Abstract
Germ-line variants in the 3′ untranslated region (3′UTR) of cancer genes disrupting microRNA (miRNA) regulation have recently been associated with cancer risk. A variant in the 3′UTR of the KRAS oncogene, referred to as the KRAS-variant, is associated with both cancer risk and altered tumor biology. Here we test the hypothesis that the KRAS-variant can act as a biomarker of outcome in epithelial ovarian cancer (EOC), and investigate the cause of altered outcome in KRAS-variant positive EOC patients. As this variant appears to be associated with tumor biology, we additionally test the hypothesis that this variant can be directly targeted to impact cell survival.
EOC patients with complete clinical data were genotyped for the KRAS-variant and analyzed for outcome (n=536), response to neoadjuvant chemotherapy (n=125), and platinum resistance (n=306). Outcome was separately analyzed for women with known BRCA mutations (n=79). Gene expression was analyzed on a subset of tumors with available tissue. Cell lines were employed to confirm altered sensitivity to chemotherapy with the KRAS-variant. The KRAS-variant was directly targeted through siRNA/miRNA oligonucleotides in cell lines and survival was measured.
Post-menopausal EOC patients with the KRAS-variant were significantly more likely to die of ovarian cancer by multivariate analysis (HR=1.67, 95% CI=1.09–2.57, p=0.019, n=279). Possibly explaining this finding, EOC patients with the KRAS-variant were significantly more likely to be platinum resistant (OR=3.18, CI=1.31–7.72, p=0.0106, n=291). Additionally, direct targeting of the KRAS-variant led to a significant reduction in EOC cell growth and survival in vitro.
These findings confirm the importance of the KRAS-variant in EOC, and indicate that the KRAS-variant is a biomarker of poor outcome in EOC likely due to platinum resistance. In addition, this work supports the hypothesis that these tumors have continued dependence on such 3′UTR lesions, and that direct targeting may be a viable future treatment approach.
Keywords: Platinum resistance, KRAS-variant, ovarian cancer, outcome
Introduction
Epithelial ovarian cancer (EOC) is the second most common female pelvic reproductive organ cancer in the United States, and carries the highest mortality in this category in the Western world. It is the fifth overall leading cause of cancer death in females in the United States, with 13,850 women dying from this disease yearly1. Despite multiple new approaches to treatment, the high rates of death from EOC have remained largely unchanged for many years, with a 5-year overall survival of only 30% to 39%2.
The standard chemotherapy regimen to treat EOC currently used is carboplatin and paclitaxel3, based on prospective randomized trials4–6. While some patients are found ultimately to be resistant to platinum-based chemotherapy (referred to as “platinum resistant”), developing recurrence within 6 months of treatment, it is initially given to all EOC patients. An improved understanding of the fundamental biological differences in EOC tumors that could explain platinum resistance amongst EOC patients would allow a more rational selection of treatments2,3,4.
miRNAs are a class of 22-nucleotide noncoding RNAs that are aberrantly expressed in virtually all cancer types, where they can function as a novel class of oncogenes or tumor suppressors5. In EOC, in addition to distinguishing normal ovarian tissue from malignant ovarian tissue6,7, miRNA expression patterns have been shown to be important in EOC pathogenesis8,9, and are associated with altered EOC patient outcome10 and response to treatment11. MiRNA expression differences have also been associated with chemotherapy and platinum resistance in EOC10,13.
Additional insight into the importance of miRNAs in cancer has come from the discovery of inherited single nucleotide polymorphisms (SNPs) that disrupt miRNA coding sequences12 and miRNA binding sites in the 3′ untranslated regions (3′UTR) of oncogenes12,13. An example of such a functional variant is rs61764370, referred to as the KRAS-variant, which is located in the KRAS 3′UTR in a let-7 miRNA complementary site (LCS). This variant has been reported to be a genetic marker of risk for lung cancer12, ovarian cancer (especially for women from hereditary breast and ovarian cancer (HBOC) families14) and triple negative breast cancer15. In addition, this variant may have an association with BRCA1, being enriched in both ovarian cancer patients from BRCA1 HBOC families14 and breast cancer patients who carry pathogenic BRCA1 mutations16. The KRAS-variant has also been shown to be associated with altered miRNA and gene expression in tumors12,17, to act as a biomarker of poor outcome in head and neck cancer17 and to be a biomarker of resistance to targeted chemotherapy in colon cancer18. These findings suggest a continued functional role of the KRAS-variant in tumors, and an association with aggressive tumor biology and poor cancer specific outcome.
Here, we evaluate the potential of the KRAS-variant to act as a biomarker of outcome in EOC in both the presence and absence of deleterious BRCA mutations. We also investigate the potential cause of altered outcome in KRAS-variant EOC patients by studying the response to neoadjuvant platinum-based chemotherapy, assessing platinum resistance, and evaluating EOC tumor gene expression. Finally, we test the hypothesis that directly targeting this gain-of-function KRAS-variant could reduce cell growth and survival in EOC cell lines with this lesion.
