genetic manipulation of the antibody variable domains to produce monomeric, single chain antibodies (scFvs), Fab fragments, bivalent diabodies and tetravalent tantabs. In order to access valencies beyond these, typically liposomes or nanoparticles are biofunctionalized with either monoclonal antibodies (MABs) or scFvs.16–20 However, valencies of these antibody conjugates can be hard to control and generally are in the range of 10’s to 1000’s. Recently, Rossi et al. described the formation of a hexavalent CD20 antibody composed of 6 Fabs and one Fc region.21 Access to intermediate valencies (i.e., >4) has been limited primarily due to the challenge of producing such large and rather complex proteins recombinantly. Nevertheless, being able to prepare antibody conjugated nanoparticles that are similar in valency and size to IgM’s, for example, would be advantegous, since they should be able to leverage the greater circulation half-lives and avidity observed for antibody-nanoparticles, but with considerably reduced molecular weight and size. In particular, higher valent antibody nanoparticles with diameters in the range of 60–80 nm, should be able to take full advantage of their targeting potential due to enhanced avidity, without reliance on the EPR effect.22, 23
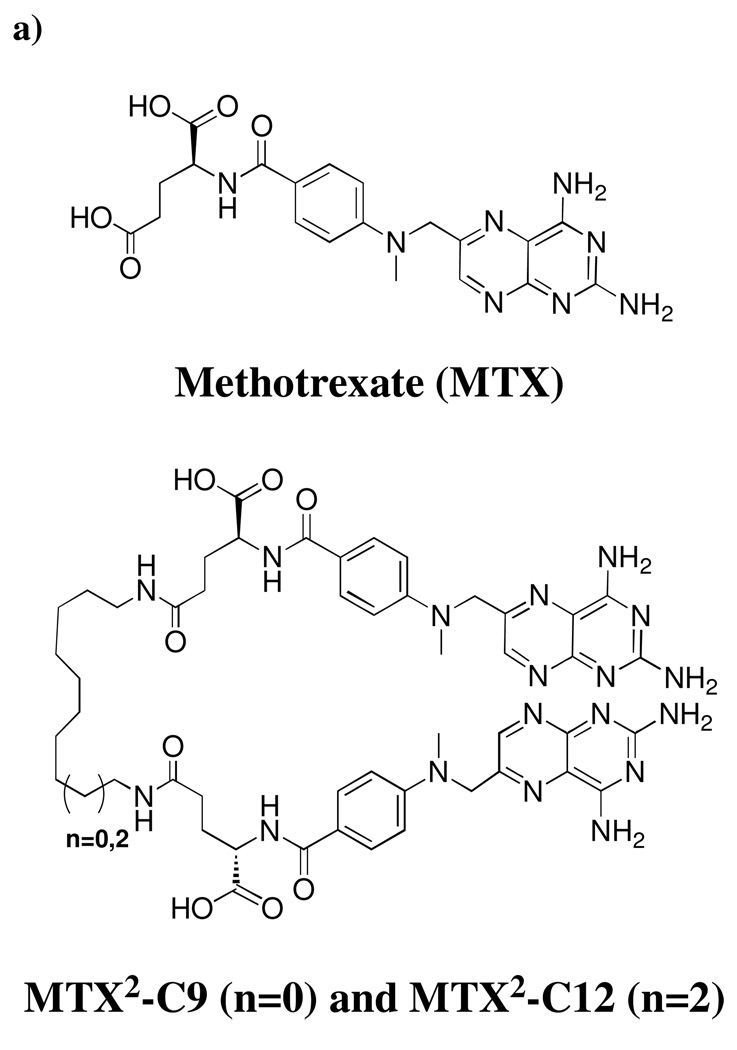
Previously, we have demonstrated that dimers of the potent inhibitor of E.coli dihydrofolate reductase (DHFR), methotrexate (Fig. 1a.), rapidly and robustly dimerize DHFR.24, 25 Taking advantage of this observation, we have shown that DHFR-DHFR (DHFR2) fusion proteins will undergo macrocyclization. The size of the DHFR nanorings was found to be dependent on the length and composition of the linker peptide between the DHFRs, resulting in the formation of DHFR nanorings composed of from 2 to 8 monomers and ranging in diameter from 7–30 nm.25
Figure 1.
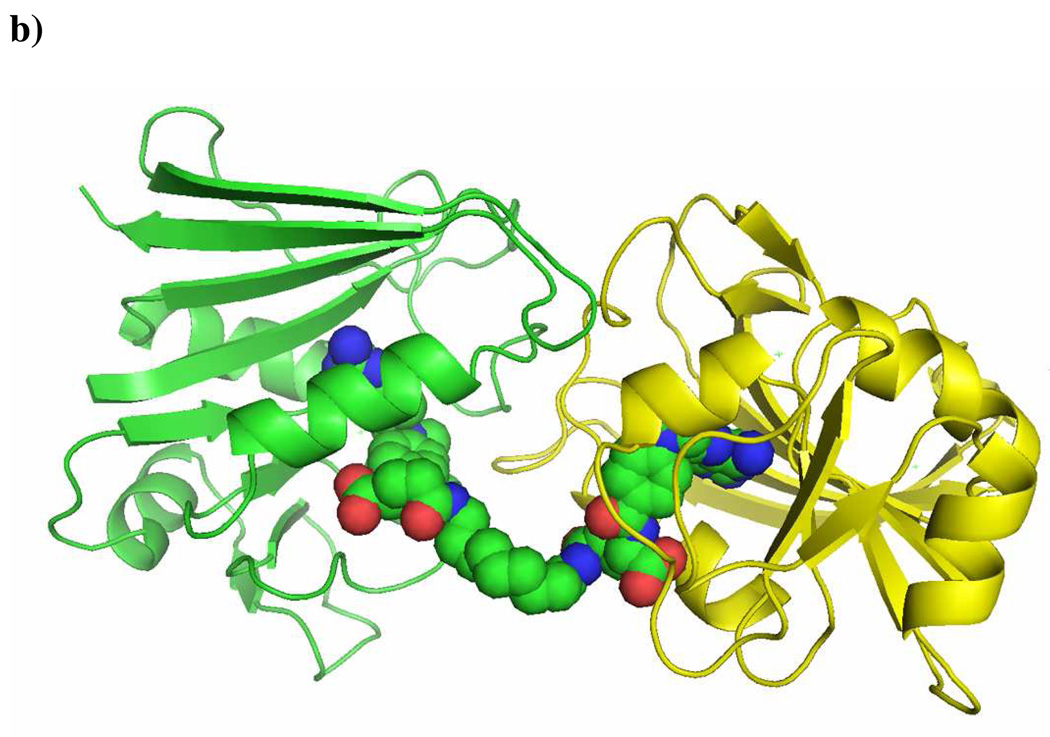
a) Structures of MTX and Bis-MTXs (MTX2-C12 & MTX2-C12).; b) Crystal structure of MTX2-C9 (space filling molecule) with ecDHFR (green and yellow molecules that form a dimer in the asymmetric unit of the hexagonal lattice. Figure drawn with PyMol.
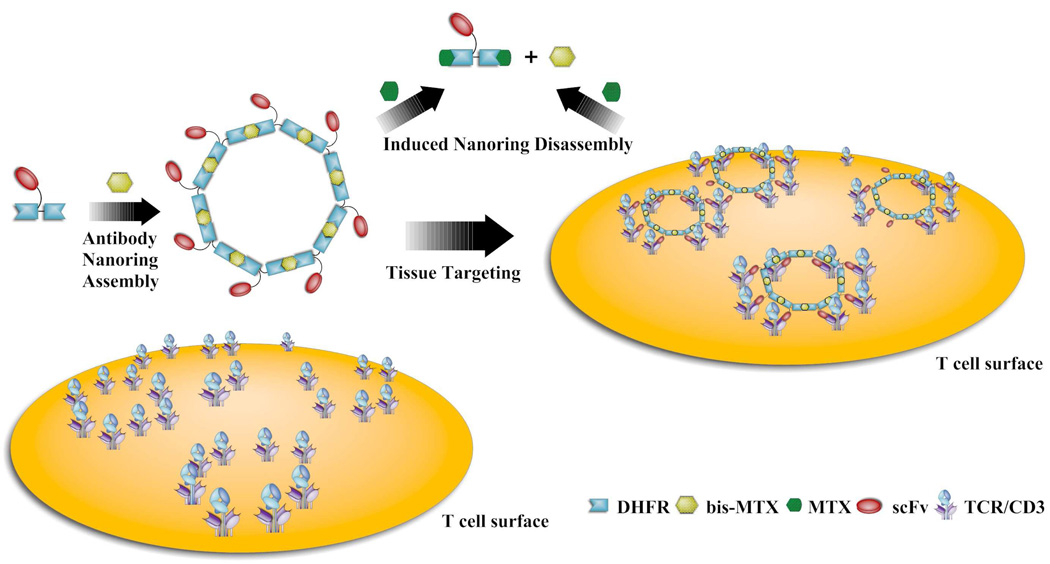
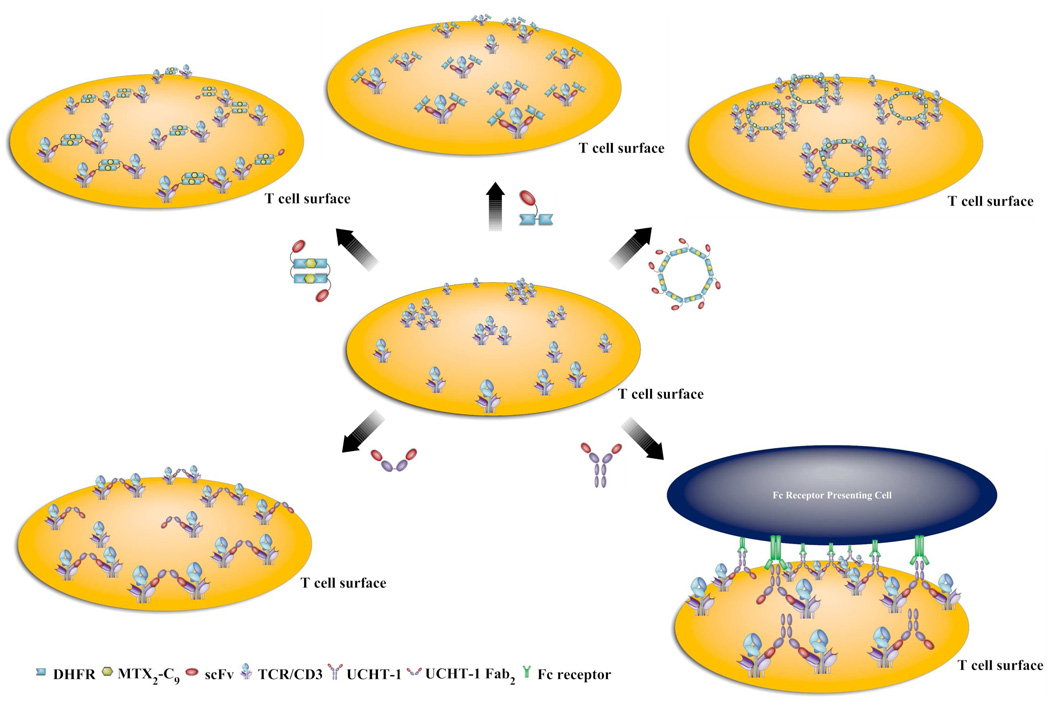
Recently, we have been able to construct bivalent chemically self-assembled antibody nanorings (CSANs) that mimic the binding and internalization behavior of the parental MAB. The bivalent CSANs were prepared by dimerizing an anti-CD3 scFv-DHFR2, fusion protein containing an internal flexible 13 amino acid linker with bisMTX.26 Our success in forming bivalent scFv-DHFR2 fusion protein nanorings that mimic IgG’s has encouraged us to further develop the antibody nanorings with higher valencies and avidities, such as tetravalent IgA and decavalent, IgM. (Scheme 1) Consequently, we engineered octavalent anti-CD3 CSANs (Scheme 1), which were characterized by size exclusion chromatography, static light scattering, and atomic force microscopy. The octavalent anti-CD3 CSANs exhibited improved affinity to CD3 expressing cells compared to the parental mAb UCHT-1. The octavalent CSANs could be successfully disassembled by treatment with excess amount of monovalent ligand. In addition, we found that anti-CD3 CSANs and the parental mAb UCHT-1 exhibited a similar cellular internalization mechanism. In contrast to the MAB, the bivalent and octavalent CSANS were unable to induce T cell proliferation, and they exhibited divergent effects on the T-cell receptor internalization and IL-2 receptor expression.
