Abstract
Germicidin synthase (Gcs) from Streptomyces coelicolor is a type III polyketide synthase (PKS) with broad substrate flexibility for acyl groups linked through a thioester bond to either coenzyme A (CoA) or acyl carrier protein (ACP). Germicidin synthesis was reconstituted in vitro by coupling Gcs with fatty acid biosynthesis. Since Gcs has broad substrate flexibility, we directly compared the kinetic properties of Gcs with both acyl-ACP and acyl-CoA. The catalytic efficiency of Gcs for acyl-ACP was 10-fold higher than for acyl-CoA suggesting a strong preference towards carrier protein starter unit transfer. The 2.9 Å germicidin synthase crystal structure revealed canonical type III PKS architecture along with an unusual helical bundle of unknown function that appears to extend the dimerization interface. A pair of arginine residues adjacent to the active site affect catalytic activity but not ACP binding. This investigation provides new and surprising information about the interactions between type III PKSs and ACPs that will facilitate the construction of engineered systems for production of novel polyketides.
Keywords: Polyketide synthase, germicidin, acyl carrier protein
INTRODUCTION
Polyketides are ubiquitous secondary metabolites produced by bacteria, fungi, plants, and animals,1–4 and constitute one of the largest sources for natural product-based pharmaceuticals (antibiotics, antiparasitics, antifungals, anticancer drugs, and immunosuppressants) and other commercial products (food additives, pigments and nutraceuticals).5–7 Most polyketides are synthesized by three broad classes of polyketide synthases (PKSs), types I, II, and III, that share a common mechanism of sequential decarboxylative condensations of a wide range of acyl-coenzyme A (CoA) substrates.3,8,9 Polyketide structural diversity is dictated by selectivity of starter and extender units, the number of condensation reactions, and the manner of off-loading or ring closure of the fully elaborated polyketide chains.10 Type I PKSs are multifunctional proteins made up of modules sub-divisible into multiple discrete catalytic domains, all of which ultimately control the size, regio- and stereochemical characteristics of the polyketide scaffolds. Type II PKS are composed of dissociable enzyme complexes where each catalytic domain is expressed from an individual gene.9 Type III PKSs differ in that a single active site is used iteratively for starter unit loading, each Claisen condensation step, and the final off-loading/cyclization of the polyketide chain. Found as homodimers of a single ~40 kDa polypeptide, conventional type III PKSs iteratively condense malonyl-CoA derivatives with acyl-CoA esters3 rather than utilizing substrates that are covalently linked to acyl carrier proteins (ACP), which is a general feature of type I and II PKSs.8 Plant type III PKSs tend to utilize a larger variety of acyl-CoAs (e.g. cinnamoyl-CoAs) while the more divergent bacterial type III PKSs usually capture acyl-CoA substrates from primary metabolism.10
Manipulation of secondary metabolic pathways has enabled the generation of libraries of unnatural compounds.11,12 For example, 50 macrolides were prepared by systematically modifying modular components of the type I PKS, 6-deoxyerythronolide B synthase.13 However, since type III PKSs contain only a single catalytic site, exchange of functional domains is not a viable strategy to introduce new functional properties. Rather, differences in the active site cavity are responsible for the diversity among type III PKSs.3 Until recently, in vitro synthesis of novel polyketides has been dependent upon the substrate flexibility of the type III PKSs.14–16In vivo combinatorial biosynthesis was shown to be another promising route.17–19 Expanding the active site pocket by amino acid substitutions of a type III PKS enabled the synthesis of more complex molecules.20,21 This characteristic makes type III PKSs excellent candidates for enzyme engineering approaches to expand the structural diversity of small molecules derived from these systems.
Although early work indicated that type III PKSs use acyl-CoA starter units exclusively, numerous exceptions have been reported since. Recently, two type III PKSs, germicidin synthase (Gcs) and SCO7671 from Streptomyces coelicolor, were shown in vitro to accept both unnatural acyl-CoAs and acyl-ACPs as starter unit substrates.22 Similarly, ArsB and ArsC, both type III PKSs from Azotobacter vinelandii, obtain substrates directly from the ACP domains of the type I fatty acid synthase (FAS), ArsA, to give phenolic lipids.23 BpsA from Bacillus subtilis is speculated to directly accept the acyl moiety of acyl-ACP that is synthesized by the type II FAS system.24 FtpA from Myxococcus xanthus was also shown to use acyl-ACPs, which involves initial activation and transfer by a fatty acid-AMP ligase.25 In addition, the architecture of Steely1, a type I FAS/type III PKS hybrid from the social amoeba, Dictyostelium discoideum, whose type III PKS domain presumably accepts a thioester directly from its ACP domain.26 The ability of type III PKSs to utilize acyl-CoA and/or acyl-ACP starter units represents a new opportunity to manipulate these systems to expand natural product chemical diversity by engineering artificial hybrid PKS systems. However, our ability to develop rational approaches to engineer this remarkable class of biosynthetic enzymes requires a deeper understanding of the molecular basis for starter unit selectivity and processing. Therefore, following our initial observation of the unique molecular recognition of Gcs for an acyl-ACP starter unit donor,22 we were motivated to further investigate the biochemical and structural details of its catalytic function and protein-protein interactions.
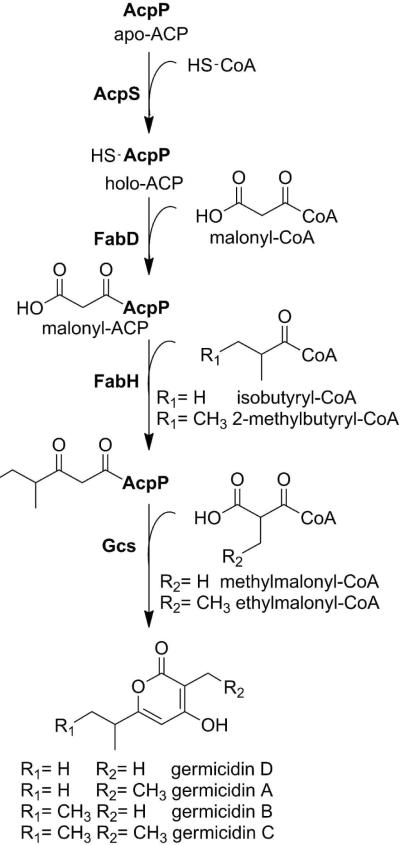
Bioinformatic analysis of the S. coelicolor genome resulted in identification and characterization of Gcs as a type III PKS responsible for producing germicidin A, B and C (Scheme 1).27,28 This investigation provided evidence that the proposed pathway for these natural products includes incorporation of branched acyl-chain starter units. The proposed β-ketoacyl thioester-linked ACPs are known intermediates in Streptomyces fatty acid biosynthesis formed by FabH- or FabF-catalyzed condensation of malonyl-ACP with 2-methylbutyryl-, isovaleryl-, isobutyryl, or n-butyryl-CoA.29 Homologs of Gcs (Figure S1) can be found in numerous Streptomyces species including S. lividans (99% identity), S. sviceus (88% identity), S. scabiei (87% identity), S. zinciresistens (84% identity) and S. viridochromogenes (87% identity), the latter being a confirmed producer of germicidins.30 The recently described S. coelicoflavus strain that produces surugapyrone A (germicidin D) likely contains a Gcs homolog as well.31
Scheme 1.