Results
Overall survival in EOC patients with the KRAS-variant
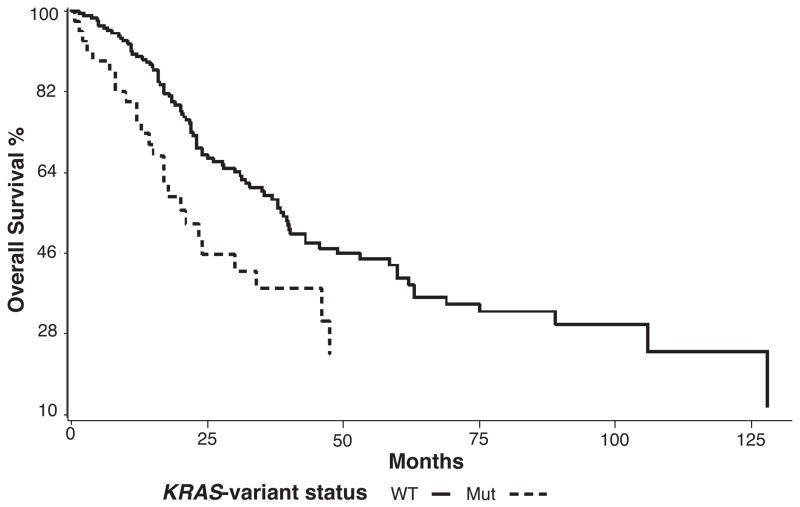
We evaluated the association of the KRAS-variant with overall survival in 454 EOC patients either tested and negative or untested for deleterious BRCA mutations. For the entire cohort the KRAS-variant did not predict worse survival by Kaplan-Meier analysis. Yet based on prior evidence that the KRAS-variant is most strongly associated with post-menopausal ovarian cancer19, we evaluated survival in women over 52 years of age (n=279), an age considered to be an appropriate surrogate for menopausal status. By Kaplan-Meier analysis, survival was significantly reduced in post-menopausal KRAS-variant EOC patients (n=59) compared to non-variant EOC patients (n=220, Figure 1, log rank p = 0.0399, non-KRAS-variant survival median 60 months, KRAS-variant survival median 34 months). When other variables including age, stage, grade, histology and treatment center were included with KRAS-variant status in a multivariate Cox proportional hazards regression model, the KRAS-variant was a statistically significant predictor of reduced overall survival for post-menopausal women with EOC (Table 1); the HR for the KRAS-variant was 1.67 (95% CI: 1.09–2.57, p = 0.019).
Figure 1. The KRAS-variant predicts significantly worse overall survival for post-menopausal ovarian cancer patients over 52 years of age.
Overall survival for ovarian cancer patients with (n=59) and without (n=220) the KRAS-variant are compared using Kaplan Meier analysis. Outcome is significantly worse for KRAS-variant positive EOC patients over 52 years of age by log-rank test (p = 0.0399).
Table 1.
The KRAS-variant is Associated with Reduced Survival in Post-menopausal (>52 years of age) Ovarian Cancer Patients (n = 279)
| Variable | HR | 95% CI | P value |
|---|---|---|---|
| KRAS status | 1.671 | 1.087 – 2.568 | 0.0192 |
| Age | 1.025 | 1.002 – 1.049 | 0.0307 |
| Stage | 1.380 | 1.185 – 1.607 | <0.0001 |
| Grade | 1.341 | 0.912 – 1.972 | 0.1360 |
| Histology | 0.970 | 0.900 – 1.045 | 0.4168 |
| Center ( Non-Yale vs. Yale) | 1.868 | 1.438 – 2.427 | <0.0001 |
HR: hazards ratio obtained from Cox proportional Hazards multivariate analysis
CI: confidence interval
Studies Included: Yale New Haven Hospital, Italy #1, Italy #2
We next evaluated the association of the KRAS-variant with survival in a separate cohort of EOC patients carrying deleterious BRCA1 or BRCA2 mutations (n=79). EOC patients carrying BRCA mutations were significantly younger than the EOC patients without BRCA mutations (52.7 vs 60.8 years of age, p<0.0001). In addition, EOC patients with BRCA mutations had a significantly longer median survival by multivariate analysis controlling for age, stage, grade and histology, than EOC patients without BRCA mutations (120 months vs 52 months, p = 0.0036). There was not a significant difference in survival between EOC patients with BRCA mutations with or without the KRAS-variant in a multivariate analysis using a multivariate Cox proportional hazards regression model (Supplemental Table 1, KRAS-variant HR = 0.75, 95% CI: 0.21–2.72, p = 0.66). There were too few patients to evaluate the impact of the KRAS-variant on survival in post-menopausal EOC patients harboring deleterious BRCA mutations.
The KRAS-variant and platinum resistance
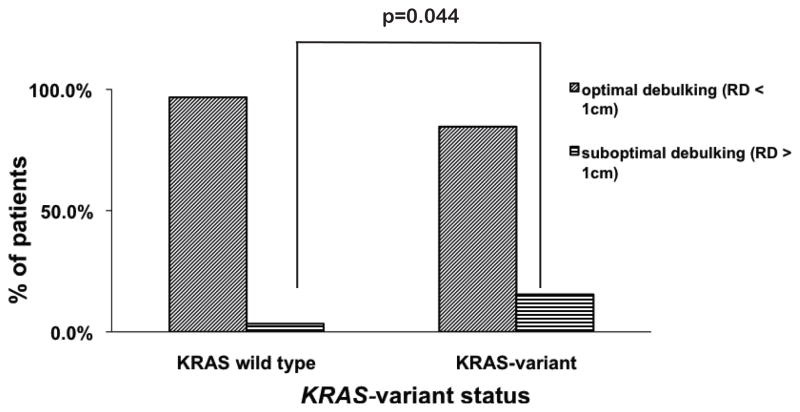
To gain insight into potential reasons for the reduced survival in post-menopausal KRAS-variant positive EOC patients, we evaluated the association of KRAS-variant positivity with response to platinum-based chemotherapy, the standard first-line chemotherapy in the treatment of EOC. We first evaluated all women with EOC who were treated at Yale-New Haven Hospital with neoadjuvant platinum-containing chemotherapy followed by surgical cytoreduction (n = 116), and used residual disease after surgery (cytoreduction) as a surrogate marker of patient response to chemotherapy. We found that 15.4% of KRAS-variant patients (n=26) were suboptimally cytoreduced (>1cm of residual disease after surgery), compared with only 3.33% of non-variant patients (n=90) (Figure 2, p=0.044). The KRAS-variant was also significantly associated with suboptimal cytoreduction after neoadjuvant chemotherapy and surgery in a multivariate logistic regression model controlling for age, stage, grade and histology (Supplemental Table 2, OR = 9.36, 95% CI: 1.34 – 65.22, p = 0.024).
