Abstract
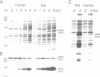
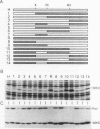
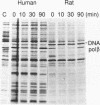
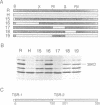
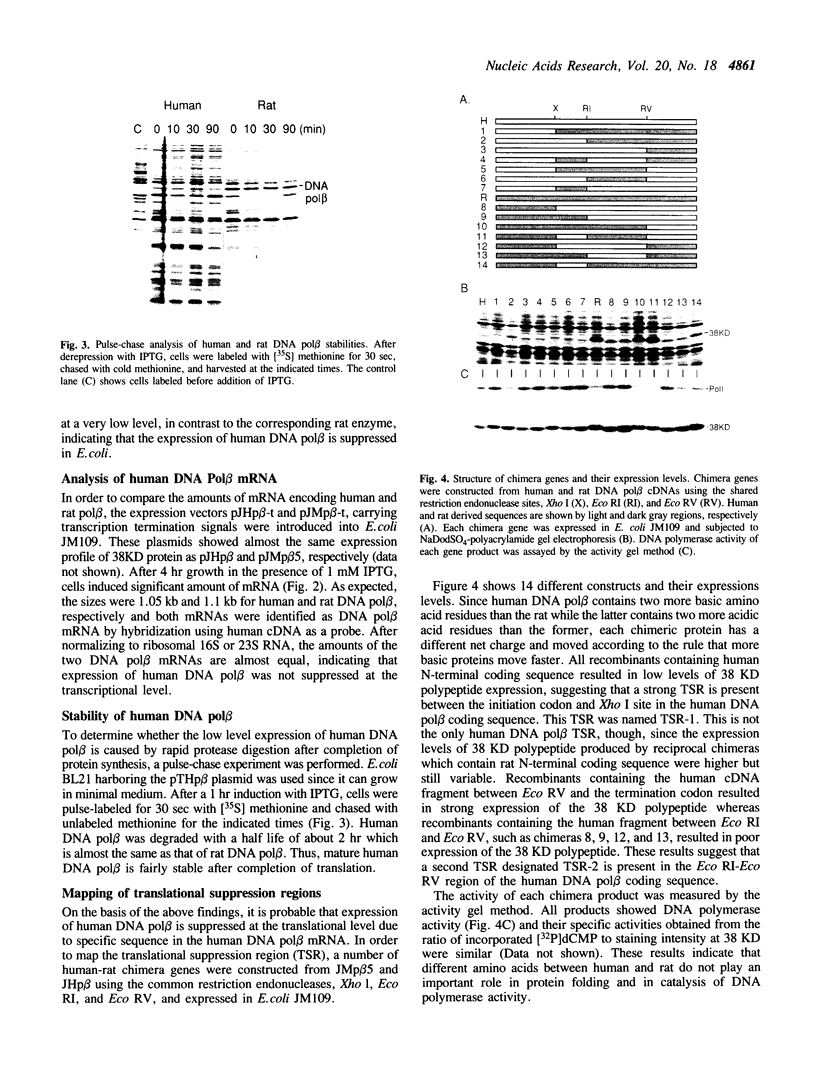
Although human DNA polymerase beta (DNA pol beta) shows 96% identity with rat DNA pol beta at the amino acid level, it is weakly expressed in Escherichia (E.) coli relative to the rat enzyme. The mechanism of this suppression was investigated. Pulse-chase protein labeling and steady state mRNA analysis showed that mature human DNA pol beta protein is relatively stable in E. coli and the levels of human and rat DNA pol beta mRNA were comparable indicating that the human DNA pol beta expression is suppressed at the translational level. By systematic expression analysis of a number of chimeric genes composed of human and rat cDNAs, two strong translational suppression regions were mapped in the human DNA pol beta mRNA; one was named TSR-1, corresponding to CGG encoding arginine (arg) at position 4 and the other, termed TSR-2, is located between codons 153 and 199. Since substitution of the rat Arg-4 codon with synonymous codons showed strong effects upon the expression level, we propose that the arg codon at the N-terminal coding region plays a role in modulating expression.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abbotts J., SenGupta D. N., Zmudzka B., Widen S. G., Notario V., Wilson S. H. Expression of human DNA polymerase beta in Escherichia coli and characterization of the recombinant enzyme. Biochemistry. 1988 Feb 9;27(3):901–909. doi: 10.1021/bi00403a010. [DOI] [PubMed] [Google Scholar]
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Andersson S. G., Kurland C. G. Codon preferences in free-living microorganisms. Microbiol Rev. 1990 Jun;54(2):198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aota S., Gojobori T., Ishibashi F., Maruyama T., Ikemura T. Codon usage tabulated from the GenBank Genetic Sequence Data. Nucleic Acids Res. 1988;16 (Suppl):r315–r402. doi: 10.1093/nar/16.suppl.r315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes H. J., Arlotto M. P., Waterman M. R. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A., Kedar P., Wilson S. H., Modak M. J. Active-site modification of mammalian DNA polymerase beta with pyridoxal 5'-phosphate: mechanism of inhibition and identification of lysine 71 in the deoxynucleoside triphosphate binding pocket. Biochemistry. 1989 Jul 25;28(15):6305–6309. doi: 10.1021/bi00441a023. [DOI] [PubMed] [Google Scholar]
- Chang L. M. Phylogeny of DNA polymerase-beta. Science. 1976 Mar 19;191(4232):1183–1185. doi: 10.1126/science.769158. [DOI] [PubMed] [Google Scholar]
- Chen G. F., Inouye M. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 1990 Mar 25;18(6):1465–1473. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date T., Tanihara K., Numura N. Construction of Escherichia coli vectors for expression and mutagenesis: synthesis of human c-Myc protein that is initiated at a non-AUG codon in exon 1. Gene. 1990 May 31;90(1):141–144. doi: 10.1016/0378-1119(90)90450-6. [DOI] [PubMed] [Google Scholar]
- Date T., Yamaguchi M., Hirose F., Nishimoto Y., Tanihara K., Matsukage A. Expression of active rat DNA polymerase beta in Escherichia coli. Biochemistry. 1988 Apr 19;27(8):2983–2990. doi: 10.1021/bi00408a048. [DOI] [PubMed] [Google Scholar]