Proposed metabolic pathway for germicidin A, B, and C in Streptomyces coelicolor. Proteins are in bold.
In this study, we sought to determine the preference of S. coelicolor Gcs for acyl-CoAs or acyl-ACPs starter unit substrates. Furthermore, the fatty acid pathway, the putative source of acyl-ACPs, was reconstituted biochemically and coupled with Gcs to generate germicidin A in vitro. Additional insights were gained by solving the crystal structure of Gcs to assess potential surface residues involved in the molecular recognition of the type II FAS ACP acyl-chain starter unit donor. Protein-protein interaction studies were conducted to analyze the binding characteristics of the Gcs•ACP complex, and to investigate the role of the surface residues. Cumulatively, these data provide new insights into a type III PKS enzyme capable of using both acyl-CoAs and acyl-ACPs for assembly of a range of bioactive and structurally diverse pyrone natural products.
EXPERIMENTAL PROCEDURES
Materials
Malonyl-CoA, methylmalonyl-CoA, isobutyryl-CoA and acetoacetyl-CoA were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ethylmalonyl-CoA was synthesized according to published procedures.32 Aliquots of acyl-CoAs were maintained at −80 °C with 0.05% formic acid. 4-Hydroxy-3,6-dimethyl-2H-pyran-2-one was purchased from Alpha Aesar for use as an authentic standard. 3-Ethyl-4-hydroxy-6-methyl-2H-pyran-2-one was prepared from dehydroacetic acid (Sigma) as previously described.33
Bacterial Strains and Protein Purification
E. coli BL21(DE3) strains transformed with either pDHS9758, pDHS10019, and pSG3045, containing Sco2389 (acyl-carrier protein, AcpP), Sco7221 (germicidin synthase, Gcs), and Sco4744 (acyl-carrier protein synthase, AcpS), respectively, were constructed as previously described.22E. coli BL21(DE3) strains harboring plasmids pLH14 and pLH16, containing S. glaucescens β-ketoacyl-(acyl carrier protein) synthase III (KAS III, FabH) and malonyl CoA:acyl carrier protein malonyltransferase (FabD), respectively, were kind gifts from the laboratory of Kevin Reynolds.29 Mutants of Gcs were constructed by mutagenesis using the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA, USA) to replace an arginine with alanine at positions Arg276, Arg277, Arg280 or Arg317 using pDHS10019 as a template. Primers used for mutagenesis are provided in Table S1. Protein expression and purification was performed as previously described.22 Approximately 50 mg Gcs could be purified from a 1 L culture. Protein concentrations were determined using absorbance at 280 nm and calculated extinction coefficient (1 A280 = 0.62 mg/mL).
Size Exclusion Chromatography
Nickel-NTA purified His6-tagged Gcs protein was loaded onto a 120 mL HiLoad 16/60 Superdex 200 (GE Healthcare Bio-Sciences Corp, Piscataway, NJ, USA) column equilibrated with storage buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 10% glycerol, 0.5 mM TCEP). Fractions were combined, concentrated, frozen, and stored at −80 °C. Calibration of the column was performed with molecular weight markers from Sigma. Gcs eluted as a single peak at 74 mL, which is consistent with a dimeric complex in solution (Figure S2).
X-ray Crystallography
Gcs crystals were grown in 3–5 days at 4 °C using the hanging-drop vapor-diffusion method from an equal mixture of protein solution (10 mg/mL, freshly dialyzed into 20 mM HEPES, pH 7.5, 1 mM TCEP) and reservoir solution (21–25% isopropanol, 300 – 400 mM ammonium acetate, 0.1 M Tris buffer pH 8.5–9.0). To protect crystals from isopropanol evaporation during mounting, a volume of glycerol equal to twice the starting drop volume was added to the drop before the crystals were harvested in loops and cryo-protected in liquid N2. Attempts to obtain co-crystals with a CoA substrate (hexanoyl- or acetoacetyl-CoA) were unsuccessful. Attempts to soak in hexanoyl- or acetoacetyl-CoA into Gcs crystals after they had formed resulted in dissolution of the crystals within seconds.
Diffraction data were collected at 100 K on GM/CA-CAT beamline 23ID-D at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL, USA). Data were processed in cubic point group 432 (space group P4132 or P4332) using the HKL2000 suite.34 The structure was solved by molecular replacement. The BALBES server35 identified a solution in space group P4132 using a homology model based on the structure of S. coelicolor tetrahydroxynaphthalene synthase (PDB code 1U0M).36 A single polypeptide occupied the asymmetric unit, which had extremely high solvent content (80% v/v, Vm = 6.1 Å3/Da). An identical molecular replacement solution was obtained from the same probe structure using PHASER.37,38 The expected physiological dimer forms on a crystallographic 2-fold axis. Initial refinement steps were performed with REFMAC539 and manual modeling was completed using COOT.40 Final rounds of refinement were performed with PHENIX41 using a translation-libration-screw (TLS) model of molecular motion with 4 TLS groups identified by the TLSMD server (Table 1).42
Table 1.