Figure 2. The KRAS-variant is associated with suboptimal debulking after neoadjuvant chemotherapy.
Surgical debulking after neoadjuvant chemotherapy is compared in ovarian cancer patients (n=116) with the KRAS-variant (n=26) or without (n=90). By chi-squared analysis the KRAS-variant patients are significantly more likely to be suboptimally debulked with greater residual disease (RD) than non-variant patients (p=0.044).
To further investigate if the cause of poor response to neoadjuvant platinum-based chemotherapy seen in KRAS-variant EOC patients was due to resistance to platinum chemotherapy, we evaluated platinum resistance in all EOC patients treated adjuvantly with platinum chemotherapy without documented BRCA mutations with available response data (n=291). We found that platinum resistance (disease recurrence within 6 months of receiving platinum-based chemotherapy) was significantly more likely in KRAS-variant positive EOC patients than in non-KRAS-variant EOC patients (16.67% vs 7.56%, p = 0.034). The KRAS-variant was a statistically significant predictor for platinum resistance for EOC patients of all ages in a multivariate logistic regression analysis controlling for residual disease remaining after cytoreductive surgery, stage, histology, age and grade (Table 2, OR = 3.18, 95% CI: 1.31 – 7.72, p = 0.0106).
Table 2.
The KRAS-variant is Associated with Platinum resistance
| KRAS-variant Genotype | Univariate | Multivariate3 | ||||
|---|---|---|---|---|---|---|
| OR1 | 95% CI2 | p | OR | 95% CI | p | |
| All | ||||||
| Non-variant (n=225) | 1.00 | 1.00 | ||||
| Variant (n=66) | 2.45 | 1.08 – 5.53 | 0.0313 | 3.18 | 1.31 – 7.72 | 0.0106 |
OR: odds ratio obtained from logistic regression
CI: confidence interval
Multivariate: adjusted for age, stage, grade, histology, residual disease after cytoreductive surgery, and treatment center
Studies: Yale, Italy #1, Italy #2
Gene Expression in KRAS-variant Ovarian Tumors
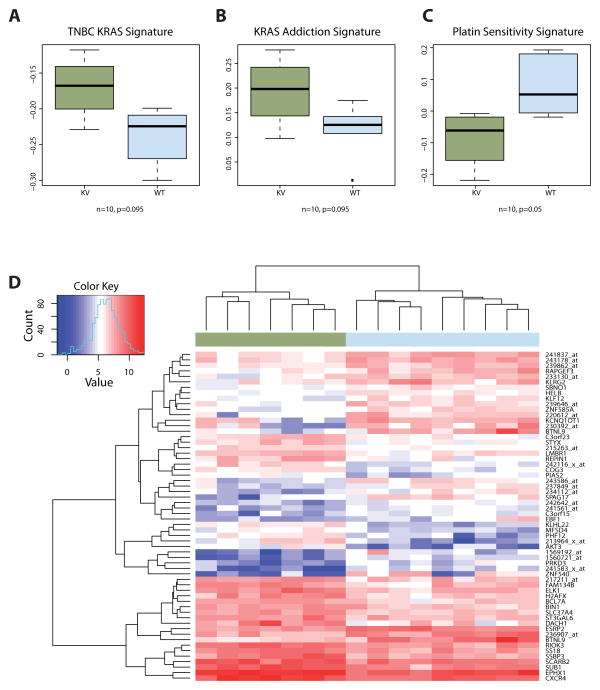
Gene expression studies were performed on a small cohort of ovarian cancer patients who had fresh frozen tissue available (Brescia cohort), and compared between seven serous EOC samples with the KRAS-variant and nine without the variant (n=16). We found that within this cohort, in post-menopausal EOC patients over 52 years of age with EOC (n=10), that a gene signature previously found associated with KRAS-variant associated triple negative breast cancer (TNBC)15 was also upregulated in KRAS-variant associated EOC (Figure 3A). In addition, again similar to the prior analysis in TNBC, we found overexpression of KRAS-associated downstream pathways in EOC KRAS-variant tumors, consistent with “KRAS addiction”20 (Figure 3B).
Figure 3. Differential Gene Expression in KRAS-variant (KV) EOC Tumors.
A. A signature of 50 differentially expression gene candidates in KV triple negative breast tumors15 shows higher scores in KV EOC samples than in non-variant samples. B. Genes associated with KRAS-addicted tumors20 were used to create a corresponding signature, which is up-regulated in KV EOC tumors. C. Re-analysis of differential gene expression in carboplatin-sensitive and resistant EOC cells21 shows differential expression of the top 20 genes in KV EOC tumors. D. Top differentially expressed genes between KV (green) and non-variant (blue) tumor samples.
Using prior analyses of gene expression data identifying platinum resistant versus sensitive signatures21, we tested and found that KRAS-variant EOC samples had a lower carboplatin sensitivity signature compared to non-variant EOC samples (Figure 3C). In agreement with findings in prior studies showing that the activation of the AKT pathway was frequently involved in platinum resistance, we found that AKT3 was one of the most significantly up-regulated transcripts in KRAS-variant EOC tumors (Figure 3D).