Scheme 1.
Scheme for the assembly and disassembly of octavalent anti-CD3 scFv antibody targeting T cell receptor.
Results and discussion
X-ray crystallographic studies of MTX2-C9 and E. coli DHFR
Previously, we have determined the structure of Bis-MTX that incorporated a linker containing 12 methylenes (MTX2-C12).24 However, the inherent flexibility of the linker did not allow electron density for the linker to be visualized. To explore the molecular basis for the induction of DHFR dimerization, we crystallized and carried out the structure determination of E. coli DHFR as a binary complex with MTX2-C9 and compared it to the MTX2-C12 complex. Diffraction data are described in the Supplemental material (Table S1 and Fig. S1). Analysis of these data revealed the same dimer orientation as observed for the E. coli DHFR MTX2-C12 complex and showed clearly interpretable density for each MTX. The density for the linker was less well defined and the higher thermal parameters for this section suggest this region is more flexible. Nevertheless, a complete trace of the linker could be made (Fig. 1b). Structural data reveal the binding of two atoms of Mn in the lattice. Coordination of Mn1 is bipyrimidal with the first shell coordination made by the carbonyl of Ser335 of monomer 2 and water molecules (three from symmetry-related packing interactions). The second coordination sphere is made with the functional groups of Glu17, Asn18, Asp342 and Ala343 from a symmetry-related molecule, and Glu334 and chloride ion from monomer 2 of DHFR (Fig. S1). These contacts help stabilize the dimerization of E. coli DHFR in this complex. The chloride ions also coordinate through a network of water molecules and the functional groups of Asp135 and Arg159. The dimer interface of E. coli DHFR is made by residues 17, 18, 19, 20, 21, 22, 144, 147 and 148 of one molecule of E. coli DHFR and their palendromic counterparts across the interface (Fig. S1). The side chain interactions of these residues with water molecules that coordinate with Mn also help stabilize dimer formation. The packing of these dimers is such that there is a large pocket near the MTX γ-glutamate site that provides sufficient space for the bis-MTX linkers (Fig. 1).
Although E coli DHFR protein was incubated with the cofactor NADPH, there was no density. The conformation of the Met 20 loop of ecDHFR is consistent with the occluded conformation that excludes NADPH binding.27
These structural data are consistent with previous studies demonstrating the ability of MTX2-C9 to efficiently form DHFR2 nanorings and further suggest that the incorporation of an additional linker conjugated at the center of the bivalent linker could be used to carry a reporter or drug molecule with little interference on protein dimerization.
Assembly of multivalent CSANS
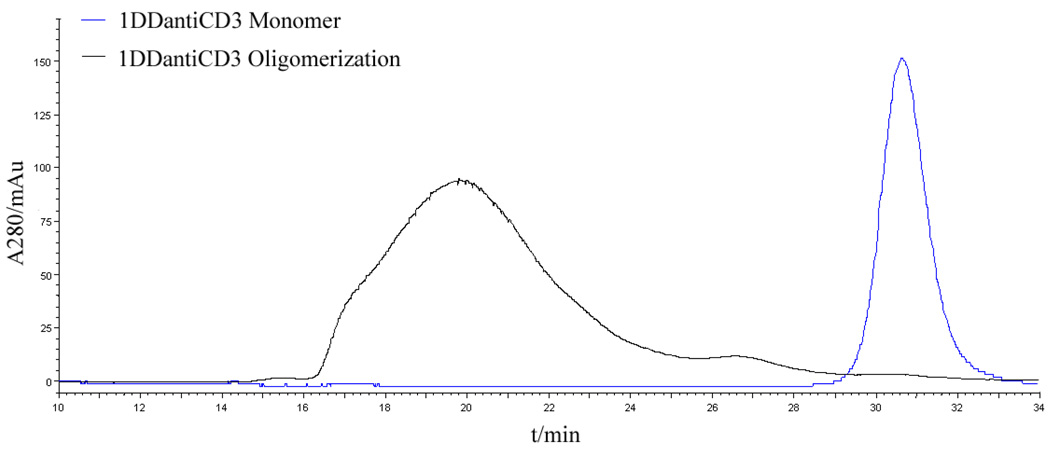
To construct the DHFR2-scFv fusion protein that is capable of forming octavalent CSAN, two copies of DHFR were fused in tandem with one amino acid linker (G), followed by a 13 amino acid linker to anti-CD3 scFv (named 1DDantiCD3). The 1DDantiCD3 fusion protein was expressed in BL21 (DE3) cells, refolded and purified as previously described.26 After incubation of the 1DDantiCD3 fusion protein with 1.1 equivalents of bisMTX for 1 hour, assemblies of multivalent scFv were characterized by size-exclusion chromatography using a Superdex G200 column (Fig. 2). The assemblies of 1DDantiCD3 and bisMTX eluted in a broad peak centered at 19.7 min with 100% oligomerization of 1DDantiCD3 fusion protein observed. In comparison, previous results showed that the dimerization of 13DDantiCD3 eluted at 27 min with the formation of a minor amount of intramolecular macrocyclized monomer.26 The inability to observe formation of this intramolecular monomer is likely due to the steric contraints imposed by reducing the length of the linker between the two DHFR proteins from thirteen amino acids to one amino acid.
Figure 2.
Characterization of 1DDantiCD3 oligomerization by Size-Exclusion Chromatography. Blue curve: 1DDantiCD3 monomer; Black curve: induced oligomerization of 1DDantiCD3 with bisMTX.
To evaluate the valency of the CSAN, static light-scattering experiments were performed to determine the molecular weight of the oligomers. Size exclusion chromatography coupled with multi-angle light scattering can be used to determine the weight-average molecular weight (Mw) and radius of gyration (Rg) of protein aggregates. Analysis of the scattered light from the broad peak as observed by UV absorbance gave an experimental Mw of 525 ± 20 kDa and a Rg of 24.8 ± 1 nm. The molecular weight of 525 ± 20 kDa is 8.2 times that of the monovalent 1DDantiCD3 protein (64 kDa) suggesting a predominantly octavalent species in solution, which would have an expected Mw of 512 kDa. This result is consistent with our previous work where unfunctionalized 1DD proteins were found to form octamers as the major species.25 Based on this analysis, the leading and trailing edges of the broad peak contain species ranging in size from 12-mers to 6-mers, respectively. Previously rings lacking the anti-CD3 scFv were shown to have a radius of gyration of 10.8 ± 1 nm while for the scFv rings the calculated rg is 24.8 ± 1 nm. This increase radius is likely due to the extended anti-CD3 scFv’s appended to the nanorings.