Crystallographic summary.
| Parameter | Native |
|---|---|
| Space Group | P4132 |
| Dimensions (Å) a,b,c | 182.70 |
| X-ray source | APS 23ID-D |
| Wavelength λ (Å) | 0.97934 |
| dmin(Å) a | 2.90 (3.00–2.90) |
| Unique observations | 23,726 |
| Rmerge (%) a,b | 6.8 (57.8) |
| 〈I/σ〉 a | 27.7 (4.3) |
| Completeness (%) a | 99.9 (100) |
| Avg. redundancy a | 9.6 (9.7) |
| R/Rfree c,d | 0.238/0.265 |
| RMSD bond length (Å) | 0.007 |
| RMSD bond angle (°) | 0.757 |
| Avg. Protein B-factor (Å2) | 63.3 |
| Wilson B (Å2) | 78.8 |
| Ramachandran plot e | |
| Favored | 97.37 |
| Allowed | 2.37 |
| Disallowed | 0.26 |
Values in parenthesis are for outer shell
Rmerge = Σ|Ii-〈I〉|/ΣIi, where Ii is the intensity of the ith observation and 〈I〉 is the mean intensity
R = Σ|Fo-|Fc∥/Σ|Fo| where Fo is the observed structure factor and Fc is the calculated structure factor used in the refinement
Rfree = Σ|Fo-|Fc∥/Σ|Fo| where Fo is the observed structure factor and Fc is the calculated structure factor from 5% of reflections not used in the refinement
From MOLProbity62
Reconstituting the Fatty Acid Biosynthesis Pathway with Gcs
In order to synthesize germicidin A, the Gcs reaction was coupled with the fatty acid biosynthesis pathway to access the proposed branched 3-oxo-4-methyl-pentyl-ACP intermediate. In a total of 2.0 mL, the reaction consisted of 50 mM HEPES, pH 7, 150 mM NaCl, 10 mM MgSO4, 2 mM TCEP, 100 μM CoA, 400 μM malonyl-CoA, 200 μM ethylmalonyl-CoA, 200 μM isobutyryl-CoA, 10 μM AcpP, 2.5 μM FabD, 2.5 μM FabH, 2.5 μM Gcs, and 2.5 μM AcpS. The reaction proceeded at room temperature overnight followed by extraction with two equal volumes of ethyl acetate. The resulting organic layer was dried under vacuum. The dried residue was re-dissolved in acetonitrile and fractionated on a semi-preparative C18 column using an isocratic gradient (25% acetonitrile in water with 0.1% formic acid). Approximately 15 μg of purified product was characterized by capillary 1H NMR and LC-ESIMS.
Synthesis of 3-Oxo-4-methyl-pentyl-CoA
A 1 mL reaction mixture containing of 50 mM HEPES, pH 7, 150 mM NaCl, 1 mM TCEP, 1 mM malonyl-CoA, 1 mM isobutyryl-CoA and 25 μM FabH was prepared. The reaction proceeded at 25 °C for three hours followed by the addition of 10 μL of formic acid. 3-oxo-4-methyl-pentyl-CoA was purified on a semi-preparative C18 column using a binary gradient starting at 95% solvent A (5 mM NH4OAc, pH 5.4) and 5% solvent B (MeOH, 0.1% formic acid) for five minutes followed by a linear gradient over 30 minutes ending with 50% solvent B. Pooled elution fractions were concentrated using rotary evaporation to remove methanol and finally dried by lyophilization. Product formation was verified by LC-MS as described in the Supporting Information. Stock solutions were quantified at 260 nm using a calibration curve of acetoacetyl-CoA.
Acylation of AcpP
A 250 μM mixture of apo- and holo-AcpP, 10 μM AcpS, 0.5 mM 3-oxo-4-methylpentyl-CoA, 10 mM MgSO4, HEPES buffer (50 mM HEPES, pH 7.0, 150 mM NaCl), in a total volume of 2 mL was incubated at 25 °C for 3 hours (of note, Sfp and Svp were both ineffective).43,44 The reaction mixture was loaded onto a 5 mL His-Trap column (GE Healthcare), washed with 5 column volumes of HEPES buffer, and finally eluted with 5 column volumes of HEPES buffer with 0.3 M imidazole. The protein was concentrated using a 3K MWCO centrifuge column (Pall Life Sciences, Ann Arbor, MI, USA) followed by buffer exchange on a PD-10 column (10% glycerol, 0.1 M NaCl, 50 mM HEPES, pH 6.8; GE Healthcare). Protein concentration was determined by using the Bradford protein assay (Bio-Rad, Hercules, CA, USA) with BSA as the standard. 3-Oxo-4-methyl-pentyl-AcpP was analyzed with electrospray mass spectrometry by using a ThermoFinnigan LTQ linear ion trap instrument (capillary temperature, 250 °C; capillary voltage, 32 V; tube lens, 95 V). Mass spectra were deconvoluted by using ProMassN for Xcalibur (Novatia, Monmouth Junction, NJ, USA).
Enzyme Kinetics
The kinetic constants of Gcs, Km and kcat, were determined by varying starter unit acyl-CoA concentrations while using a fixed extender unit concentration of either methylmalonyl-CoA or ethylmalonyl-CoA. The enzyme reaction solution consisted of 100 mM Tris-HCl, pH 7.8, 150 mM NaCl, 1.0 mM methylmalonyl-CoA or ethylmalonyl-CoA, and various concentrations of 3-oxo-4-methyl-pentyl-CoA in a total volume of 40 μL. The reaction solution was pre-warmed at 30 °C for five minutes before adding 0.2 μM Gcs. The reaction proceeded for one minute before being quenched with the addition of 10 μL of 6.05 M HCl in methanol and frozen at −80 °C until analysis. Reactions with acetoacetyl-CoA used 3 μM Gcs and the reaction time was extended to five minutes. Reactions were analyzed by injecting 25 μL onto a Beckman HPLC with a XBridge C18 column (5 μm particle size, 4.6 × 250 mm) using an isocratic gradient of 25% or 30% acetonitrile in water with 0.1% formic acid. Authentic standards were used to quantify product formation at 290 nm. Reactions with varying concentrations of 3-oxo-4-methyl-pentyl-AcpP (a mixture of 33% apo-, 31% holo-, 36% 3-oxo-4-methyl-pentyl-AcpP as determined by MS) were conducted in the same fashion. Steady-state parameters were determined by fitting to the equation, ν = [E]kcat[S]/([S]+Km) using GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA).
