Abstract
Membrane type-1 matrix metalloproteinase (MT1-MMP) is a promising drug target in malignancy. The structure of MT1-MMP includes the hemopexin domain (PEX) that is distinct from and additional to the catalytic domain. Current MMP inhibitors target the conserved active site in the catalytic domain and, as a result, repress the proteolytic activity of multiple MMPs instead of MT1-MMP alone. In our search for non-catalytic inhibitors of MT1-MMP, we compared the pro-tumorigenic activity of wild-type MT1-MMP with a MT1-MMP mutant lacking PEX (ΔPEX). In contrast to MT1-MMP, ΔPEX did not support tumor growth in vivo, and its expression resulted in small fibrotic tumors that contained increased levels of collagen. Because these findings suggested an important role for PEX in tumor growth, we performed an inhibitor screen to identify small molecules targeting the PEX domain of MT1-MMP. Using the Developmental Therapeutics Program (NCI/NIH) virtual ligand screening compound library as a source and the X-ray crystal structure of PEX as a target, we identified and validated a novel PEX inhibitor. Low dosage, intratumoral injections of PEX inhibitor repressed tumor growth and caused a fibrotic, ΔPEX-like tumor phenotype in vivo. Together, our findings provide a preclinical proof-of-principle rationale for the development of novel and selective MT1-MMP inhibitors that specifically target the PEX domain.
Keywords: MT1-MMP, hemopexin domain, small molecule, tumor growth, migration, type I collagen
Introduction
Membrane type-1 matrix metalloproteinase (MT1-MMP) is an archetype of membrane-type MMPs (1). MT1-MMP expression is associated with multiple pathophysiological conditions, including tumor cell invasion (2–6). In migrating cells, MT1-MMP accumulates at the leading and trailing edges to contribute efficiently to matrix cleavage and cell locomotion (7, 8). MT1-MMP is regulated both as a proteinase and as a membrane protein by coordinated mechanisms including activation of the MT1-MMP proenzyme, inhibition by TIMPs, self-proteolytic inactivation, homodimerization, trafficking throughout the cell to the plasma membrane, the internalization into the transient compartment inside the cells and recycling back to the plasma membrane [reviewed in (9, 10). These events stimulate intracellular cytoskeleton rearrangements and set in motion cellular invasion machinery.
There is a consensus among researchers that invasion-promoting MT1-MMP plays the pivotal and multiple roles in tumor progression and metastasis, especially in cell invasion and directional cell motility (6). The MT1-MMP multi-domain structure includes the N-terminal prodomain, the catalytic domain (CAT), the hinge linker, the hemopexin domain (PEX), the transmembrane and the C-terminal cytoplasmic domains (11). MT1-MMP is synthesized as a latent zymogen that requires proteolytic processing of the N-terminal inhibitory prodomain to generate the mature enzyme (12, 13).
CAT is the most important domain for the pro-tumorigenic MT1-MMP function (14–16). PEX, however, is also crucial for many specific MMP functions (17–19). In MMPs, including MT1-MMP, the PEX structure is a four-bladed β-propeller (20). PEX is essential for the association, the lateral diffusion and the proteolysis of the underlying collagen substratum by MT1-MMP (19, 21). PEX is directly involved in MT1-MMP homodimerization and interactions with CD44, TIMP-2 and MMP-2 (22–24). According to the results by others, blade IV of PEX is necessary for MT1-MMP homodimerization while blade I is required for heterodimerization with CD44 (20, 25). In the symmetrical MT1-MMP homodimer, blades II and III of the first monomer seems to interact with blades III and II of the second monomer while in the asymmetrical homodimer the contacts involve blades I/II and I/II/III/IV of the monomers (20). The requirement of PEX in the MMP-2 activation process also remains insufficiently understood. In a few studies MT1-MMP lacking PEX was capable of accomplishing MMP-2 activation (26, 27). Other studies suggested that homodimerization and MMP-2 activation were interconnected (20, 28, 29).