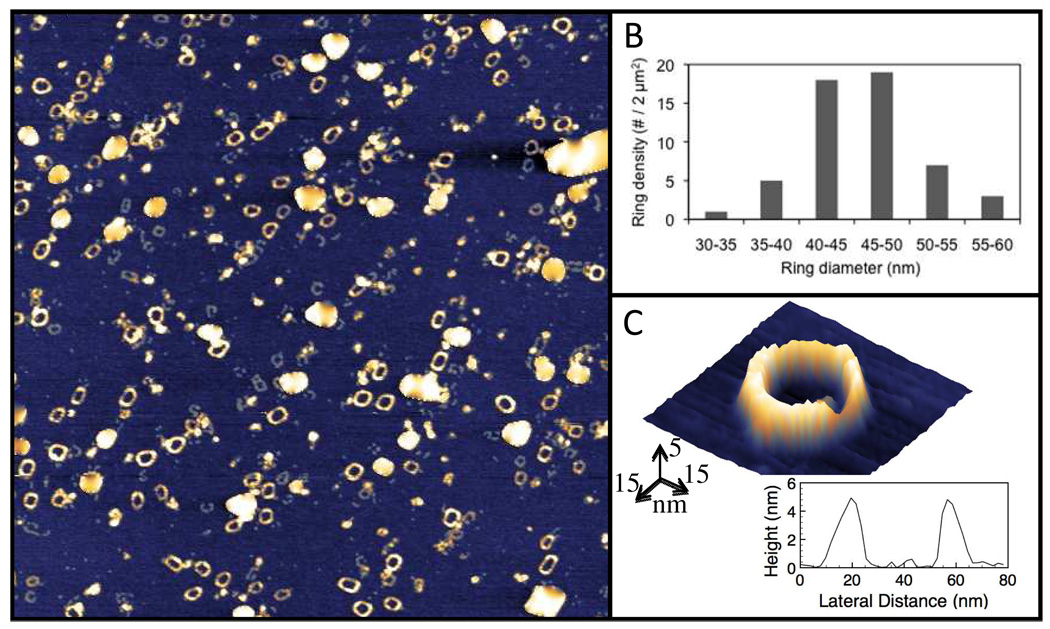
To characterize both the size and morphology of the constructed CSANs, high resolution intermittent contact mode atomic force microscopy (AFM) experiments were carried out under ambient conditions. As shown in Fig. 3, the rings appear homodisperse on the mica surface in open ring form. Previous measurements of heteromeric nanorings containing the enzyme Hint1 were measured by dynamic light scattering as well as AFM and found to be ~50 nm in size.28 Consistent with results from light scattering experiments, a purified sample of the octavalent scFv rings was shown to have an average diameter of 45–55 nm (Fig. 3).
Figure 3.
Characterization of octavalent anti-CD3 CSANs (1DDantiCD3 octamer) formation by Atomic Force Microscopy. (A) 2 µm × 2 µm AFM topograph showing homodispersed octavalent CSANs on a prepared atomically flat mica surface. Maximum height (white) is 5 nm. (B) Quantification of 55 individual ring diameters using 5 nm binning, showing the average ring sizes to exist as ~40–50 nm across. (C) Pseudo-3D representation of an individual nanoring, showing a peak-to-peak diameter of 45 nm. Large white particles in A are salt crystals, artifacts due to the short sample rinse time required to keep the nanorings on the surface for study.
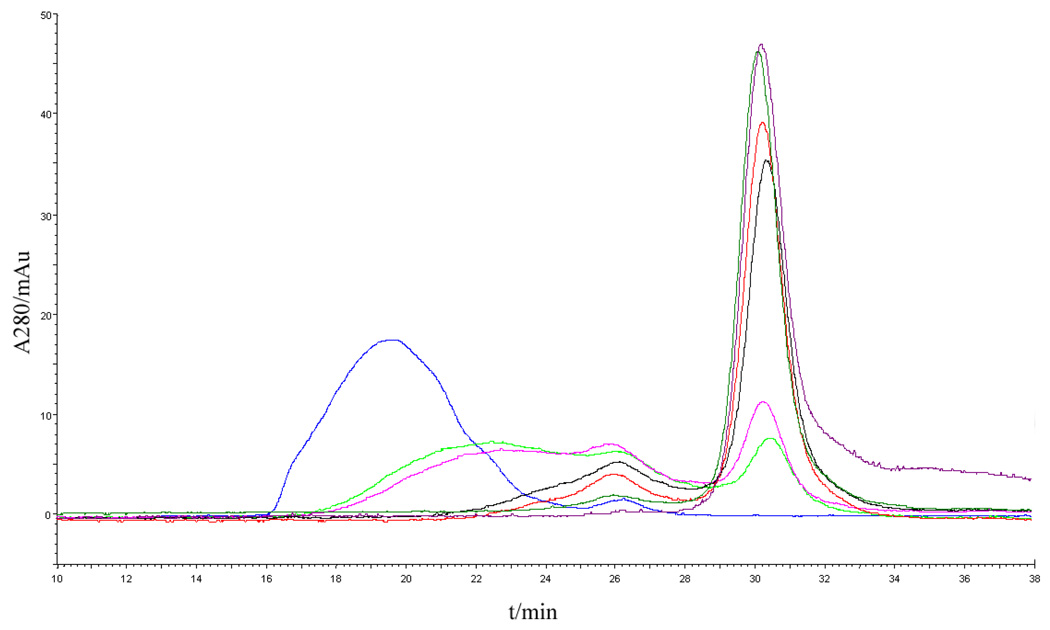
An advantage of this chemically controlled assembly method is the ability to controllably disassemble CSANs with excess amount of monovalent ligand. Disassembly of octavalent CSANs was examined by incubating the CSANs with a 10-fold excess competitive monovalent methotrexate at room temperature. The rate of disassembly to intermediate oligomers and monomer was probed at various time points by size-exclusion chromatography (Fig. 4). Octavalent CSANs rapidly disassembled into smaller oligomers and monomer within minutes. After 40 minutes about 75% monomer was observed while complete disassembly occurred within 3 hours. The half life for the disassembly with a 10-fold excess of competing methotrexate was determined to be 28 mins, which corresponds to a rate of 0.025 min−1 (Fig. S2). Considering the potential of scFv nanorings as therapeutic agents and the renal filtration threshold of 65 kDa, the fast disassembly process of the nanoring disassembly may provide opportunities to tune the in vivo pharmacokinetics of the CSANs.
Figure 4.
Characterization of induced octavalent anti-CD3 CSANs disassembly with methotrexate by Size-Exclusion Chromatography. Blue curve: CSANs; light green, pink, black, red, dark green, and purple curves: disassembled oligomers after incubation with methotrexate for 5, 20, 40, 80, 150, and 240 min, respectively.
Binding of anti-CD3 CSANS to cells
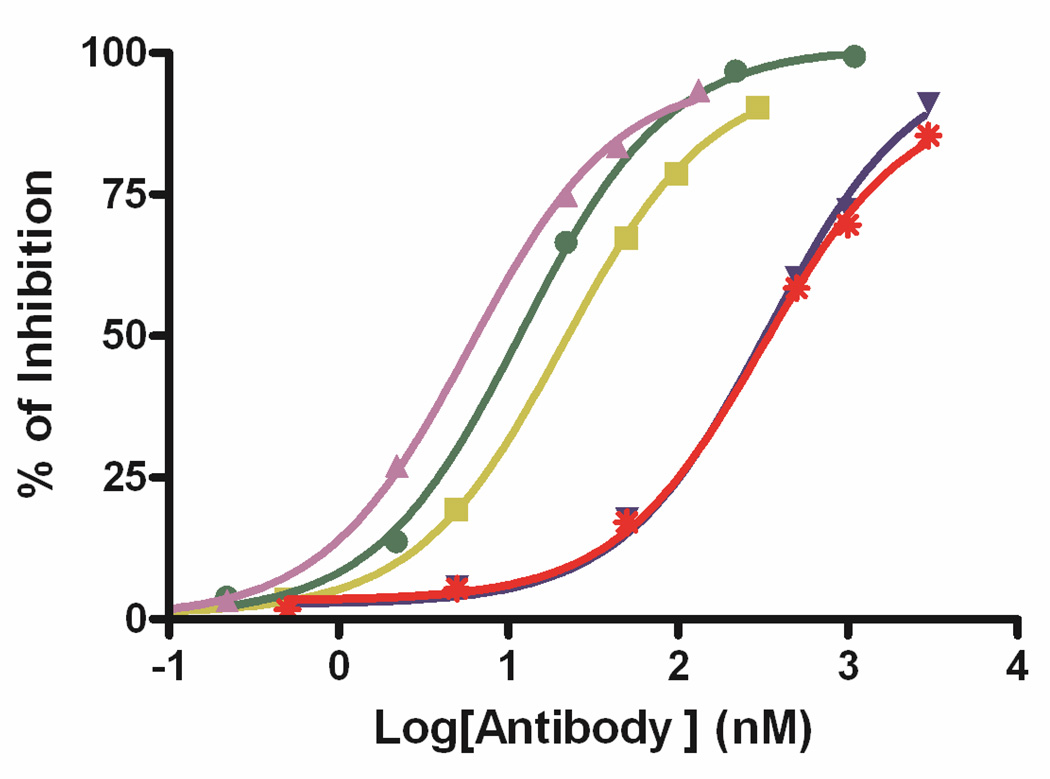
The octavalent anti-CD3 CSANs were evaluated for the binding affinity to the CD3+ T leukemia cells HPB-MLT by determining the dissociation constant with a flow cytometric competitive binding assay. Binding of non-labeled CSAN to T leukemia cells was competed with sub-saturated amount of FITC labeled mAb anti-CD3 UCHT-1, followed by quantitating the fluorescence intensities of cell bound FITC labeled UCHT-1 with flow cytometry (Fig. 5). The octavalent anti-CD3 CSANs were found to have a Kd value of 0.93± 0.2 nM, which is 52-fold lower than the Kd for the monovalent 1DDantiCD3 (Kd = 49 ± 6 nM), 4-fold lower than the Kd for the divalent anti-CD3 CSANs (Kd = 3.51 ± 0.2 nM),26 and 2-fold lower than the Kd for the parental mAb UCHT-1(Kd = 1.8 ± 0.2 nM). The inability of the octavalent CSANs to fully realize the maximum theoretical binding affinity to the cell surface is likely due to several factors, including the fact that CD3 is not displayed on 100% of the cellular surface. In addition, there is likely to steric constrains on the number of CD3s, particularly in the context of being associated with the TCR, that can be oligomerized on the surface.