Although miRNA expression data was not available on tumor samples, we did compare expression of the let-7b miRNA that had previously been shown to be altered in KRAS-variant positive lung tumors 12 and triple negative breast tumors 15, in two cell lines with the KRAS-variant (BG-1 and IGROV1) compared to a non-variant line (CAOV3). We found that let-7b was significantly lower in the cells with the KRAS-variant (Supplemental Figure 1), consistent with our prior results, and a recent publication showing the association of low levels of this miRNA and poor ovarian cancer outcome 22.
The KRAS-variant and chemosensitivity in ovarian cancer cell lines
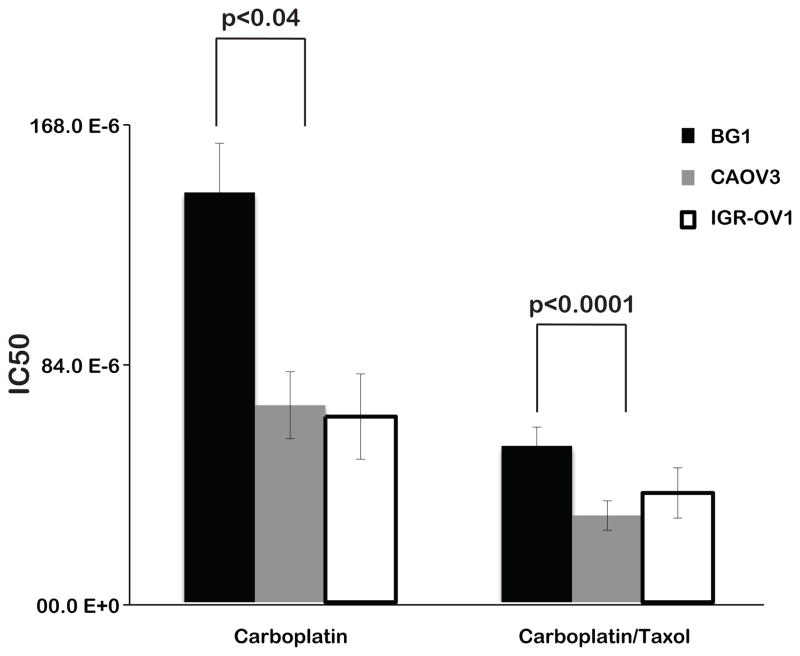
To confirm altered chemosensitivity in the presence of the KRAS-variant, we utilized EOC cell lines with and without the KRAS-variant and tested their sensitivity to different chemotherapeutic agents. We tested a cell line that is KRAS-variant positive/BRCA wild-type (BG1), a non-variant/BRCA wild-type cell line (CAOV3), and a cell line KRAS-variant positive/BRCA1 mutant (IGR-OV1). We found that the KRAS-variant line, BG1, was significantly resistant to carboplatin (p<0.04) and carboplatin/paclitaxel combination chemotherapy (p<0.0001) compared to CAOV3, the cell line without the KRAS-variant. In contrast, IGROV1, the cell line with the KRAS-variant and a deleterious BRCA1 mutation, was not resistant to these agents compared to CAOV3 (Figure 4). These findings are in agreement with our clinical findings that the KRAS-variant is associated with platinum resistance, but perhaps not in the presence of deleterious BRCA mutations.
Figure 4. The KRAS-variant is associated with resistance to Carboplatin and Carboplatin/Taxol chemotherapy in cell lines.
Cell lines with the KRAS-variant (BG1) and without the KRAS-variant (CAOV3) were treated with chemotherapy and half maximal inhibitory concentration (IC50) is shown on the Y-axis, and chemotherapeutic agent on the X-axis. Higher IC50 represents resistance to the tested chemotherapeutic agent. BG1 = KRAS-variant/BRCA wild-type cell line; CAOV3 = non-variant/BRCA wild-type cell line; IGR-OV1 = KRAS-variant/BRCA1 mutant cell line. Error bars are RSE.
We additionally evaluated agents frequently used as second-line therapy for patients who have failed carboplatin/paclitaxel chemotherapy, including doxorubicin, topotecan and gemcitabine. We found that the KRAS-variant line, BG1, was significantly resistant to each of these agents compared to CAOV3, the non-variant cell line (Table 3).
Table 3.
Chemosensitivity in a KRAS-variant cell line (BG1) versus a non-variant line (CAOV3)
| Gemcitabine | Doxorubicin | Topotecan | RSE | |
|---|---|---|---|---|
| BG1 | 30.4 10^6 | 307.5 10^9 | 161.8 10^9 | 21.69 |
| CAOV3 | 2.2 10^9 | 75.9 10^9 | 30.8 10^9 | 19.67 |
Numbers are IC50 values from a minimum of 4 separate experiments.
RSE= relative standard error which is the standard error divided by the mean and expressed as a percentage. Differences are statistically significant (p<0.01) indicating that the KRAS-variant line is more resistant to these agents.