Binding Kinetics
All biolayer interferometry measurements were made using an Octet RED instrument (ForteBio, Menlo Park, CA, USA) using streptavidin (SA) biosensors. Assays were performed in 96-well black microplates at 25 °C and 1000 rpm. All volumes were 200 L. AcpP was biotinylated using EZ-Link NHS-LC-LC-biotin (succinimidyl-6-[biotinamido]-6-hexanamidohexanoate) (Thermo Scientific Pierce, Rockford, IL, USA) at a 5:1 molar ratio of biotin to protein for 30 min at 25 °C followed by dialysis into phosphate buffered saline (10 mM Phosphate, pH 7.4, 150 mM NaCl) using a 3K MWCO Slide-A-Lyzer (Thermo Scientific Pierce). All Gcs proteins were similarly dialyzed. Biotinylated AcpP (50 μg/mL) was loaded onto the sensors for 600 s. After a baseline in 1× PBS kinetics buffer (ForteBio) was established, tethered AcpP was exposed to Gcs protein at concentrations between 0.3 and 10 μM. Association was monitored for 1800 s followed by dissociation in 1× PBS kinetics buffer for 1800 s. A reference sensor (only tethered AcpP) was subtracted from each data set. Shift data were analyzed with ForteBio Analysis software (version 7.0). Kinetic parameters (kon and koff) and affinity (KD) were determined from a global non-linear regression of association and dissociation binding kinetics using a 1:1 Langmuir binding model.
Relative Enzyme Activity Assay
The relative activities of Gcs wild-type and each Gcs mutant were determined. Reactions preceded similarly as described above using 0.2 μM Gcs wild-type or mutant, 1.0 mM ethylmalonyl-CoA and either 100 μM 3-oxo-4-methyl-pentyl-CoA or 5 μM 3-oxo-4-methyl-pentyl-AcpP. In the case of the R276/317A double mutant, for which no activity was detected, the reaction time was subsequently extended to two hours (with wild-type enzyme as a control) to confirm the absence of product (see main text).
RESULTS AND DISCUSSION
Germicidin Biosynthesis Pathway
Starter units for the germicidin pathways were predicted to originate from fatty acid biosynthesis with Gcs utilizing S. coelicolor endogenous fatty acid intermediates as reaction substrates.22,27 A previous study revealed that a S. coelicolor ΔScFabH∷EcFabH mutant produced substantially less germicidin metabolites than the wild-type strain27 because E. coli FabH preferentially accepts straight-chain acyl-CoAs.45 The fatty acid acyl carrier protein, AcpP, is adjacent to FabH within the S. coelicolor type II FAS operon. Thus, we reasoned that AcpP was the most likely acyl carrier protein starter unit donor for Gcs. The proposed pathway (Scheme 1) begins with AcpS, which converts AcpP from the apo into the holo form. FabD employs malonyl-CoA to convert holo-AcpP into malonyl-AcpP and FabH follows by catalyzing the condensation of acyl-CoAs (e.g. 2-methylbutyryl-CoA, isobutyryl-CoA, and acetyl-CoA) with malonyl-AcpP to generate the corresponding β-ketoacyl-AcpPs. For example, we reasoned that 3-oxo-4-methyl-pentyl-AcpP would form in a FabH-catalyzed reaction of isobutyryl-CoA with malonyl-AcpP and serve as the starter unit for germicidin biosynthesis.29 We tested this model by reconstituting the fatty acid-coupled germicidin pathway in vitro. The reaction mixture consisted of FabD, FabH, AcpS, apo/holo-AcpP, and Gcs. When co-administered with ethylmalonyl-CoA, germicidin A is the predicted product. The mass of the major peak formed was consistent with germicidin A and the 1H NMR confirmed the assignment based on comparison with published data (Figure S3).30
Surprisingly, in control experiments, where one of the enzymes was omitted from the reaction, we observed that just FabH and Gcs were sufficient to form germicidin A in vitro (Figure S3). We hypothesized that the condensation of isobutyryl-CoA with malonyl-CoA was catalyzed by FabH to form 3-oxo-4-methyl-pentyl-CoA. This acyl-CoA substrate could then be converted by Gcs directly to germicidin A. A reaction consisting of only FabH with isobutyryl-CoA and malonyl-CoA was tested for the production of 3-oxo-4-methyl-pentyl-CoA. Indeed, this reaction produced both free coenzyme A along with a new product bearing a molecular mass matching the predicted 3-oxo-4-methyl-pentyl-CoA (LC-ESIMS: [M−H]− = 878.25 m/z, and [M+H]+ = 880.35; calculated 878.16 and 880.17, respectively) (Figure S4). Upon purification, Gcs was able to convert the new FabH product along with methylmalonyl-CoA or ethylmalonyl-CoA into germicidin D and germicidin A, respectively. The ability of FabH to act independent of malonyl-AcpP was surprising, but not without precedent in bacterial anabolism. FabH from E. coli can catalyze formation of acetoacetyl-CoA from malonyl-CoA and acetyl-CoA.46 Additionally, an acetoacetyl-CoA synthase from a soil-isolated Streptomyces sp. strain, which shares homology to KAS III was recently described.47
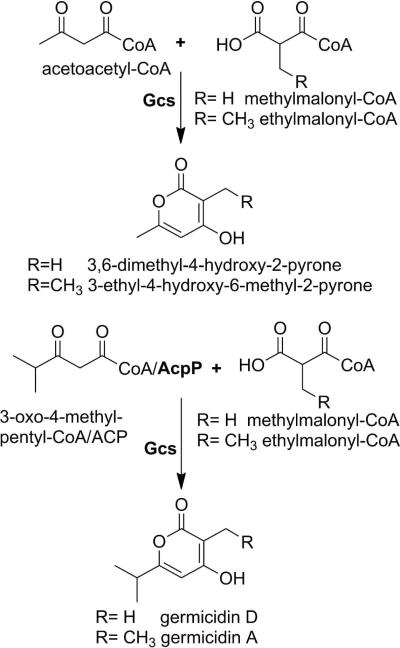
Enzyme Kinetics
The experiments above demonstrated that two parallel paths to germicidin products are possible, involving acyl-AcpP and acyl-CoA starter units. In order to assess which route might be favored, we determined the kinetic parameters for both types of reactions (Scheme 2). Many bacterial type III PKS enzymes use only malonyl-CoA as both starter and extender units. For example, 1,3,6,8-tetrahydroxynaphthalene (THN) synthase from S. griseus,48 PhlD from Pseudomonas fluorescens,49 DpgA from Amycolatopsis orientalis,50 use five, four and three molecules of malonyl-CoA, respectively. Other bacterial type III PKSs including ArsB and ArsC from Azotobacter vinelandii and SrsA from S. griseus use long chain acyl-thioesters as a starter unit.23,51,52 Like SrsA, Gcs can use additional extender units such as methylmalonyl- and ethylmalonyl-CoA.22,27 Furthermore, we have previously demonstrated that Gcs and SCO7661 are able to catalyze reactions using a range of unnatural acyl-CoA and acyl-ACP-tethered starter units.22
Scheme 2.