Figure 5.
Flow cytometric competitive binding assay was applied to determine the disassociation constant of 1DDantiCD3 monomer (red stars), 13DDantiCD3 monomer (blue downward triangles), divalent CSANs (yellow squares), octavalent CSANs (purple upward triangles), and UCHT-1 (green dots) to T-leukemia cells HPB-MLT.
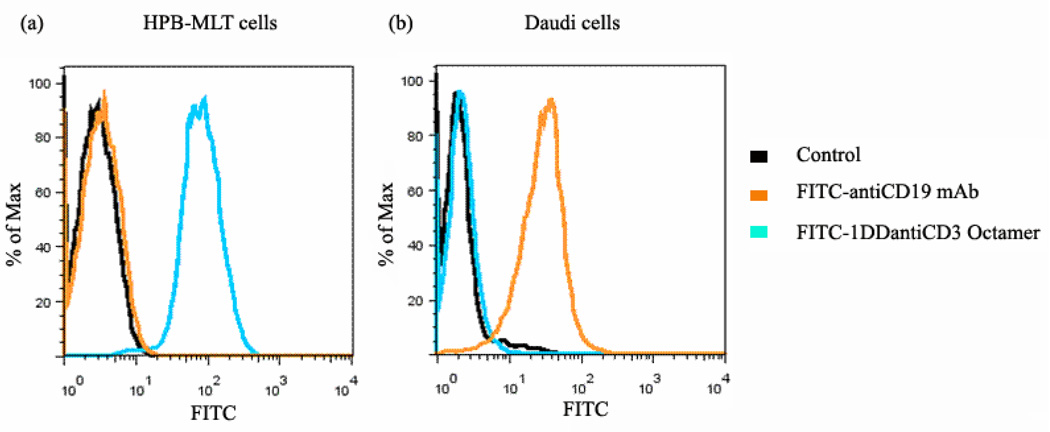
To demonstrate the specific binding ability of the engineered anti-CD3 CSANs to cells that only express CD3 receptor, FITC-octavalent anti-CD3 CSANs, or FITC-antiCD19 mAb, were incubated with Daudi cells (CD3−/CD19+ B lymphoma cell line). FITC-antiCD19 mAb was used as positive control. After washing with FACS buffer, bound cell fluorescence was analyzed by flow cytometry. When HPB-MLT cells (CD3+/CD19− T leukemia cell line) were treated with FITC-octavalent anti-CD3 CSANs or FITC-antiCD19 mAb, only FITC-octavalent anti-CD3 CSANs exhibited strongly bound fluorescence (Fig. 6a). While FITC-antiCD19 mAb exhibited obvious bound fluorescence on Daudi cells, no specific fluorescence was detected on cells treated with FITC-octavalent anti-CD3 CSANs (Fig. 6b). Therefore, octavalent anti-CD3 CSANs were able to specifically bind cells that express CD3 receptor.
Figure 6.
FITC-octavalent anti-CD3 CSANs bound to CD3+ HPB-MLT cells (a) but not to CD3- Daudi cells (b). HPB-MLT cells or Daudi cells were incubated with FITC-antiCD19 mAb or FITC-octavalent anti-CD3 CSANs at 37°C for 30 mins. FITC-antiCD19 mAb was used as positive control bound to Daudi cells. The fluorescence of bound FITC-antiCD19 mAb or FITC- octavalent anti-CD3 CSANs was analyzed by flow cytometry. The horizontal axis of the diagram represents the fluorescent intensity, and the vertical axis shows the percentage of maximum cell number.
Mechanism of anti-CD3 CSANs cellular uptake
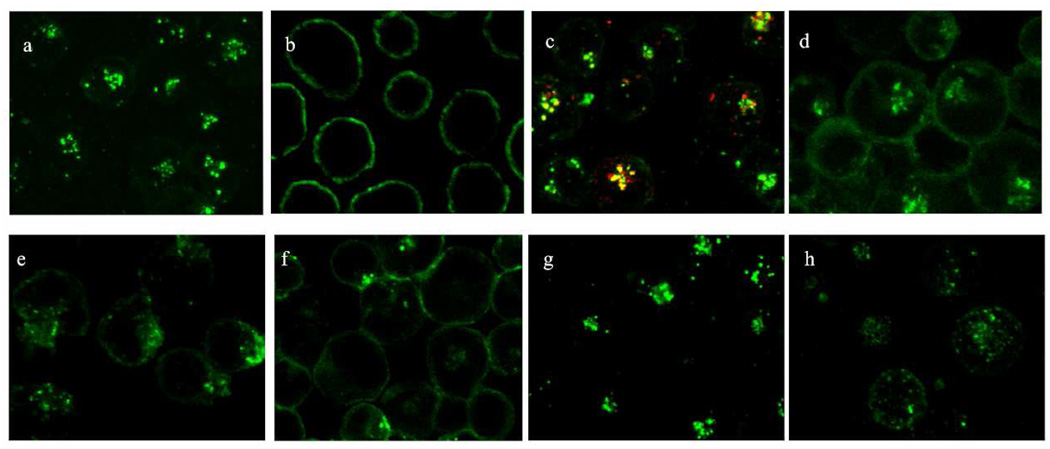
Internalization of fluorescently labeled octavalent anti-CD3 CSANs to T leukemia cells HPB-MLT was characterized by confocal laser scanning microscopy. After incubation with HPB-MLT at 37 °C, intracellular punctuates harboring the fluorescent octavalent anti-CD3 CSANs could be observed (Fig. 7a). In contrast, intracellular uptake was not observed when HPB-MLT cells were treated with octavalent anti-CD3 CSANs at 4 °C, strongly suggesting that the internalization mechanism relies on an energy dependent process (Fig. 7b). Co-localization studies at 37 °C with transferring, a marker of receptor-mediated endocytosis, suggested that octavalent CSANs localize to endosomes and follow an internalization mechanism similar to transferrin, which relies on clathrin-dependent endocytosis29 (Fig. 7c). This was further confirmed by the use of a lysosomal specific fluorescent dye, LysoTracker Red DND-99, which identified the intracellular punctate location of CSANs as late endosomes (Fig. S3).
Figure 7.
Internalization studies of FITC labeled octavalent anti-CD3 CSANs (green) to HPB-MLT T-leukemia cells. (a) Incubation at 37 °C or (b) 4 °C. (c) Colocalization with Alexa Fluor 594-labeled transferrin (red) after incubation at 37 °C. Incubation in the presence of clathrin-dependent endocytosis inhibitor chlorpromazine (d), actin polymerization inhibitor cytochalasin D (e), protein tyrosine kinase inhibitor genistein (f), caveolin-dependent endocytosis inhibitor nystatin (g), or cholesterol depletion agent methyl-β-cyclodextrin (h) at 37 °C. All incubations were for 30 min.
To further characterize the mechanism of octavalent anti-CD3 CSANs cellular internalization, we performed the internalization experiments in the presence of known inhibitors of components of the endocytosis pathway. The treatment with chlorpromazine, a molecule known to inhibit clathrin-mediated endocytosis,30 partially inhibited the cell penetration of octavalent anti-CD3 CSANs, suggesting that the octavalent anti-CD3 CSANs partially follow a clathrin-dependent endocytotic pathway (Fig. 7d). Consistent with the dependence of clathrin-dependent endocytosis on actin mobilization,31 treatment with an actin polymerization inhibitor,32 cytochalasin D, significantly blocked the intracellular uptake of octavalent anti-CD3 CSANs (Fig. 7g).
Since phosphorylation of clathrin and the CD3 ζ chain by the protein tyrosine kinase, Lck, appears to be required for TCR internalization, we examined the effect of the tyrosine kinase inhibitor, genistein, on antibody internalization.33 Consistent with the known mechanism of TCR/CD3 internalization, treatment of HPB-MLT T leukemia cells with genistein completely blocked the intracellular uptake of octavalent anti-CD3 CSANs (Fig. 7h) as well as, monovalent 1DDantiCD3, divalent CSANs and mAb UCHT-1. (Fig. S4–6)
Nevertheless, the inability to completely block octavalent anti-CD3 CSANs internalization suggests that other uptake mechanisms, such as caveolin-dependent processes, may coexist. However, pretreatment of HPB-MLT cells with nystatin, an inhibitor of caveolin-dependent endocytosis,34 failed to block endocytosis (Fig. 7e). Partial inhibition of CSANs internalization was observed after treatment with methyl-β-cyclodextrin, which has been shown to deplete cellular membranes of cholesterol;thus blocking lipid-raft mediated endocytosis.35 Methyl-β-cyclodextrin treatment also resulted in the loss of defined punctates and generally dispersal of labeled octavalent anti-CD3 CSANs within the cell, which is consistent with the importance of cholesterol in maintaining the integrity of endosome and lysosome membranes36 (Fig. 7f).