To shed more light on PEX’s function, we compared the pro-invasive, pro-tumorigenic capacity of the wild-type MT1-MMP and MT1-MMP mutant lacking PEX. Because our results pointed to an important, albeit underestimated, role of PEX in the MT1-MMP-dependent tumorigenesis, we also identified a small-molecule, PEX-targeting inhibitor. This inhibitor selectively targeted PEX and, as a result, repressed the pro-tumorigenic function of MT1-MMP.
Materials and Methods
General reagents and antibodies
Reagents were purchased from Sigma–Aldrich unless indicated otherwise. Murine monoclonal and rabbit polyclonal MT1-MMP antibodies (3G4 and Ab8345, respectively) and a hydroxamate inhibitor (GM6001) were from Chemicon. Rat tail type I collagen (COL-I) was from BD Biosciences. Rabbit polyclonal antibodies to Ki-67 and COL-I were from Thermo Scientific and Novus Biologicals, respectively. A murine monoclonal V5 antibody was from Invitrogen.
Cells
All of the original cell lines were obtained from ATCC in 2011 (Manassas, VA). Human breast carcinoma MCF-7 and MDA-MB-231 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM)-10% FBS. Human mammary epithelial 184B5 cells were maintained in mammary epithelial cell growth medium (MEGM; Invitrogen). 184B5 cells stably transfected with the original pLenti6/V5-D-TOPO lentiviral vector (184B5 cells) or the lentiviral vector encoding the MT1-MMP C-terminally tagged with a V5 tag (185B5-MT cells) were constructed earlier (30). MCF-7 cells stably expressing the wild type MT1-MMP (MCF7-MT cells), MT1-MMP C-terminally tagged with a V5 tag (MCF7-MT-V5 cells) or the β3 integrin subunit (MCF7-β3/zeo cells) alone or co-expressing the latter with MT1-MMP (MCF7-β3/MT cells) or the MT1-MMP mutant lacking the 319–508 PEX sequence (MCF7-β3/ΔPEX cells) were obtained earlier (12, 27, 30–32).
Cleavage of fluorescent peptide
CAT was expressed in E. coli, purified from the inclusion bodies and refolded to restore its native conformation (33). The activity assay was performed at 37°C in triplicate in wells of a 96-well plate in 0.2 ml 50 mM HEPES, pH 7.5 containing 1 mM CaCl2, 50 μM ZnCl2, 20% glycerol, the (7-methoxycoumarin-4-yl)acetyl-Pro-Leu-Gly-Leu-(3-[2,4-dinitrophenyl]-L-2,3-diaminopropionyl)-Ala-Arg-NH2 fluorescent substrate (10 μM; Bachem) and CAT (10 nM) (34). Reaction velocity was monitored at λex=320 nm and λem=400 nm. The increasing concentrations of the compounds were co-incubated for 30 min with CAT at ambient temperature. The residual activity of CAT was measured. IC50 values were calculated by determining the compound concentrations which inhibited the cleavage activity by 50%.
Cell viability assays
Assays were performed in wells of a 96-well flat bottom, white-wall plates. 184B5-MT and MCF7-β3/MT cells (5×104) were grown for 16 h in MEGM-10% FBS and DMEM-10% FBS, respectively. 184B5-MT cells were replenished with fresh MEGM (0.1 ml/well) and incubated for an additional 24 h in the presence of the compounds (100 μM) or vehicle (1% dimethyl sulfoxide; DMSO). MCF7-β3/MT cells were replenished with fresh DMEM-10% FBS (0.1 ml/well) and incubated for an additional 6 h in the presence of the compounds (400 μM) or vehicle (2% DMSO). The viable cells were counted using a luminescent ATP-Lite assay (PerkinElmer). Each datum point represented the results of at least three independent experiments performed in triplicate.