Targeting the KRAS-variant
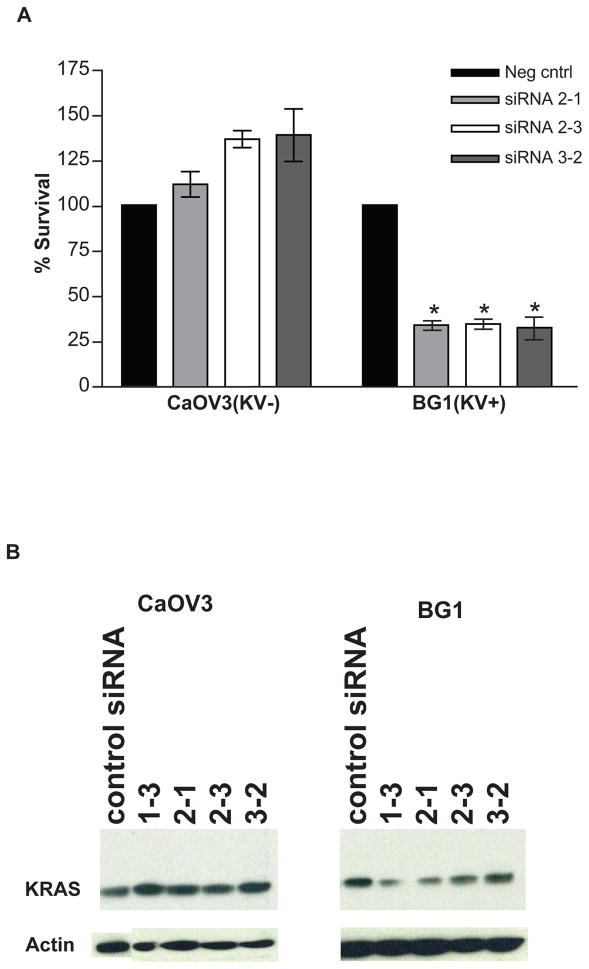
Because prior 15 and current gene expression findings suggested a continued use of KRAS signaling in KRAS-variant associated tumors, we evaluated the impact of directly targeting this 3′UTR lesion. We designed siRNA/miRNA-like complexes that could directly bind the altered allele in KRAS-variant transcripts, but not non-KRAS-variant transcripts (Supplemental Figure 2). We found that transfecting these oligonucleotide duplexes that target the KRAS-variant caused a significant decrease in cell survival in the KRAS-variant carrying BG1 cell line (p<0.001), but had no effect in CAOV3 (Figure 5A) or SKOV3 (data not shown), two non-variant EOC cell lines. This finding was concordant with a moderate decrease in KRAS protein levels by western blot in BG1, but not in CAOV3 (Figure 5B) or SKOV3 (data not shown) after treatment.
Figure 5. Targeting the KRAS-variant impacts cell survival.
Cell lines with (BG1) and without (CAOV3) the KRAS-variant were treated with siRNA/miRNA combinations that bind selectively to the variant allele. A. Decreased cell survival in the KRAS-variant line, BG1 (p<0.001), with no effect on the non-variant line, CAOV3. B. Decreased KRAS protein expression in BG1 (right) concordant with the decrease in cell survival, with no effect on CAOV3 (left). Different siRNAs are denoted by numbers.
Discussion
Our data indicate that the KRAS-variant is a biomarker of poor outcome for post-menopausal women (over 52 years of age) with epithelial ovarian cancer. The poor outcome in KRAS-variant associated ovarian cancer may be due to the association of the KRAS-variant with resistance to platinum-based chemotherapy, based on a worse response to neoadjuvant platinum-based chemotherapy, and significantly increased platinum resistance in adjuvantly treated EOC patients with the KRAS-variant.
The biological differences between KRAS-variant EOC and non-variant EOC tumors are supported by gene expression data done on available tumors, which suggests KRAS addiction and AKT-mediated platinum resistance in KRAS-variant EOC. Platinum resistance was further confirmed in vitro in an ovarian cancer cell line with the KRAS-variant as compared to a non-variant line. Finally, evidence for the continued dependence of KRAS-variant-associated EOC on this germ-line lesion was shown through direct targeting of this mutation, which led to significant growth and survival inhibition in a KRAS-variant EOC cell line versus non-variant EOC lines.
The association of the KRAS-variant with poor survival specifically for post-menopausal women could be due to underlying biology associated with this variant, or confounding effects in these studies. Supporting the hypothesis that this finding reflects underlying biology, the KRAS-variant is associated with post-menopausal ovarian cancer14 (Supplemental Table 3), with a median age of diagnosis near 59 years of age. It is known that relative survival varies by age, with older women twice as likely to die within 5 years of diagnosis of EOC, supporting the hypothesis that post-menopausal women may have biologically different tumors then younger women23. In addition, the KRAS-variant has been shown to be a biomarker of triple negative breast cancer risk specifically in pre-menopausal women, less then 52 years of age15. This may indicate that the role of the KRAS-variant in cancer risk and biology in different tissues depends on the patient’s hormonal environment, i.e. menopausal status, and that women with the KRAS-variant may be first at risk for pre-menopausal breast cancer and subsequently post-menopausal ovarian cancer. Studies are ongoing to test this hypothesis.
However, it is equally plausible that younger women diagnosed with EOC in our cohort were more likely to have undocumented deleterious BRCA mutations, as BRCA mutation carriers were significantly younger in these studies. Our findings that the KRAS-variant does not predict for poor outcome in our cohort of EOC patients with known deleterious BRCA mutations may be partially explained by the fact that BRCA mutations are associated with platinum-sensitivity, and this effect may act downstream of any resistance caused or exacerbated by the KRAS-variant to platinum agents. The younger patients in our cohorts may also have had other subtypes of ovarian cancer seen more frequently in younger women, such as borderline tumors, and were perhaps misdiagnosed. Although our data sets were extremely well clinically annotated, a weakness of our study is that BRCA status was not obtained on all of our EOC patients, and although pathology reports were available, tumor tissue was not available for re-review. This does highlight the importance of using clinically well-annotated data sets to study functional markers such as the KRAS-variant, to allow appropriate interpretation of the results. A recent study that failed to find the association of the KRAS-variant with poor outcome and resistance to therapy in EOC24 used ovarian collections used for genome wide association studies (GWAS) that had very limited clinical information, and important factors such as BRCA status and ovarian cancer specific survival were not available nor included in their analyses.