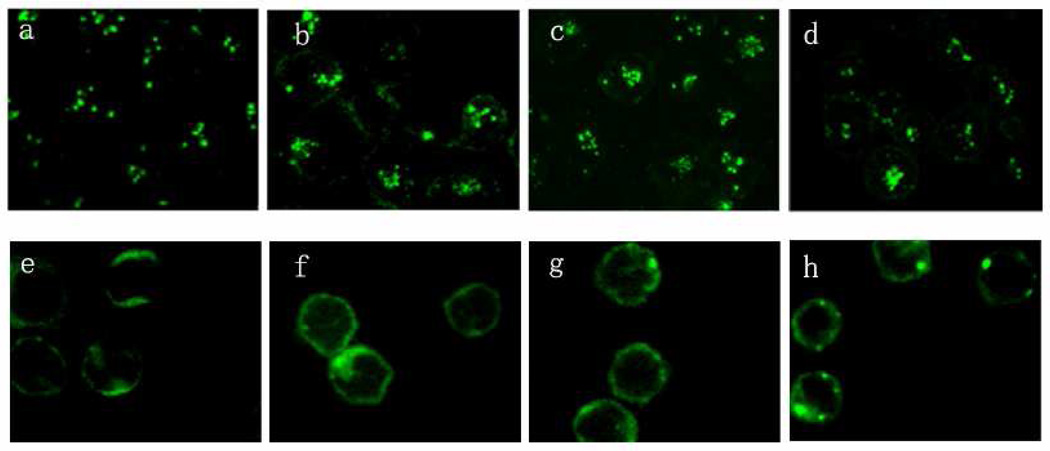
Since both T-leukemia cells, such as HPB-MLT cells and normal T-cells found in peripheral blood mononuclear cells (PBMCs) display CD3 on their surfaces, we also investigated the abilty of the anti-CD3 CSANs to be internalized by PBMCs (Fig. 8). In contrast to the rapid internalization observed for octavalent anti-CD3 CSANs by HPB-MLT cells, very little of the recombinant multivalent antibody was internalized by unstimulated PBMCs at 37 °C. Similar results were found for the monovalent DHFR2-antiCD3, bivalent CSANs and parental antibody. Taken together, these results demonstrate that anti-CD3 CSANs predominately enter the cell through a clathrin-dependent endocytosis pathway that requires cholesterol, actin polymerization and protein tyrosine kinase activation Several other mechanisms, including lipid-raft dependent endocytosis or other non-clathrin dependent endocytosis, may simultaneously be minor contributors to the internalization process of anti-CD3 CSANs. Valency and avidity of the anti-CD3 appear to have little impact on the cell internalization mechanism. It is evident from the binding assay and endocytosis studies that anti-CD3 CSANs specifically bind the T-cell leukemia cell-line, HPB-MLT cells, followed by rapid internalization, while avoiding uptake by normal PBMCs.
Figure 8.
Comparing the internalization studies of anti-CD3 antibodies to HPB-MLT T leukemia cells and normal T cells. Confocal images of HPB-MLT T leukemia cells incubated with FITC-labeled monovalent 1DDantiCD3 (a) or FITC-labeled divalent anti-CD3 CSANs (b) or FITC-labeled octavalent anti-CD3 CSANs (c) or FITC-labeled UCHT-1 for 30 mins at 37°C. Confocal images of PBMC incubated with FITC-labeled monovalent 1DDantiCD3 (a) or FITC-labeled divalent anti-CD3 CSANs (b) or FITC-labeled octavalent anti-CD3 CSANs (c) or FITC-labeled UCHT-1 for 30 mins at 37°C.
Activation of T-cells by Anti-CD3 CSANs
A limitation to the clinical use of monoclonal antibodies against the TCR/CD3 complex is the potential for severe adverse effects due to cross-linking of the TCR/CD3 complex. This occurs when cells displaying Fc receptors bind to the Fc regions of the monoclonal antibodies causing cross-linking of TCR/CD3 complex, which is followed by T-cell activation resulting in expansion/proliferation of the T cell population.37
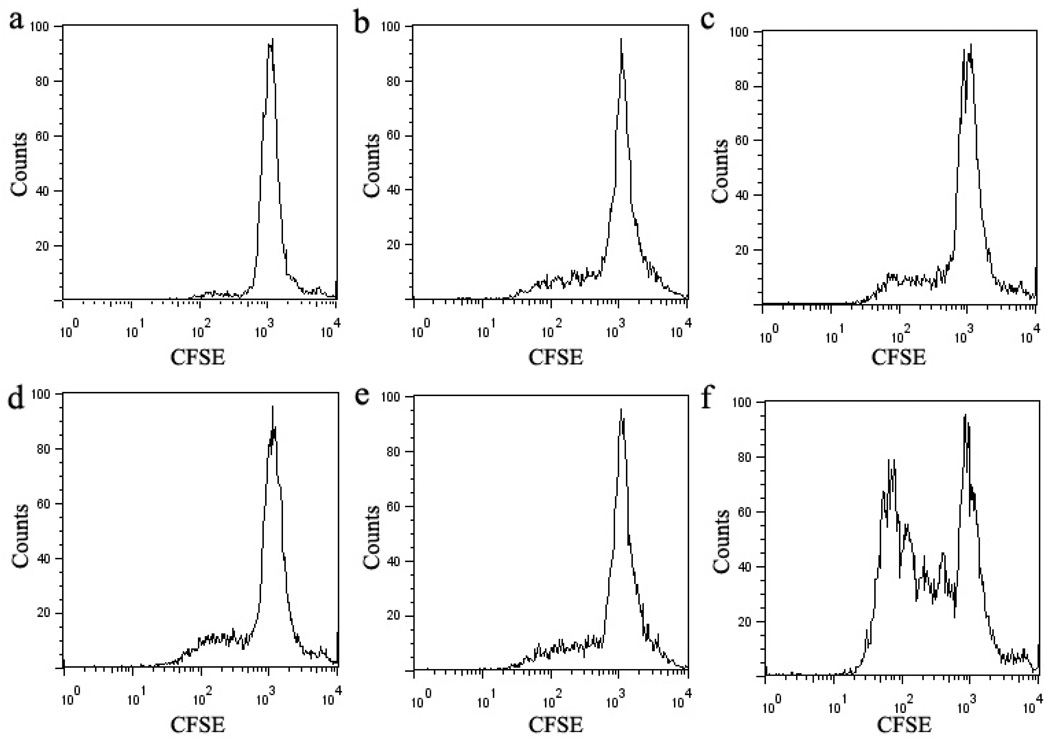
To address the effects of the activation of T cell by the engineered anti-CD3 CSANs (Scheme 2), we examined the ability of anti-CD3 CSANs to induce proliferation of PBMCs by the CFSE (carboxyfluorescein succinimidyl ester) based assay, which relies on the dilution of the fluorescent cell staining dye CFSE to monitor the divided cell cycles.38 After incubation of CFSE-labeled PBMCs with the anti-CD3 antibodies at 37 °C for 5 days, the extent of CFSE dilution was determined by flow cytometry (Fig. 9). The anti-CD3 MAB, UCHT-1, induced significant PBMC proliferation, with 66 ± 8% of the cells undergoing between 2 and 5 divisions. In contrast, when cells were treated with monovalent 1DDantiCD3, divalent CSANs, octavalent CSANs, or UCHT-1 F(ab')2 only 19 ± 3%, 20 ± 3%, 21 ± 4% and 19 ± 3%, respectively, of the cells were observed to undergo between 2 and 4 divisions. Thus, although TCR/CD3 oligomerization is required for T-cell activation, neither treatment with the bivalent nor the octavalent anti-CD3 CSANs is apparently able to induce T-cell activation by sufficient aggregation. In contrast, given that the F(ab')2 fragment of UCHT-1 only induces 19% proliferation, as compared to 65% for the full MAB, it is very likely that accessory cell Fc receptor induced oligomerization requiring cell-cell interaction is responsible for the observed UCHT-1 induced PBMC proliferation.39
Scheme 2.
Comparison of the triggering of PBMCs by a panel of anti-CD3 (monovalent 1DDantiCD3, divalent CSANs, octavalent CSANs, divalent UCHT-1 F(ab')2, and mAb anti-CD3 UCHT-1).
Figure 9.
PBMC Proliferation assay. CFSE-labeled PBMCs were stimulated with a panel of anti-CD3 for 5 days. A representative experiment of 3 is shown in a–f. (a) unstimulated PBMCs, (b) monovalent 1DDantiCD3 stimulated PBMCs, (c) divalent CSANs stimulated PBMCs, (d) octavalent CSANs stimulated PBMCs, (e) UCHT-1 F(ab')2 stimulated PBMCs, (f) UCHT-1 stimulated PBMCs.