In vitro reactions catalyzed by Gcs. Proteins are in bold.
Our initial in vitro experiments with Gcs used acetoacetyl-CoA and 3-oxo-4-methyl-pentyl-CoA as starter units to determine substrate preference and whether the extender unit choice (methylmalonyl-CoA or ethylmalonyl-CoA), plays a significant role in product profile (Figure S5a and Table 2). When using acetoacetyl-CoA, Gcs displayed typical saturation kinetics with methylmalonyl-CoA as the extender unit but not with ethylmalonyl-CoA, for which the apparent Km was greater than 1 mM. In contrast, enzymatic activity was substantially higher for 3-oxo-4-methyl-pentyl-CoA when comparing specificity constants (kcat/Km). With the branched acyl-CoA starter unit, Gcs did not display any extender unit preference (Figure S5b and Table 2). The kinetic parameters of Gcs using 3-oxo-4-methyl-pentyl-CoA were comparable to other bacterial type III PKS enzymes that employ a long chain acyl-CoA starter unit (ArsB from Azotobacter vinelandii for n-behenyl-CoA: kcat = 0.931 min−1, Km = 4.86 μM, compared to Gcs for 3-oxo-4-methyl-pentyl-CoA with saturating amounts of methylmalonyl-CoA: kcat = 12.97 min−1, Km = 12.12 μM).52
Table 2.
Enzyme kinetics constants determined for the different starter unit substrates catalyzed by Gcs. Standard deviation calculated from triplicates.
| Starter unit | Extender unit | Km (μM) | kcat (min−1) | kcat/Km (s−1 M−1) |
|---|---|---|---|---|
| acetoacetyl-CoA | methylmalonyl-CoA | 75.17 ± 8.45 | 2.91 ± 0.08 | 645 |
| 3-oxo-4-methyl-pentyl-CoA | 12.12 ± 0.31 | 12.97 ± 1.46 | 17840 | |
| 3-oxo-4-methyl-pentyl-AcpP | 1.65 ± 0.22 | 16.49 ± 0.70 | 167000 | |
| acetoacetyl-CoA | ethylmalonyl-CoA | 1244 ± 148 | 9.89 ± 0.58 | 133 |
| 3-oxo-4-methyl-pentyl-CoA | 39.25 ± 3.36 | 31.26 ± 0.96 | 13270 | |
| 3-oxo-4-methyl-pentyl-AcpP | 1.56 ± 0.20 | 14.19 ± 0.56 | 152000 | |
Gcs was then characterized using 3-oxo-4-methyl-pentyl-AcpP as a substrate to generate either germicidin A or germicidin D (Figure S5c and Table 2). The catalytic efficiency using 3-oxo-4-methylpentyl-AcpP along with either methylmalonyl-CoA or ethylmalonyl-CoA was an order of magnitude higher compared to the corresponding acyl-CoA analog. For comparison, the apparent Km value of Staphylococcus aureus FabH for malonyl-AcpP was 1.76 ± 0.40 μM53 and the Km value of Gcs for 3-oxo-4-methyl-pentyl-AcpP was 1.6 ± 0.2 μM. This data suggests that Gcs can preferentially select acyl-AcpP over acyl-CoA to produce germicidins.
X-ray Crystal Structure of Gcs
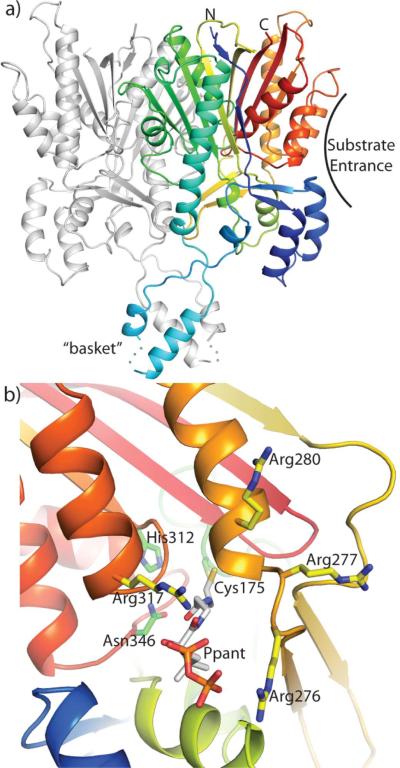
To gain fundamental information about type III PKS•ACP interactions, we determined the crystal structure of recombinant His6-Gcs at 2.9 Å resolution (Figure 1). All previously reported type III PKS structures are dimeric, and size exclusion chromatography indicated that two Gcs molecules associated to form a homodimer in solution (Figure S2). The asymmetric unit of Gcs crystals contains a single protein molecule. However, the presumed physiologically relevant dimer is formed by a crystallographic 2-fold contact that buries 2300 Å2 of protein surface area per monomer (~13.6% of the monomer surface area).
Figure 1.