The association of the KRAS-variant with resistance to platinum-based chemotherapy for non-BRCA mutant EOC patients is perhaps not surprising, as KRAS pathway disruption has been associated with platinum resistance in ovarian cancer21,25 as well as several other cancers26,27. The KRAS-variant has been previously shown to lead to KRAS and associated downstream pathway overexpression in triple negative breast cancer15, which concords with our gene expression findings in this study in EOC, even with a small number of tumors available for study. It is interesting that similar gene mis-expression patterns were found in two different types of KRAS-variant associated tumors, suggesting that these tumors, regardless of tissue of origin, perhaps utilize similar pathways in oncogenesis. It would be important and interesting to validate these findings in larger gene expression data sets as well as additional tumor types, work that is ongoing.
Perhaps most intriguing is the finding that direct targeting of the KRAS-variant lesion in KRAS-variant associated EOC cell lines leads to significantly enhanced cell death and a reduction in KRAS levels. While there is considerable work to be done to understand the mechanisms behind these findings, they do suggesting continued critical dependence of these tumors on this single, non-coding germ-line lesion. While there has been a significant effort to tailor cancer treatment by measuring tumor gene expression and determining tumor acquired mutations, there are few, if any, germ-line variants that have previously been shown to be critical targets for therapy in cancer.
There is still work to be done to better define the mechanisms of KRAS-variant associated platinum resistance and its potential as a future target for therapy in EOC. However, this work supports the conclusions that the KRAS-variant is a functional cancer mutation that is important in ovarian cancer, and that the KRAS-variant appears to allow meaningful sub-classification of the ovarian tumors it is associated with. Hopefully these findings can ultimately be used to improve ovarian cancer patient outcome.
Materials and Methods
Overall survival analysis cohorts
Complete clinical data and DNA from women diagnosed with EOC without known BRCA mutations were included from the following three institutions under individual IRB approvals. All protocols accrued patients prospectively at the time of their diagnosis to avoid selection bias. References indicate previous detailed descriptions of these patients: 1) Turin, Italy #1 (n=197)28. 2) Brescia, Italy #2 (n=59)14. 3) Yale New Haven Hospital (n=198). The Yale patients were collected prospectively on two clinical trials at Yale Medical School of newly diagnosed EOC patients diagnosed between 2000 and 2009. Supplemental Table 3.
Documented BRCA mutant EOC cases with known outcome were collected from the following two institutions: 1) Yale New Haven Hospital (n=17). 2) City of Hope Comprehensive Cancer Center (n=62). Supplemental Table 4.
As not all stage I ovarian cancer patients receive adjuvant chemotherapy, when substage information was not available for patients with Stage 1 tumors, these patients were excluded from the analysis. Otherwise Stage 1B and 1C tumors were included with stage 2–4. To minimize inadvertent inclusion of borderline tumors, tumors with an unknown grade were excluded from this analysis. For women treated with neoadjuvant chemotherapy, the date of pathological diagnosis was considered the start date of treatment. For women treated with adjuvant chemotherapy the date of surgery was considered the start date of treatment. A total of 386 patients with wild-type BRCA or not tested for BRCA mutations and 79 patients with documented BRCA mutations fit the above described parameters and were included in the two survival analyses.
Neoadjuvant chemotherapy cohort
Women with EOC who received neoadjuvant platinum-based chemotherapy followed by cytoreductive surgery at Yale New Haven Hospital between 1996 and 2010 were identified on an IRB approved protocol (n=125). Supplemental Table 5. This cohort of patients received chemotherapy as a primary treatment due to tumor burden that was too extensive for optimal surgical debulking at presentation. Following chemotherapy, patients underwent cytoreductive surgery and additional adjuvant treatment. Only patients treated with four or more cycles of neoadjuvant platinum-containing combinations were included in this analysis (n = 116). Optimal cytoreduction was defined as residual disease measuring less than 1cm remaining after surgery, while suboptimal cytoreduction was defined as residual disease measuring greater than or equal to 1cm at the completion of surgery. Only women operated on at Yale by the same group of surgeons were included, to avoid bias in surgical skill as a factor impacting residual disease.
Patients for analysis of platinum resistance
Platinum resistance was defined as progression-free survival (PFS) of less than 6 months from the completion of platinum containing adjuvant chemotherapy to the date of recurrence. The progression-free survival interval was available from women from Italy #1, Italy #2, and Yale-New Haven Hospital patients (n = 291). Supplemental Table 6 describes the clinicopathologic parameters of these patients.
Detection of the KRAS-variant
DNA was isolated using standard methods from tumor, blood or saliva. As previously shown19, the KRAS-variant does not appear to be somatically acquired nor does it require a loss of heterozygosity, so each of these tissues are appropriate to test and the results are identical regardless of the tissue tested19. The variant allele was detected using a primer specific to the KRAS-variant and a TaqMan (Applied Biosystems) PCR assay on all samples. Genotyping was done at YNHH except for on samples from COH, for which the genotyping was done in their facility. Less than 3% of populations carry two copies of the variant12. As such, patients who carried at least one copy of the variant allele were classified as KRAS-variant carriers.