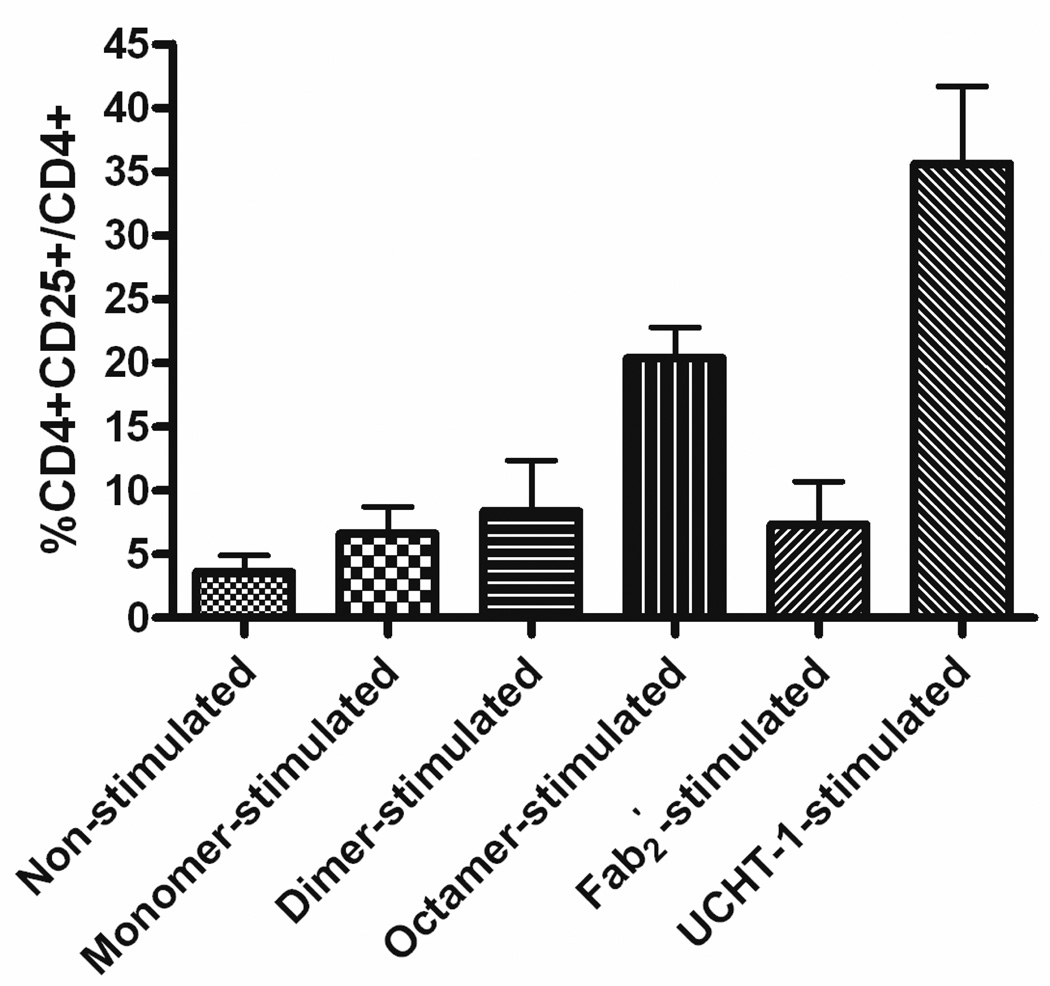
To further delineate the effect of the anti-CD3 CSANs on T-cell activation. the up-regulation of CD25 (IL-2R) on CD4+ cells was investigated. The degree of CD25 (IL-2R) regulation on CD4+ cells by engineered anti-CD3 CSANs was assessed by incubation of PBMCs with the indicated anti-CD3 species at 37 °C for 20 hours. The results of flow cytometric assay, shown in Fig. 10 (raw data, Fig S7), illustrated that there was a certain correlation between the oligomerization degree of CD3 and early stage T cell stimulation. After treatment with octavalent CSANs, the percentage of CD4+CD25+ cells over the total CD4+ cells (%CD4+CD25+/CD4+) was increased from 3.6 ± 2.2% for non-stimulated cells to 20.3 ± 4.2%, while %CD4+CD25+/CD4+ was elevated to 6.6 ± 3.0% for monovalent 1DDantiCD3 treated cells and 8.4 ± 5.6% for divalent CSANs treated cells. Compared to divalent anti-CD3 CSANs, treatment of PBMCs with UCHT-1 F(ab')2 where found to have a similar effect on IL-2R expression levels (7.3 ± 4.7%) while treatment with the UCHT-1 mAb dramatically increased the level of CD4+CD25+ cells (35.6 ± 10.5%). Therefore, although the octavalent anti-CD3 CSANs is able to induce 50% more IL2-R expression, which is with in 15% of the levels exhibited by the the UCHT-1 monoclonal antibody, the levels of induced IL-2-R apparently do not reach the threshold necessary for T-cell activation.
Figure 10.
Effects of IL-2R (CD25) expression level on CD4+ cells by triggering PBMCs with a panel of anti-CD3. PBMCs were incubated with anti-CD3 for 20 hours, and then stained with mAbs of CD4 and CD25. Percentage of CD4+CD25+ cells over CD4+ cells were analyzed by FACS. (a) Unstimulated PBMCs, (b) monovalent 1DDantiCD3 stimulated PBMCs, (c) divalent CSANs stimulated PBMCs, (d) divalent CSANs stimulated PBMCs, (e) UCHT-1 F(ab')2 stimulated PBMCs, and (f) UCHT-1 stimulated PBMCs.
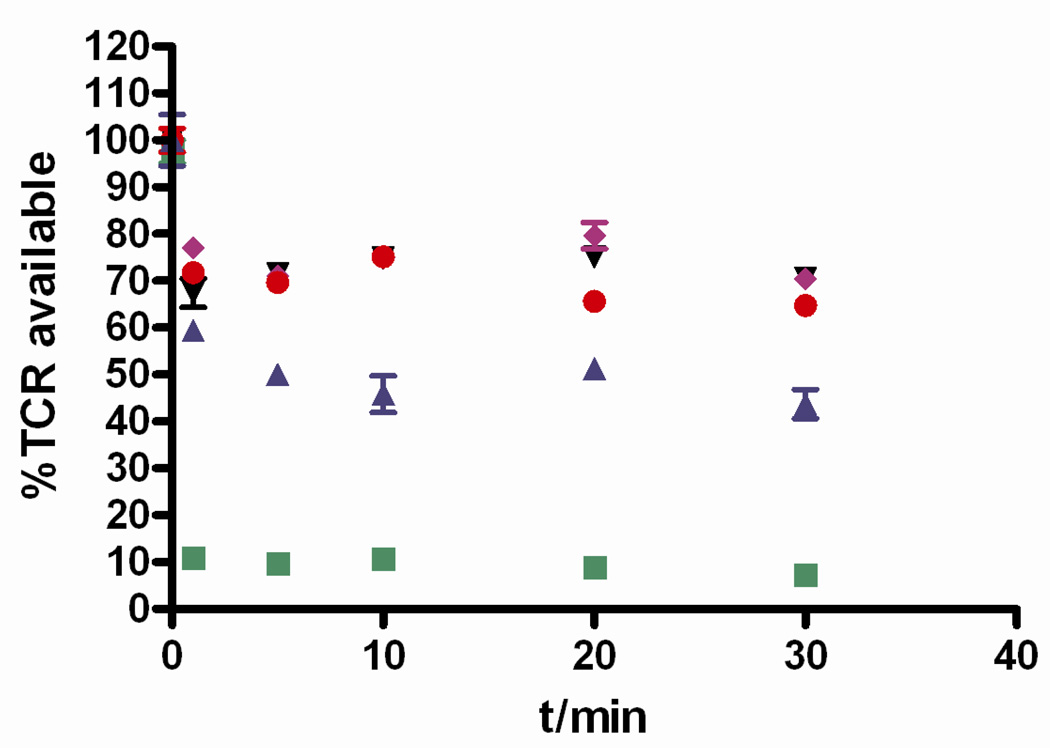
Although the physiological role of TCR down-regulation is not fully understood, the regulation of TCR expression levels play an important role in modulating T cell activation.40 To study the effect of engineered anti-CD3 CSANs on the down-regulation of TCR cell surface expression, PBMCs were incubated with monovalent 1DDantiCD3, divalent CSANs, octavalent CSANs, divalent UCHT-1 F(ab')2, and mAb anti-CD3 UCHT-1 at 37 °C for various times. After blocking receptor uptake by cooling cells to 4 °C, TCRs available on the cell surface were labeled with FITC-anti-human αβTCR followed by determining the level of labeled TCR with flow cytometry (Fig. 11). Monovalent 1DDantiCD3, divalent CSANs, and octavalent CSANs showed similar effect on TCR modulation, resulting in a loss within minutes of approximately 30% of cell surface displayed TCR. In contrast, about 90% of the TCR was down-regulated by the treatment of PBMCs with UCHT-1 mAb. This result suggested that although the binding affinity of anti-CD3 CSANs is higher than UCHT-1 mAb, the valency is still below the threshold required for maximal TCR modulation by the T cells and therefore activation. The slight difference of TCR down-regulated response induced by UCHT-1 F(ab')2 when compared to the divalent CSANs may be result from inherent differences in the cell surface interactions between the highly conformationally mobile CSANs relative to the more rigid F(ab')2.
Figure 11.
TCR modulation induced by a panel of anti-CD3 (monovalent 1DDantiCD3: black downward triangles; divalent CSANs: purple diamonds; octavalent CSANs: red dots; divalent UCHT-1 F(ab')2: blue upward triangles; and mAb anti-CD3 UCHT-1: green squares). PBMCs were treated with indicated anti-CD3 for 1, 5, 10, 20, and 30 min, and T cell receptors on cell surface were then stained with FITC-anti-human αβTCR. TCR modulation was analyzed by flow cytometry. %TCR available represents percentage of TCR on cell surface treated with anti-CD3 as a fraction of surface TCR on control cells.
Summary
By using our ability to prepare DHFR2 nanorings by chemically induced dimerization, we have developed a protocol for the reversible preparation of CSANs with valencies similar to IgMs, ranging from 8–10 scFvs. These anti-CD3 CSANs have been used to discretely probe the role of valency on T cell activation. Moreover, we have demonstrated that although the anti-CD3 CSANs are able to bind to normal unactivated T-cells, they do not activate T-cells nor are they apparently internalized. In contrast, they are able to undergo clathrin-dependent endocytosistarget and be internalized by T-leukemia cells. We believe that this approach is applicable to other scFvs and can be used to further elucidate the role of valency and receptor cross-linking on cell responses. In addition, the engineered CSANs show great potential for use as multivalent nano-vehicles for drug delivery. The future development of MTX dimerizers fitted with a third arm that can be reversibly conjugated to either a reporter group or small molecule drug will facilitate their therapeutic and chemical biological utility. Currently work is underway in to explore the use of CSANs for targeted drug delivery and imaging applications.
Methods
Construction of p1DD13CD3 Plasmid
We constructed the p1DDantiCD3 plasmid from the p13DD13CD3.2 plasmid template previously made in our lab.26 The p13DD13CD3.2 plasmid encodes a fusion protein containing a cysteine-free ecDHFR coupled by a 13 amino acid linker (GLGGGGGLVPRGT) to a second cysteine-free ecDHFR, followed by a second 13 amino acid linker to the anti-CD3 scFv gene. Plasmid p1DD13CD3 was obtained by shortening the first linker from 13 amino acids to 1 amino acid (G) using the Stratagene QuickChange Site-directed Mutagenesis Kit, following the standard protocol as described by the manufacturer. The primers used for the deletion were: forward 5’-CTGGAGCGGCGGGGTATGATCAGTCTGATTGCG-3’ and reverse 5’-CGCAATCAGACTGATCATACCCCGCCGCTCCAG-3’. The DNA sequence was verified (University of Minnesota, Advanced Genetic Analysis Center) to confirm the required deletion.
Protein Expression, Refolding and Purification
1DDantiCD3 protein was expressed, refolded and purified by our previously reported method.26 Briefly, the protein was overexpressed in the Escherichia coli strain BL21 (DE3). The inclusion bodies were obtained by sonicating cells in lysis buffer, followed by refolding the protein with the sodium N-lauroyl-sarcosine (SLS) air oxidation method. The refolded protein was purified by MTX affinity column and DEAE column.