Overall structure and active site of Gcs. a) A view of the Gcs dimer; one monomer is colored blue to red from N- to C- terminus; the second monomer is grey. b) Close up of the active site viewed from the substrate entrance. Side chains for surface arginines at the proposed ACP docking site are indicated in yellow and for the Cys-His-Asn catalytic triad in green. The phosphopantethiene arm is modeled in this figure (white C atoms) based on a FabH complex with CoA (PDB: 2GYO).61
Similar to other type III PKS structures,1,54 Gcs contains a Cys-His-Asn catalytic triad within a deep active site cavity that is accessible to the surrounding solvent. Apart from a few exterior loops, there are few major differences in the conserved αβαβα-fold or dimer interface compared with S. coelicolor THNS.36 Curiously, Gcs contains a long insertion between residues 61 and 100, which has not been observed in other type III PKSs (Figure S1). Examination of sequence alignments fails to shed light on the function of this insertion. A structural role is implied because the large insertion protrudes from the core of Gcs and folds into a four-helix bundle formed by two helices from each monomer. The shape of the helical bundle resembles a “basket” hanging from the catalytic core of the protein (Figure 1a). Electron density is especially poor for the basket, likely because it extends into the large solvent channels of the crystal and makes no contacts with other molecules within the lattice. As the basket is not in proximity to the active site entrance (Figure 1a), we do not expect that it interacts with the carrier protein. It is possible that this insertion reinforces the dimer interface. Attempts to produce Gcs mutants in which the basket insert was removed (GcsΔ63–96, GcsΔ63–97, and GcsΔ63–98) resulted only in insoluble protein (data not shown).
Identifying Putative Residues for Gcs•AcpP Binding
Based upon sequence alignments, bacterial type III PKSs, and not plant type III PKSs, were predicted to accept starter units from ACPs.1 Direct biochemical evidence was obtained when Gcs was shown to accept acyl groups carried by either CoA or ACP.22 Although the bacterial and plant enzymes have strong structural similarities, only bacterial PKSs have highly conserved residues aligned with a cationic/hydrophobic patch determined to be crucial for FabH•ACP binding.46 Similarly, other FAS complexes (ACP with FabD, FabG, and FabI) have cationic/hydrophobic patches in close proximity to their active sites that when mutated affect ACP binding.55–58
Based on the Gcs crystal structure, a cationic patch including four arginine residues (Arg276, Arg277, Arg280 and Arg317) was identified adjacent to the type III catalytic triad active site (Figure 1b). Moreover, Gcs position Arg317 aligns with the E. coli FabH Arg249 residue responsible for KAS III•ACP binding (Figure S1a).46 The arginine residues in the cationic patch were each replaced with alanine to determine the importance of these position(s) for promoting Gcs•AcpP molecular recognition. Perturbations to these interactions were predicted to affect binding and/or catalysis by affecting protein-protein contacts and/or pantetheinate arm-protein interactions. For example, a decrease in protein-protein interactions may also result in attenuated catalysis while changes to pantetheinate binding could result in attenuated catalysis and may or may not affect protein-protein interactions.
Effects of Surface Mutations on Gcs•AcpP Binding and Activity
Two assays were employed to evaluate the effects of surface residue mutations on the Gcs•AcpP specificity. The first assay was a direct measurement of AcpP binding to Gcs using biolayer interferometry.59 The second assay was a measurement of catalytic activity of Gcs mutants with either acyl-AcpP or acyl-CoA. We reasoned that any mutation that interferes with the Gcs•AcpP interaction could also affect AcpP-dependent Gcs catalytic activity.
Binding of Gcs to the AcpP immobilized onto the streptavidin biosensor led to a measured increase in biolayer thickness in real time to provide the association rate constant (kon), the dissociation rate constant (koff) and the dissociation constant (KD) (Table 3). The curve fits for the wild-type and mutant forms of Gcs closely followed a 1:1 binding model (Figure S6). However, none of these mutants significantly disrupted the specific binding to AcpP. Overall, wild-type Gcs has greater affinity for ACP than do type II FAS enzymes, based on reported KDs; for instance, S. coelicolor AcpP was reported to bind to FabD with a KD of 1.9 ± 0.3 μM,58 whereas a KD of 0.2 μM was observed for the Gcs•AcpP interaction.
Table 3.
Binding kinetics for Gcs•AcpP interaction. Standard deviation calculated from duplicates.
| Mutant | kon (M−1 s−1) | koff (s−1 * 10−4) | KD (μM) |
|---|---|---|---|
| Wild type | 1188.0 ± 46.7 | 2.203 ± 0.049 | 0.225 ± 0.015 |
| R276A | 627.6 ± 2.8 | 2.529 ± 0.837 | 0.185 ± 0.003 |
| R277A | 694.9 ± 95.5 | 1.281 ± 0.221 | 0.403 ± 0.132 |
| R280A | 769.1 ± 92.8 | 1.220 ± 0.138 | 0.184 ± 0.006 |
| R317A | 1812.5 ± 16.3 | 4.067 ± 0.301 | 0.161 ± 0.037 |
| R276/317A | 891.9 ± 69.1 | 1.703 ±0.002 | 0.192 ± 0.015 |
The mutants and wild-type Gcs were compared in their ability to produce germicidin A. Two of the mutants (R276A and R317A) had significantly impaired activity with the acyl-CoA substrate analog when compared to the wild-type Gcs protein (Table 4). Similarly, these two mutants had somewhat reduced activity with acyl-AcpP substrates (Table 4). Therefore, these residues are not likely to be responsible for the observed Gcs selectivity for AcpP. In contrast, a cationic patch residue of FabH, Arg249 (sequence aligned with Gcs Arg317), was shown to affect both binding and catalysis of malonyl-AcpP substrate while not affecting catalysis with malonyl-CoA,46 a result that is inconsistent with structures of FabH co-crystallized with CoA and related analogs where the pantetheinate arm is in direct contact with Arg249.60,61 When the pantetheinate arm was modeled directly into the Gcs structure using the coordinates from a FabH structure (PDB: 2GYO),61 the model suggests that Arg276 and Arg317 both make direct contact with the phosphoryl groups (Figure 1b). Consistent with this hypothesis, the double mutant (R276/317A) had no detectable activity for either acyl-CoA or acyl-AcpP, implying that phosphopantetheine interaction with either Arg276 or Arg317 is essential to Gcs activity. Our results suggest that the major role of residues Arg276 and Arg317 involves pantetheinate binding, and that AcpP selectivity resides elsewhere on Gcs.
Table 4.