Migration assays
Assays were performed in wells of a 24-well, 8 μm pore size Transwell plate (Corning Costar). A 6.5 mm insert membrane was coated with 0.1 ml COL-I (300 μg/ml in MEGM) and then air dried for 16 h. The collagen coating was rehydrated for 1 h in 0.2 ml MEGM. The inner chamber contained MEGM-10% FBS as a chemoattractant. The compounds (10–100 μM) or DMSO (0.1–1%) were added to both inner and outer chambers. Prior to plating in the outer chamber, cells (5×104) were co-incubated for 20 min with the compounds or DMSO in MEGM. Cells were allowed to migrate for 16–18 h. The cells remaining on the top surface of the membrane were removed with a cotton swab. The cells on the bottom surface of the membrane were fixed and stained (0.2% Crystal Violet). The incorporated dye was extracted using 1% SDS and the A570 was measured. Data are means ±SE from three individual experiments performed in triplicate.
Adhesion assays
Assays were performed in wells of a 96-well clear-bottom well plate. Wells were coated for 16–18 h at 4°C using 150 μl COL-I, Matrigel and BSA solutions (50 μg/ml, 100 μg/ml and 1%, respectively, in PBS). Wells were blocked for 1 h using 1% BSA. Cells were co-incubated for 15 min at 37°C with the compounds (25–100 μM). Cells (1×105/0.1 ml) were then allowed to adhere to the wells for 1 h at 37°C. Unattached cells were removed by washings. Attached cells were fixed and stained (0.2% Crystal Violet). The incorporated dye was extracted using 1% SDS and the A570 was measured. Data are means ±SE from three individual experiments performed in triplicate.
Collagen degradation assays
Assays were performed in wells of a 24-well culture plates. Wells were coated for 1 h at 37°C with neutralized, chilled rat tail COL-I (0.2 mg/ml, 0.25 ml in DMEM). Cells were co-incubated for 20 min at 37°C with the compounds (100 μM) or 1% DMSO. Cells (2.5×104/well) were then seeded on COL-I and cultured for 3 days. Cells were detached using 2 mM EDTA. Collagen was fixed using 4% paraformaldehyde, stained with Coomassie Blue and the images were captured using a Nikon TE-2000 microscope with a 20× objective and a CCD camera.
Pull-down and Western blotting
Cells grown in wells of a 12-well plate were lysed using 50 mM N-octyl-β-D-glucopyranoside in TBS supplemented with 1 mM phenylmethylsulphonyl fluoride, 10 mM EDTA and a protease inhibitor cocktail set III (Buffer A). Insoluble material was removed by centrifugation (14,000×g; 15 min). The supernatant aliquots (4 μg total protein) were analyzed by Western blotting with the MT1-MMP, FLAG, V5 and actin antibodies followed by the secondary horseradish peroxidase-conjugated antibody (Jackson ImmunoResearch) and a SuperSignal West Dura Extended Duration Substrate kit (Pierce). Where indicated, GM6001 (50 μM) was added to the cells 24 h prior to cell lysis.
For pull-down experiments, MCF7-MT and MCF7-MT-V5 cells were transiently transfected with the pcDNA3.1-zeo plasmid encoding the FLAG-tagged MT1-MMP construct (32). Cells were co-incubated for 24 h with the compounds (100 μM) or DMSO (1%). In 48 h post-transfection, the cells were lysed using Buffer A. Insoluble material was removed by centrifugation (14,000×g; 15 min). The supernatant aliquots (200 μg total protein) were co-incubated for 16 h with the FLAG M2 antibody-beads (10 μl; 50% slurry). The beads were collected by centrifugation and washed in Buffer A. The FLAG construct was eluted from the beads using the FLAG peptide (20 μl; 150 μg/ml; 1 h). The beads were removed by centrifugation. The supernatant samples were then analyzed by Western blotting with the V5 antibody.
Gelatin zymography
Cells (2×105) were grown for 24 h in DMEM-10% FBS in wells of a 24-well plate. The medium was replaced with fresh DMEM (0.2 ml/well) and cells were incubated for 16–18 h. Where indicated, the medium was supplemented with the MMP-2 proenzyme (0.5 nM). In 24 h, medium aliquots (20 μl) were analyzed by gelatin zymography.