Crystallization and x-ray structure determination
Recombinant ecDHFR was washed in a Centricon-10 with 20 mM Imidazole buffer, pH 6.6, and concentrated to 18.7 mg/mL for crystallization. The ecDHFR protein was incubated with NADPH and an excess of MTX-C9 for 1hr over ice prior to crystallization using the hanging drop vapor diffusion method at 4° C. The reservoir contained 14–24% PEG 6K, 200 mM MnCl2 and 18–24 mM KPO4, pH 6.6. Crystals of ecDHFR grew over several weeks time and were hexagonal, space group P61 and were cryo protected with ethylene glycol gradient of 8, 16, 24%, dip for 30 seconds at each step. Data were collected to 1.5Ǻ resolution at MacCHESS at the Cornell Synchrotron facility using beamline A-1. The data were processed using DENZO.41 The diffraction statistics are shown in Table 1 (Supplementary material) for the MTX-C9 binary complex with ecDHFR.
The structure was solved by molecular replacement methods using the coordinates for ecDHFR (1rh3) in the program Molref.42 Inspection of the resulting difference electron density maps made using the program COOT43 running on a MacG5 workstation revealed density for two molecules in the asymmetric unit with electron density for the complete inhibitor in the binary complex. The final cycles of refinement were carried out using the program Refmac5 in the CCP4 suite of programs. To monitor the refinement, a random subset of all reflections was set aside for the calculation of Rfree (5%). The models and parameter files for the inhibitors were prepared using the Dundee PRODGR44 Server website (http://davapc1.bioch.dundee.ac.uk/programs/prodrg). The Ramachandran conformational parameters from the last cycle of refinement generated by RAMPAGE45 showed that more than 98% of the residues have the most favored conformation and none are in the disallowed regions. Coordinated for these structures have been deposited with the Protein Data Bank (3OCH).
Size Exclusion Chromatography (SEC)
After incubation at room temperature with 1.1 equivalents of MTX2-C9 dimerizer (Fig. 1 a) in P500 buffer (0.5M NaCl, 50mM KH2PO4, 1mM EDTA, pH 7) with 5% (v/v) glycerol for 1 hour, the protein solution was injected onto a Superdex G200 size exclusion column (Amersham Biosciences, USA), and eluted with P500 buffer at 0.5 ml/min. Relative peak areas where quantitated by integrating the absorbance at 280nm.
Static Light Scattering Analysis
Static light scattering experiments were conducted with a multi-angle light scattering (MALS) detector (DAWN EOS, Wyatt Technology Corp.) connected in line with the SEC, using UV absorption as the concentration detector. The analysis was carried out at a flow rate of 0.5 ml/min at room temperature. The DAWN EOS was calibrated with 0.02 µM filtered toluene and further normalized with BSA using standard procedures as described by Wyatt Technology Corp. The collected data was analyzed using Astra V software (Wyatt Technology Corp.) to solve the equation relating scattered light at several angles, concentration, and weight-average molar mass by using the Zimm method to determine the average molecular weight and radius of gyration for each sample. The extinction coefficient for the 1DDantiCD3 bis-MTX complex was determined experimentally to be 2.305×103 ml/(g.cm). Experiments were repeated in duplicate with the error the standard deviation of the two runs.
Atomic Force Microscopy Analysis
Sample Preparation
Mica surfaces (Ted Pella, CA) were prepared by first immersion in DI Water (18.2 mΩ) for 10 minutes to dissociate surface cations for a homogeneous net negative surface. Then, stripped surfaces are exposed to MgCl2 solution (1mM) for 10 minutes to create a homogeneous net positively charged surface for the immobilization of the CSANs molecules. 2 µL of the protein nanorings (0.1 µM in P500 Buffer) was drop coated by pipette over the cationic mica surface for 10 minutes and immediately rinsed with 100 µL DI water and dried under Argon for imaging.
AFM imaging
A Digital Instruments (Santa Barbara, CA) Multimode Nanoscope IIIa scanning probe microscope equipped with high frequency (~300 kHz) NanoSensors PPP-NCHR (NanoandMore USA, Lady’s Island, SC) Tapping mode probes with a 42 N/m spring constant were used to scan the mica surfaces. All imaging was carried out under tapping mode, with 512×512 data acquisitions at a scan speed of 0.8 Hz at room temperature under positive N2 pressure and acoustic isolation. Supplier-provided software (Nanoscope, V5.3r1, Veeco) was utilized for extracting quantitative data such as surface cross sections from AFM images. For high-resolution imaging, only tips with observed radii nominally 5 nm were used on the atomically flat mica surfaces to perform first order XY plane fitting.
Competitive Disassembly of Anti-CD3 CSANs
Competitive disassembly experiments were performed by mixing the preformed anti- CD3 CSANs with a 10-fold excess of methotrexate in P500 buffer and incubating at room temperature for various lengths of time. Disassembly of the nanorings was visualized via SEC, as described above.
In Vitro Competitive Binding Assay
Purified 1DDantiCD3 or CSANs at varying concentrations was mixed with a subsaturating concentration of FITC-labeled mAb UCHT-1 (eBioscience Inc.) for 10 minutes. The subsaturation level of antibody was pre-determined experimentally for the HPB-MLT cells by incubating increasing concentrations of labeled antibody with the cells and visualizing binding by flow cytometry. HPB-MLT T leukemia cells were grown in RPMI-10 medium (RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 2mM L-glutamine, 100ug/ml penicillin, and 100ug/ml streptomycin) at 37 °C in 5% CO2 atmosphere. 106 cells were incubated with the antibody mixture in PBS supplemented with 1% BSA and 0.1% sodium azide for 30 minutes at room temperature. The cells were then washed and fluorescence intensities of cell bound FITC-labeled antibodies were quantitated using a FACSCalibur flow cytometer (BD Biosciences). % inhibition of binding was calculated by subtracting the mean fluorescence observed at a given competitor concentration from 100% binding (defined as mean fluorescence observed in the absence of competition) then divided by 100% binding. Relative affinities were calculated from the corresponding IC50 values according to the equation: KD(I)=IC50/(1+[FITC-UCHT1]/KD(FITC-UCHT1)), where I is the unlabeled inhibitor, [FITCUCHT1] is the concentration of FITC-UCHT1 used in the competitive reaction, KD(FITCUCHT1) is the binding affinity of FITC-UCHT1, IC50 is the concentration of the inhibitor that yields 50% inhibition of binding. KD(FITC-UCHT1) was determined as previously described by fitting mean fluorescences of cell bound FITC-labeled UCHT-1 observed at different concentrations of FITC-labeled UCHT-1 to the Lineweaver-Burk equation: 1/F=1/Fmax+(KD/Fmax)(1/ [FITC-UCHT1]).26
Specific binding of octavalent anti-CD3 CSANs to cells that express CD3 receptor
HPB-MLT cells (CD3+ T leukemia cell line) and Daudi cells (CD3- B lymphoma cell line) were cultured in RPMI 1640 containing 10% fetal bovine serum, 100 units/ml penicillin, 100 ug/ml streptomycin, and 100 mmol/l L-glutamine. Octavalent anti-CD3 CSANs was fluorescein labeled using EZ-Label™ FITC protein labeling kit (Pierce Biotechnology). FITC-antiCD19 mAb, a gift from Dr. Vallera, was used as positive control bound to Daudi cells. Cells were incubated with FITC-antiCD19 mAb or FITC-octavalent anti-CD3 CSANs at 37°C for 30 mins. Cells were washed with FACS buffer twice and then resuspended with 500ul FACS buffer. Fluorescence intensities of cell bound FITC-labeled antibodies were quantitated using FACSCalibur flow cytometer (BD Biosciences).
Preparation of Human PBMC
After approval of the protocol by the Institutional Review Board of University of Minnesota and obtaining the written informed consent form from donors, human PBMCs were isolated from the heparinized peripheral blood of healthy volunteers by density gradient centrifugation. After a 2-fold dilution with PBS, the blood samples were slowly layered on top of Ficoll-Hypaque solution and centrifuged at 400xg for 30 mins with the brake off. PBMCs were collected from the cloudy interface, and washed three times with PBS before use.
Confocal Microscopy
Anti-CD3 CSANs were firstly fluorescein labeled using the EZ-Label™ FITC protein labeling kit (Pierce Biotechnology). HPB-MLT T leukemia cells or PBMCs (5×105) were incubated with FITC-labeled anti-CD3 CSANs, or UCHT-1 in PBS at 37 °C, in 5% CO2 for 30 min on Poly Prep Slides™ coated with poly-L-lysine (Sigma). For experiments at 4 °C the cells were incubated on the slides with the appropriate antibody at 4 °C for 30 min. All incubations were carried out in the dark to prevent dye bleaching.
As for the colocalization studies, cells were incubated with FITC labeled protein and LysoTracker Red DND-99 (molecular probes) or Transferrin-Alexa Fluor 594 conjugate (invitrogen) in PBS at 37 °C for 30 min. The inhibitors used in the endocytosis mechanism studies included chlorpromazine HCl (10ug/ml), nystatin (25ug/ml), cytochalasin D (10ug/ml), methyl-β-cyclodextrin (4.3mg/ml), and genistein (10.8ug/ml). Before the incubation with the FITC-labeled antibody, HPB-MLT T leukemia cells (5×105) were pretreated with the appropriate inhibitor for one hour at 37 °C.
Following the incubation with the FITC-labeled antibody, cells were then washed twice with PBS and fixed with 2% paraformaldehyde in PBS for 10 mins. After washing twice with PBS, cells were embedded in prolong gold antifade mounting medium (Invitrogen). Images were taken within inner sections of the cells by sequential scanning using a confocal microscope (Olympus, Fluoview™ FV1000 Laser Scanning Confocal Microscope).