Relative activities of Gcs wild-type and mutants for 3-oxo-4-methyl-pentyl-CoA or -AcpP starter unit and ethylmalonyl-CoA as extender unit. Standard deviation calculated from triplicates.
| Starter unit: | 3-oxo-4-methyl-pentyl-CoA | 3-oxo-4-methyl-pentyl-AcpP |
|---|---|---|
| Activity (%) | Activity % | |
| WT | 100 ± 6 | 100 ± 5 |
| R276A | 25 ± 6 | 46 ± 12 |
| R277A | 75 ± 3 | 106 ± 4 |
| R280A | 103 ± 4 | 100 ± 5 |
| R317A | 24 ± 6 | 64 ± 7 |
| R276/317A | No activity | No activity |
CONCLUSION
PKSs are responsible for making a vast range of natural products with diverse biological activities. Harnessing the biosynthesis of polyketides has the potential to open up new sources of valuable small molecules for pharmaceutical development. Re-engineering of PKSs demands an in-depth understanding of the multiple proteins involved and how they interact for functional catalysis. In this study we investigated the Gcs type III PKS, which selectively employs acyl-ACPs as starter unit donor, a role previously limited to type I and II PKSs. Towards this goal, we set out to characterize the specificity of the protein-protein interaction, and to initiate efforts to probe the key features that govern ACP recognition by Gcs. We first reconstituted germicidin synthesis by coupling Gcs with the endogenous S. coelicolor fatty acid pathway. This study revealed that Gcs functions with a 10-fold higher activity towards acyl-AcpP compared to the corresponding acyl-CoA, which suggests the predominance of this pathway in vivo.27 The Gcs crystal structure revealed canonical type III PKS architecture except that the dimer interface was extended by a 40-residue insertion. Similar to type II FAS enzymes, Gcs has a cationic patch surrounding the entrance to its catalytic pocket, a feature considered pivotal for FAS•ACP docking.1,46,55–58 However, upon changing the putative surface arginine residues to alanine, the Gcs mutant proteins retained high affinity for AcpP. Differences between the Gcs mutants were revealed when comparing relative activities, with two alanine variants showing attenuated catalytic function against both acyl-AcpP and acyl-CoA and the double mutant having no activity. Based on homology modeling, the two surface arginine residues are likely to directly interact with the phosphate group of pantetheinate, and the synergistic effect of the two partially inactivating mutations implies that at least one of these arginines is essential. However, the amino acids that affect Gcs•AcpP binding remain to be identified. This work has highlighted the role of cationic residues surrounding the active site of a bacterial type III PKS capable of accepting acyl-ACPs. Further study into the type III PKS•AcpP interaction may lead to engineered type I/III PKS hybrid proteins capable of creating novel polyketide structures.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by the National Institute of Health Grants DK42303 (to J. L. S), GM076477 (to D. H. S. and J. L. S.) and the Hans W. Vahlteich Professorship (D. H. S.). Beamline 23ID-D is supported by the National Institutes of Health, National Institute of General Medicine Sciences (Y1-GM-1104) and National Cancer Institute (Y1-CO-1020) through the GM/CA Collaborative Access Team at APS, which is supported by the United States Department of Energy. Special thanks to Frank E. Bartley III for help with crystallization experiments.
Footnotes
Supporting Information. Figures S1–S6 and Tables S1. This material is available free of charge via the Internet at http://pubs.acs.org.
The atomic coordinates and structure factors (code XXXX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
REFERENCES
- (1).Austin MB, Noel AJP. Nat. Prod. Rep. 2003;20:79. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- (2).Walsh CT. Science. 2004;303:1805. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- (3).Abe I, Morita H. Nat. Prod. Rep. 2010;27:809. doi: 10.1039/b909988n. [DOI] [PubMed] [Google Scholar]
- (4).Saruwatari T, Praseuth AP, Sato M, Torikai K, Noguchi H, Watanabe K. J. Antibiot. 2011;64:9. doi: 10.1038/ja.2010.130. [DOI] [PubMed] [Google Scholar]
- (5).Newman DJ, Cragg GM. J. Nat. Prod. 2007;70:461. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- (6).Hill AM. Nat. Prod. Rep. 2006;23:256. doi: 10.1039/b301028g. [DOI] [PubMed] [Google Scholar]
- (7).Staunton J, Weissman KJ. Nat. Prod. Rep. 2001;18:380. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- (8).Fischbach MA, Walsh CT. Chem. Rev. 2006;106:3468. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- (9).Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Nat. Prod. Rep. 2007;24:162. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- (10).Hertweck C. Angew. Chem., Int. Ed. 2009;48:4688. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- (11).Walsh CT. Chembiochem. 2002;3:125. doi: 10.1002/1439-7633(20020301)3:2/3<124::AID-CBIC124>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- (12).Olano C, Mendez C, Salas JA. Microbial Biotech. 2011;4:144. doi: 10.1111/j.1751-7915.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Xue Q, Ashley G, Hutchinson CR, Santi DV. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11740. doi: 10.1073/pnas.96.21.11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Abe I, Morita H, Nomura A, Noguchi H. J. Am. Chem. Soc. 2000;122:11242. [Google Scholar]
- (15).Morita H, Noguchi H, Schroder J, Abe I. Euro. J. Biochem. 2001;268:3759. doi: 10.1046/j.1432-1327.2001.02289.x. [DOI] [PubMed] [Google Scholar]
- (16).Morita H, Yamashita M, Shi SP, Wakimoto T, Kondo S, Kato R, Sugio S, Kohno T, Abe I. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13504. doi: 10.1073/pnas.1107782108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chemler JA, Yan YJ, Leonard E, Koffas MAG. Org. Lett. 2007;9:1855. doi: 10.1021/ol0703736. [DOI] [PubMed] [Google Scholar]
- (18).Werner SR, Chen H, Jiang HX, Morgan JA. J. Mol. Catal. B: Enzym. 2010;66:257. [Google Scholar]
- (19).Katsuyama Y, Funa N, Miyahisa I, Horinouchi S. Chem. Biol. 2007;14:613. doi: 10.1016/j.chembiol.2007.05.004. [DOI] [PubMed] [Google Scholar]