Gene expression analysis of EOC with and without the KRAS-variant
Gene expression in fresh frozen tumor samples from 16 patients (9 non-variant, 7 KRAS-variant) was profiled on the Affymetrix GeneChip Human Genome U133 Plus 2.0 platform. All samples were from high-grade serous epithelial ovarian tumors that were stage IIIC or IV. Images were processed with the MAS5 algorithm and probes that were judged absent in at least 75% of the samples were removed. Intensity values were log-transformed and quantile-normalized. Differential gene expression was assessed in samples from patients over 52 years of age (n=6 non-variant, 4 KRAS-variant) using a linear model and empirical Bayesian error moderation as implemented in the LIMMA package for R statistical software29.
Association of previously published results with the KRAS-variant in our data set was assessed using a signature approach in order to reduce cross-platform effects15. Briefly, signature scores were computed as Pearson correlation between the respective signature vector of gene contributions and each sample’s expression profile for these genes. Differences between signature scores in KRAS-variant and non-variant EOC samples were assessed using paired Kolmogorov-Smirnov test. Unless otherwise indicated, gene lists from the respective publications were used as signature vectors. Data from Peters and colleagues21 was obtained from Gene Expression Omnibus (GSE1926) and reanalyzed to generate a signature from the 50 most significantly differentially expressed genes between platinum-sensitive and resistant samples.
Chemosensitivity/cell viability assays
The activity of drugs alone or in combination was determined by a high-throughput CellTiter-Blue cell viability assay. For these assays, 1.2×103 cells were plated in each well of 384-well plates using a Precision XS liquid handling station (Bio-Tek Instruments, Inc., Winooski, VT) and allowed to attach overnight with incubation at 37°C, 5% CO2. Using the liquid handling station, all drugs were serially diluted 2:3 or 1:2 in media and 5 μl of these dilutions were added to appropriate wells at indicated times. Four replicate wells were used for each drug concentration and an additional four control wells received a diluent control without drug. At the end of the incubation period with drugs, 5 μl CellTiter-Blue reagent (Promega Corp., Madison, WI) was added to each well. Cell viability was assessed by the ability of the remaining viable cells to bioreduce resazurin to resorufin. The fluoresence of resorufin (579nm Ex/584nm Em) was measured with a Synergy 4 microplate reader (Bio-Tek Instruments, Inc.). The fluorescence data was transferred to Microsoft Excel to calculate the percent viability relative to the four replicate cell wells that did not receive drug. IC50s were determined using a sigmoidal equilibrium model regression using XLfit version 5.2 (ID Business Solutions Ltd.). The IC50 was defined as the concentration of drug required for a 50% reduction in growth/viability. All experiments were carried out a minimum of three times.
Targeting the KRAS-variant
SiRNA sequences were designed to target the KRAS-variant sequence by placing the SNP at varying positions of the 6 nucleotides at the 5′ end of the siRNA guide strand corresponding to the so-called “seed sequence”. Blast searches were performed to minimize cross-reactivity. In some of the siRNA sequences DNA nucleotides were introduced in order to optimize thermoenergetic features for preferred incorporation of the guide strand into the argonaute effector complex or to increase specificity for the variant. SiRNA guide strand sequences used in the experiments are as follows (lower case = RNA, upper case = DNA; GS = guide strand, PS = passenger strand):
2-1 GS ugcaucacuugaggucaggag
2-1 PS ccugaccucaagugaugcacc
2-3 GS TGCATCACuugaggucaggag (passenger strand same as 2-1)
3-2 GS ucaucacuugaggucaggagu
3-2 PS uccugaccucaagugaugcac
The negative control used was purchased from Qiagen (AllStars Negative Control siRNA). Knockdown efficiency and specificity to the KRAS-variant of these sequences were confirmed using a dual luciferase assay (data not shown). Oligonucleotide combinations were annealed using standard conditions and then transfected into cells using standard protocols. Cell survival was assayed using MTT assays and experiments were done in quadruplicate, and repeated in four independent experiments for all lines. Cell lysates were collected 72 hours after transfection and KRAS protein levels measured by Western analysis using a probe specific to KRAS as previously described19.
Statistics
To assess the significance of demographic variables, a χ2 test or a two-sided Fisher’s exact test was used for categorical variables. A t test was used for continuous variables, such as age.
The overall survival time of KRAS-variant and wild-type patients was compared using the Kaplan-Meier method30, and the statistical significance of the survival curves was determined by the log-rank test31. A Cox proportional hazards regression model32 was used to assess the impact of the KRAS-variant and demographic and prognostic variables (age, stage, grade, and histology) on overall survival.
Multivariate logistic regression analyses33 were used to determine the impact of the KRAS-variant and other demographic and prognostic factors on the probability of suboptimal cytoreduction. Multivariate logistic regression analyses33 were used to assess the association of the KRAS-variant and other prognostic factors on the probability of platinum resistance.
All statistical analyses were performed using SAS 9.1.3 (SAS Institute Inc., Cary, NC) and in R 2.12.1 (R Foundation for Statistical Computing).
Supplementary Material
Acknowledgments
Thanks to JL and his group for supplying the cell line associated chemosensitivity data. Appreciation to Greg Wilhoite for performing the taqman assay and Josef Herzog for processing samples for the City of Hope. Appreciation to Discovery to Cure at Yale University for samples. Appreciation to Jeff Boyd for supplying the BG1 cell line for targeting studies. FS and JW were supported by a Mary Kay Foundation Grant, and an R01 from the NCI (CA131301-01A1). JW was supported by a K08 grant [CA124484]. AS was supported by R01CA122728. SLN is partially supported by the Morris and Horowitz Families Endowed Professorship and NIH R01CA74415. JNW and SS are supported by RC4CA153828 from the National Cancer Institute and the Office of the Director, National Institutes of Health, and also by funding from the Markel Foundation. Appreciation to the Shannon Family Foundation Trust, the Segesta family, and donations made in memory of Jennifer DeGroff.