PBMC Proliferation Assay
PBMCs were stained with 5 uM CFSE (carboxyfluorescein succinimidyl ester, Invitrogen) in PBS for 15 minutes at 37 °C, then washed and incubated in RPMI-10 medium for an additional 30 minutes. Labeled cells were washed and resuspended in RPMI-10 medium. 2×105 CFSE-labeled PBMCs were then cultured with 10ug/ml of the indicated antiCD3 sample in a 96-well flat-bottom plate for 5 days, followed by analysis with the FACScalibur flow cytometer.
Analysis of Expression of IL-2R (CD25)
2×105 PBMCs were incubated with the indicated anti-CD3 samples (5ug/ml) in RPMI- 10 medium at 37 °C for 20 hours. After transferring incubated cells from the plate to tubes by pipet, cells were pelleted by centrifugation. The supernatant was discarded, and cell pellet was washed with diluent (PBS with 1% BSA, 0.1% sodium azide). Cells were then stained with Alexa Fluor 488-antiCD25, and APC-antiCD4 (eBiosciences Inc.) at the concentration recommended by the manufacturer at 4 °C for 30 mins. Cells were then washed, and analyzed with the FACScalibur flow cytometer.
T Cell Receptor Modulation
2×105 PBMCs were incubated with the indicated antibody samples (5ug/ml) in RPMI- 10 medium at 37 °C for the desired time. T cell receptor modulation was stopped by cooling cells at 4 °C for 10 mins, followed by washing cells twice with ice-cold diluent (PBS with 1% BSA, 0.1% sodium azide). After staining with FITC-anti-human αβTCR (eBiosciences Inc.), cells were analyzed with the FACScalibur flow cytometer.
Supplementary Material
Acknowledgements
Financial support for these studies through NIH grants CA120116 (CRW) and CA125360 (CRW) is gratefully acknowledged.
CS was supported by the grant T32CA138312 from the National Cancer Institute.
REFERENCES
- 1.Carter PJ. Nat. Rev. Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 2.Holliger P, Hudson PJ. Nat. Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 3.Jain M, Kamal N, Batra SK. Trends Biotechnol. 2007;25:307–316. doi: 10.1016/j.tibtech.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Nelson AL, Reichert JM. Nat. Biotechnol. 2009;27:331–337. doi: 10.1038/nbt0409-331. [DOI] [PubMed] [Google Scholar]
- 5.Kipriyanov SM, Le Gall F. Mol. Biotechnol. 2004;26:39–60. doi: 10.1385/MB:26:1:39. [DOI] [PubMed] [Google Scholar]
- 6.Holliger P, Prospero T, Winter G. Proc. Nat. Acad. Sci. U.S.A. 1993;90:6444–6448. doi: 10.1073/pnas.90.14.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iliades P, Kortt AA, Hudson PJ. FEBS Lett. 1997;409:437–441. doi: 10.1016/s0014-5793(97)00475-4. [DOI] [PubMed] [Google Scholar]
- 8.Pack P, Pluckthun A. Biochemistry. 1992;31:1579–1584. doi: 10.1021/bi00121a001. [DOI] [PubMed] [Google Scholar]
- 9.Pack P, Muller K, Zahn R, Pluckthun A. J. Mol. Biol. 1995;246:28–34. doi: 10.1006/jmbi.1994.0062. [DOI] [PubMed] [Google Scholar]
- 10.Olafsen T, Cheung CW, Yazaki PJ, Li L, Sundaresan G, Gambhir SS, Sherman MA, Williams LE, Shively JE, Raubitschek AA, Wul AM. Protein Eng. Des. Sel. 2004;17:21–27. doi: 10.1093/protein/gzh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller K, Meng G, Liu J, Hurst A, Hsei V, Wong WL, Ekert R, Lawrence D, Sherwood S, DeForge L, Gaudreault J, Keller G, Sliwkowski M, Ashkenazi A, Presta L. J. Immunol. 2003;170:4854–4861. doi: 10.4049/jimmunol.170.9.4854. [DOI] [PubMed] [Google Scholar]
- 12.Wu CB, Ying H, Grinnell C, Bryant S, Miller R, Clabbers A, Bose S, McCarthy D, Zhu RR, Santora L, Davis-Taber R, Kunes Y, Fung E, Schwartz A, Sakorafas P, Gu JJ, Tarcsa E, Murtaza A, Ghayur T. Nat. Biotechnol. 2007;25:1290–1297. doi: 10.1038/nbt1345. [DOI] [PubMed] [Google Scholar]
- 13.Zhang JB, Tanha J, Hirama T, Khieu NH, To R, Hong TS, Stone E, Brisson JR, MacKenzie CR. J. Mol. Biol. 2004;335:49–56. doi: 10.1016/j.jmb.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 14.Rossi EA, Goldenberg DM, Cardillo TM, Stein R, Chang CH. Blood. 2009;113:6161–6171. doi: 10.1182/blood-2008-10-187138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deyev SM, Waibel R, Lebedenko EN, Schubiger AP, Pluckthun A. Nat. Biotechnol. 2003;21:1486–1492. doi: 10.1038/nbt916. [DOI] [PubMed] [Google Scholar]
- 16.Debbage P. Curr. Pharm. Des. 2009;15:153–172. doi: 10.2174/138161209787002870. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Daryl C, Zou H, Hayes ME, Adams GP, Kirpotin DB, Marks JD. J. Mol. Biol. 2007;371:934–947. doi: 10.1016/j.jmb.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fadel TR, Steenblock ER, Stern E, Li N, Wang XM, Haller GL, Pfefferle LD, Fahmy TM. Nano Lett. 2008;8:2070–2076. doi: 10.1021/nl080332i. [DOI] [PubMed] [Google Scholar]
- 19.McCarron PA, Marouf WM, Quinn DJ, Fay F, Burden RE, Olwill SA, Scott CJ. Bioconjugate Chem. 2008;19:1561–1569. doi: 10.1021/bc800057g. [DOI] [PubMed] [Google Scholar]
- 20.Ackerson CJ, Jadzinsky PD, Jensen GJ, Kornberg RD. J. Am. Chem. Soc. 2006;128:2635–2640. doi: 10.1021/ja0555668. [DOI] [PubMed] [Google Scholar]
- 21.Rossi EA, Goldenberg DM, Cardillo TM, Stein R, Wang Y, Chang CH. Cancer Res. 2008;68:8384–8392. doi: 10.1158/0008-5472.CAN-08-2033. [DOI] [PubMed] [Google Scholar]
- 22.Iyer AK, Khaled G, Fang J, Maeda H. Drug Discovery Today. 2006;11:812–818. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt MM, Wittrup KD. Mol. Cancer Ther. 2009;8:2861–2871. doi: 10.1158/1535-7163.MCT-09-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson JCT, Kanter A, Thuduppathy GR, Cody V, Pineda PE, McIvor RS, Wagner CR. J. Am. Chem. Soc. 2003;125:1501–1507. doi: 10.1021/ja026264y. [DOI] [PubMed] [Google Scholar]
- 25.Carlson JCT, Jena SS, Flenniken M, Chou TF, Siegel RA, Wagner CR. J. Am. Chem. Soc. 2006;128:7630–7638. doi: 10.1021/ja060631e. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Hapka D, Chen H, Vallera DA, Wagner CR. Angew. Chem., Int. Ed. 2008;47:10179–10182. doi: 10.1002/anie.200803507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawaya MR, Kraut J. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 28.Chou TF, So C, White BR, Carlson JCT, Sarikaya M, Wagner CR. ACS Nano. 2008;2:2519–2525. doi: 10.1021/nn800577h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanover JA, Beguinot L, Willingham MC, Pastan IH. J. Biol. Chem. 1985;260:5938–5945. [PubMed] [Google Scholar]
- 30.Wang LH, Rothberg KG, Anderson RGW. J. Cell Biol. 1993;123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson AS, Smythe E, Ayscough KR. Cell. Mol. Life Sci. 2009;66:2049–2065. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampath P, Pollard TD. Biochemistry. 1991;30:1973–1980. doi: 10.1021/bi00221a034. [DOI] [PubMed] [Google Scholar]
- 33.Luton F, Buferne M, Davoust J, Schmittverhulst AM, Boyer C. J. Immunol. 1994;153:63–72. [PubMed] [Google Scholar]
- 34.Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RGW. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 35.Kabouridis PS, Janzen J, Magee AL, Ley SC. Eur. J. Immunol. 2000;30:954–963. doi: 10.1002/1521-4141(200003)30:3<954::AID-IMMU954>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 36.Deng D, Jiang N, Hao SJ, Sun H, Zhang GJ. Biochim. Biophys. Acta Biomembr. 2009;1788:470–476. doi: 10.1016/j.bbamem.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 37.Choi I, De Ines C, Kurschner T, Cochlovius B, Sorensen V, Olafsen T, Sandlie I, Little M. Eur. J. Immunol. 2001;31:94–106. doi: 10.1002/1521-4141(200101)31:1<94::aid-immu94>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 38.Bronnerfraser M. J. Cell Biol. 1985;101:610–617. [Google Scholar]
- 39.Parren P, Warmerdam PAM, Boeije LCM, Capel PJA, Vandewinkel JGJ, Aarden LA. J. Immunol. 1992;148:695–701. [PubMed] [Google Scholar]
- 40.Geisler C. Crit. Rev. Immunol. 2004;24:67–85. doi: 10.1615/critrevimmunol.v24.i1.30. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology. Part A. Vol. 276. New York: Academic Press; 1997. pp. 224–225. [DOI] [PubMed] [Google Scholar]
- 42.Collaborative Computational Project, Number 4. Acta Crystallogr. 1994;D50:760–763. [Google Scholar]
- 43.Emsley P, Cowtan K. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Schuettelkopf AW, van Aalten DMF. Acta Cryst. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 45.Lovell SC, Davis IW, Arendell WB, III, de Baker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins Struct. Funct. Gen. 2002;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.