- Date T., Yamamoto S., Tanihara K., Nishimoto Y., Liu N., Matsukage A. Site-directed mutagenesis of recombinant rat DNA polymerase beta: involvement of arginine-183 in primer recognition. Biochemistry. 1990 May 29;29(21):5027–5034. doi: 10.1021/bi00473a005. [DOI] [PubMed] [Google Scholar]
- Date T., Yamamoto S., Tanihara K., Nishimoto Y., Matsukage A. Aspartic acid residues at positions 190 and 192 of rat DNA polymerase beta are involved in primer binding. Biochemistry. 1991 May 28;30(21):5286–5292. doi: 10.1021/bi00235a023. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Gutman G. A., Hatfield G. W. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci U S A. 1989 May;86(10):3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose F., Hotta Y., Yamaguchi M., Matsukage A. Difference in the expression level of DNA polymerase beta among mouse tissues: high expression in the pachytene spermatocyte. Exp Cell Res. 1989 Mar;181(1):169–180. doi: 10.1016/0014-4827(89)90191-2. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Ito K., Date T., Wickner W. Synthesis, assembly into the cytoplasmic membrane, and proteolytic processing of the precursor of coliphage M13 coat protein. J Biol Chem. 1980 Mar 10;255(5):2123–2130. [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Kumar A., Widen S. G., Williams K. R., Kedar P., Karpel R. L., Wilson S. H. Studies of the domain structure of mammalian DNA polymerase beta. Identification of a discrete template binding domain. J Biol Chem. 1990 Feb 5;265(4):2124–2131. [PubMed] [Google Scholar]
- Kurland C. G. Codon bias and gene expression. FEBS Lett. 1991 Jul 22;285(2):165–169. doi: 10.1016/0014-5793(91)80797-7. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta D. N., Zmudzka B. Z., Kumar P., Cobianchi F., Skowronski J., Wilson S. H. Sequence of human DNA polymerase beta mRNA obtained through cDNA cloning. Biochem Biophys Res Commun. 1986 Apr 14;136(1):341–347. doi: 10.1016/0006-291x(86)90916-2. [DOI] [PubMed] [Google Scholar]
- Spanos A., Sedgwick S. G., Yarranton G. T., Hübscher U., Banks G. R. Detection of the catalytic activities of DNA polymerases and their associated exonucleases following SDS-polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Apr 24;9(8):1825–1839. doi: 10.1093/nar/9.8.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Sørensen M. A., Kurland C. G., Pedersen S. Codon usage determines translation rate in Escherichia coli. J Mol Biol. 1989 May 20;207(2):365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- Tobias J. W., Shrader T. E., Rocap G., Varshavsky A. The N-end rule in bacteria. Science. 1991 Nov 29;254(5036):1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wada K., Wada Y., Doi H., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):1981–1986. doi: 10.1093/nar/19.suppl.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood C. R., Boss M. A., Patel T. P., Emtage J. S. The influence of messenger RNA secondary structure on expression of an immunoglobulin heavy chain in Escherichia coli. Nucleic Acids Res. 1984 May 11;12(9):3937–3950. doi: 10.1093/nar/12.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zmudzka B. Z., SenGupta D., Matsukage A., Cobianchi F., Kumar P., Wilson S. H. Structure of rat DNA polymerase beta revealed by partial amino acid sequencing and cDNA cloning. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5106–5110. doi: 10.1073/pnas.83.14.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]