- (20).Abe I, Watanabe T, Morita H, Kohno T, Noguchi H. Org. Lett. 2006;8:499. doi: 10.1021/ol052912h. [DOI] [PubMed] [Google Scholar]
- (21).Shi SP, Wanibuchi K, Morita H, Endo K, Noguchi H, Abe I. Org. Lett. 2009;11:551. doi: 10.1021/ol802606w. [DOI] [PubMed] [Google Scholar]
- (22).Gruschow S, Buchholz TJ, Seufert W, Dordick JS, Sherman DH. Chembiochem. 2007;8:863. doi: 10.1002/cbic.200700026. [DOI] [PubMed] [Google Scholar]
- (23).Miyanaga A, Funa N, Awakawa T, Horinouchi S. Proc. Natl. Acad. Sci. U.S.A. 2008;105:871. doi: 10.1073/pnas.0709819105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Nakano C, Ozawa H, Akanuma G, Funa N, Horinouchi S. J. Bacteriol. 2009;191:4916. doi: 10.1128/JB.00407-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hayashi T, Kitamura Y, Funa N, Ohnishi Y, Horinouchi S. Chembiochem. 2011;12:2166. doi: 10.1002/cbic.201100344. [DOI] [PubMed] [Google Scholar]
- (26).Austin MB, Saito T, Bowman ME, Haydock S, Kato A, Moore BS, Kay RR, Noel JP. Nat. Chem. Biol. 2006;2:494. doi: 10.1038/nchembio811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Song LJ, Barona-Gomez F, Corre C, Xiang LK, Udwary DW, Austin MB, Noel JP, Moore BS, Challis GL. J. Am. Chem. Soc. 2006;128:14754. doi: 10.1021/ja065247w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Aoki Y, Matsumoto D, Kawaide H, Natsume M. J. Antibiot. 2011;64:607. doi: 10.1038/ja.2011.59. [DOI] [PubMed] [Google Scholar]
- (29).Han L, Lobo S, Reynolds KA. J. Bacteriol. 1998;180:4481. doi: 10.1128/jb.180.17.4481-4486.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Petersen F, Zahner H, Metzger JW, Freund S, Hummel RP. J. Antibiot. 1993;46:1126. doi: 10.7164/antibiotics.46.1126. [DOI] [PubMed] [Google Scholar]
- (31).Sugiyama Y, Oya A, Kudo T, Hirota A. J. Antibiot. 2010;63:365. doi: 10.1038/ja.2010.60. [DOI] [PubMed] [Google Scholar]
- (32).Taoka S, Padmakumar R, Lai MT, Liu HW, Banerjee R. J. Biol. Chem. 1994;269:31630. [PubMed] [Google Scholar]
- (33).Kappe T, Aigner R, Roschger P, Schnell B, Stadlbauer W. Tetrahedron. 1995;51:12923. [Google Scholar]
- (34).Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- (35).Long F, Vagin AA, Young P, Murshudov GN. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008;64:125. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Austin MB, Izumikawa M, Bowman ME, Udwary DW, Ferrer JL, Moore BS, Noel JP. J. Biol. Chem. 2004;279:45162. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- (37).Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:432. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- (38).McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2005;61:458. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- (39).Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- (40).Emsley P, Cowtan K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- (41).Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002;58:1948. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- (42).Painter J, Merritt EA. J. Appl. Crystallogr. 2006;39:109. [Google Scholar]
- (43).Quadri LEN, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Biochem. J. 1998;37:1585. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- (44).Sanchez C, Du LC, Edwards DJ, Toney MD, Shen B. Chem. Biol. 2001;8:725. doi: 10.1016/s1074-5521(01)00047-3. [DOI] [PubMed] [Google Scholar]
- (45).Li YL, Florova G, Reynolds KA. J. Bacteriol. 2005;187:3795. doi: 10.1128/JB.187.11.3795-3799.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhang YM, Rao MS, Heath RJ, Price AC, Olson AJ, Rock CO, White SW. J. Biol. Chem. 2001;276:8231. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- (47).Okamura E, Tomita T, Sawa R, Nishiyama M, Kuzuyama T. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11265. doi: 10.1073/pnas.1000532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Funa N, Ohnishi Y, Ebizuka Y, Horinouchi S. J. Biol. Chem. 2002;277:4628. doi: 10.1074/jbc.M110357200. [DOI] [PubMed] [Google Scholar]
- (49).Zhao HM, Zha WJ, Rubin-Pitel SB. J. Biol. Chem. 2006;281:32036. doi: 10.1074/jbc.M606500200. [DOI] [PubMed] [Google Scholar]
- (50).Chen HW, Tseng CC, Hubbard BK, Walsh CT. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14901. doi: 10.1073/pnas.221582098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Funabashi M, Funa N, Horinouchi S. J. Bio. Chem. 2008;283:13983. doi: 10.1074/jbc.M710461200. [DOI] [PubMed] [Google Scholar]
- (52).Funa N, Ozawa H, Hirata A, Horinouchi S. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6356. doi: 10.1073/pnas.0511227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).He X, Reynolds KA. Antimicrob. Agents Chemother. 2002;46:1310. doi: 10.1128/AAC.46.5.1310-1318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Piel J. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14002. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Zhang YM, Wu BN, Zheng J, Rock CO. J. Biol. Chem. 2003;278:52935. doi: 10.1074/jbc.M309874200. [DOI] [PubMed] [Google Scholar]
- (56).Parris KD, Lin L, Tam A, Mathew R, Hixon J, Stahl M, Fritz CC, Seehra J, Somers WS. Structure. 2000;8:883. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- (57).Rafi S, Novichenok P, Kolappan S, Zhang XJ, Stratton CF, Rawat R, Kisker C, Simmerling C, Tonge PJ. J. Biol. Chem. 2006;281:39285. doi: 10.1074/jbc.M608758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Arthur CJ, Williams C, Pottage K, Ploskon E, Findlow SC, Burston SG, Simpson TJ, Crump MP, Crosby J. ACS Chem. Biol. 2009;4:625. doi: 10.1021/cb900099e. [DOI] [PubMed] [Google Scholar]
- (59).Concepcion J, et al. Comb. Chem. High Throughput Screening. 2009;12:791. doi: 10.2174/138620709789104915. [DOI] [PubMed] [Google Scholar]
- (60).Qiu X, Janson CA, Smith WW, Head M, Lonsdale J, Konstantinidis AK. J. Mol. Biol. 2001;307:341. doi: 10.1006/jmbi.2000.4457. [DOI] [PubMed] [Google Scholar]
- (61).Alhamadsheh MM, Musayev F, Komissarov AA, Sachdeva S, Wright HT, Scarsdale N, Florova G, Reynolds KA. Chem. Biol. 2007;14:513. doi: 10.1016/j.chembiol.2007.03.013. [DOI] [PubMed] [Google Scholar]
- (62).Davis IW, Murray LW, Richardson JS, Richardson DC. Nucleic Acids Res. 2004;32:W615. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.