Footnotes
Conflict of Interest
JW and FS are co-founders of a company that has licensed IP regarding the KRAS-variant from Yale University. They both own stock in this company.
References
- 1.Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Parmar M, et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet. 2003;361:2099–2106. doi: 10.1016/s0140-6736(03)13718-x. [DOI] [PubMed] [Google Scholar]
- 3.Pfisterer J, et al. Gemcitabine plus carboplatin compared with carboplatin in patients with platinum-sensitive recurrent ovarian cancer: an intergroup trial of the AGO-OVAR, the NCIC CTG, and the EORTC GCG. J Clin Oncol. 2006;24:4699–4707. doi: 10.1200/JCO.2006.06.0913. [DOI] [PubMed] [Google Scholar]
- 4.Herzog T, Pothuri B. Ovarian cancer: a focus on management of recurrent disease. Nat Clin Pract Oncol. 2006;3:604–611. doi: 10.1038/ncponc0637. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack F. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Iorio M, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mezzanzanica D, Bagnoli M, De Cecco L, Valeri B, Canevari S. Role of microRNAs in ovarian cancer pathogenesis and potential clinical implications. The International Journal of Biochemistry & Cell Biology. 2010;42:1262–1272. doi: 10.1016/j.biocel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 9.van Jaarsveld M, Helleman J, Berns E, Wiemer E. MicroRNAs in ovarian cancer biology and therapy resistance. Int J Biochem Cell Biol. 2010;42:1282–1290. doi: 10.1016/j.biocel.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Eitan R, et al. Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecologic Oncology. 2009;114:253–259. doi: 10.1016/j.ygyno.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 11.Lu L, et al. MicroRNA let-7a: A potential marker for selection of paclitaxel in ovarian cancer management. Gynecological Oncology. 2011 doi: 10.1016/j.ygyno.2011.04.033. in press. [DOI] [PubMed] [Google Scholar]
- 12.Chin L, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen K, et al. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306–1311. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 14.Ratner E, et al. A KRAS-variant in Ovarian Cancer Acts as a Genetic Marker of Cancer Risk. Cancer Research. 2010;15:6509–6515. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paranjape T, et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncology. 2011 doi: 10.1016/S1470-2045(11)70044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollestelle A, et al. Prevalence of the variant allele rs61764370 T>G in the 3′UTR of KRAS among Dutch BRCA1, BRCA2 and non-BRCA1/BRCA2 breast cancer famlies. Breast Cancer Res Treat. Jul 30; doi: 10.1007/s10549-010-1080-z. [Epub ahead of print](2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen B, et al. A let-7 microRNA binding site polymorphism in the KRAS 3′UTR is associatied with reduced survival in oral cancers. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp099. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graziano F, et al. Genetic modulation of the let-7 microRNA binding to KRAS 3′-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan. The Pharmacogenomics Journal. doi: 10.1038/tpj.2010.9. Epub ahead of print, 1–7 (2010) [DOI] [PubMed] [Google Scholar]
- 19.Chin L, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters D, Freund J, Ochs R. Genome-wide trascriptional analysis of carboplatin response in chemosensitive and chemoresistant ovarian cancer cells. Mol Cancer Ther. 2005;4:1605–1616. doi: 10.1158/1535-7163.MCT-04-0311. [DOI] [PubMed] [Google Scholar]
- 22.Helland A, et al. Deregulation of MYCN, LIN28B and LET7 in a Molecular Subtype of Aggressive High-Grade Serous Ovarian Cancers. PLoS One. 6 doi: 10.1371/journal.pone.0018064. epub before print (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ACS. Cancer Facts & Figures; Society, A.C. Cancer Facts & Figures. 2010. pp. 1–56. [Google Scholar]
- 24.Pharoah P, Palmieri R, Ramus S, et al. The role of KRAS rs61764370 in invasive epithelial ovarian cancer: implications for clinical testing. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-10-3405. online March 8th(2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, et al. Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer. BMC Medical Genomics. 2009;2:1755–1758. doi: 10.1186/1755-8794-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youn C, et al. Oncogenic H-Ras up-regulates expression of ERCC1 to protect cells from platinum-based anticancer agents. Cancer Res. 2011;64:4849–4857. doi: 10.1158/0008-5472.CAN-04-0348. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki H, et al. Evaluation of Kras gene mutation and copy number gain in non-small cell lung cancer. J Thorac Oncol. 2011;6:15–20. doi: 10.1097/JTO.0b013e31820594f0. [DOI] [PubMed] [Google Scholar]
- 28.Lu L, Katsaros D, Rigault de la Longrais I, Sochirca O, Yu H. Hypermethylation of let-7a-3 in Epithelial Ovarian Cancer Is Associated with Low Insulin-like Growth Factor-II Expression and Favorable Prognosis. Cancer Research. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- 29.Smyth G Limma. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. Gentleman R, Dudoit V, Irizarry SR, Huber W, editors. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 30.Kaplan E, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 31.Mantel N. Evaluation of survival data and two new rand order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- 32.Cox D. Regression models and life tables. J R Stat Soc. 1972;34:187–220. [Google Scholar]
- 33.Cox D. The Analysis of Binary Data. Methuen; London: 1970. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.