To analyze the status of MMP-2 in tumor xenografts, snap-frozen tumors (~0.1 mg tissue) were extracted in 1 ml 20 mM Tris-HCl, pH 7.4, supplemented with 150 mM NaCl, 1% deoxycholate, 1% IGEPAL, a protease inhibitor cocktail set III, 1 mM phenylmethylsulfonyl fluoride and 10 mM EDTA. The pellet was removed by centrifugation (14,000×g; 30 min). The protein concentration was then equalized among the samples (1 mg/ml). To determine the status of MMP-2, the samples (5 μg total protein) were analyzed by gelatin zymography.
Tumor xenografts
On day 1, MCF7-β3/WT and MCF7-β3/ΔPEX cells (4×105) were injected subcutaneously in 0.1 ml DMEM containing 3 mg/ml Matrigel into the mammary tissue of athymic BALB/c nu/nu 4-week-old female mice (Charles River). Because young, 4-week, female mice produce high levels of estrogen, no additional estrogen supplementation is required to support the growth of estrogen-dependent MCF7 cells. Tumors were measured weekly by caliper measurements of two perpendicular diameters of xenografts (D1 and D2). Tumor volume was calculated using the formula: V = π/6 (D1 × D2)3/2. Starting on day 29, mice received intratumoral injections of compound 9 (0.5 mg/kg; three times/week). Control animals received vehicle injections (2% DMSO). On day 46, animals were sacrificed according to the NIH guidelines. Tumors were excised and either freeze-molded in Optimum Cutting Temperature (OCT) compound (Sakura Finetek) or embedded in paraffin. Tumor sections were stained with hematoxylin-eosin (H&E) and with the antibodies to MT1-MMP, Ki-67 and COL-I followed by the secondary antibody conjugated with red Alexa Fluor 594 (Molecular Probes). Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). The fluorescence images were acquired using an Olympus BX51 fluorescence microscope equipped with a MagnaFire camera.
RT-PCR
To measure the expression levels of human MT1-MMP, Ki-67 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; loading control), total RNA was extracted from cultured cells and tumors using TRIzol reagent and additionally purified using the RNeasy columns (Qiagen). The RNA purity was estimated by measuring the A260/280 and the A260/230 ratios. The integrity of the RNA samples was validated using an Experion automated electrophoresis system (Bio-Rad). The RNA templates (50 ng) were used in the 25-μl RT-PCR reactions supplemented with the corresponding forward and reverse primers (0.6 μM each) using the OneStep RT-PCR system (Qiagen) (12). After the completion of the first strand synthesis, RT-PCR was performed for 35 cycles (denaturation at 95°C, annealing at 55°C and elongation at 72°C; 30, 30 and 60 sec, respectively). The amplified products were separated using agarose gel electrophoresis.
Protein-ligand docking
The protein-ligand docking simulations were performed using the MT1-MMP crystal structure coordinates from PDB 3C7X as a target and a ~275,000 compound library of the Developmental Therapeutics Program (DTP) NCI/NIH as a source of small-molecule ligands. The docking into the druggable pocket was performed using Q-MOL modeling package (35). The PDB 3C7X molecule preparation included adding of hydrogen atoms and the assignment of the standard Optimized Potential for Liquid Simulations (OPLS) atom types (36). The ligand structures were prepared for docking using the Q-MOL small-molecule minimization protocol. Polyphenols and compounds with either a molecular mass below 220 Da or chlorine atoms attached to aliphatic carbons and compounds which failed minimization were discarded. The crystal structure was treated as a set of grid-based potentials. The minimized ligand structures were docked using a Monte Carlo simulation in the internal coordinate space of the crystal structure. The identified hits were ranked according to their binding energy. The resulting hits with the lowest binding energy were selected for our further studies.
Compound databases
The compound databases were obtained from “The DTP/NCI Open Chemical Repository”. The compounds were >95% pure as certified by the DTP/NCI Services. The ligands were dissolved in 100% DMSO and stored at −20°C.
Structure modeling
The predicted binding mode of compound 9 in a complex with PEX (PDB 3C7X) was built using the Q-MOL full-atom flexible protein-ligand docking in the internal coordinates. The protein ψ, ϕ and χ angles of all of the amino acid residues, which are within a 7.5Å distance from ligand atoms, were allowed to change while the positional and rotable torsion variables of a ligand molecule were unfixed.
Results
Role of PEX in tumor growth
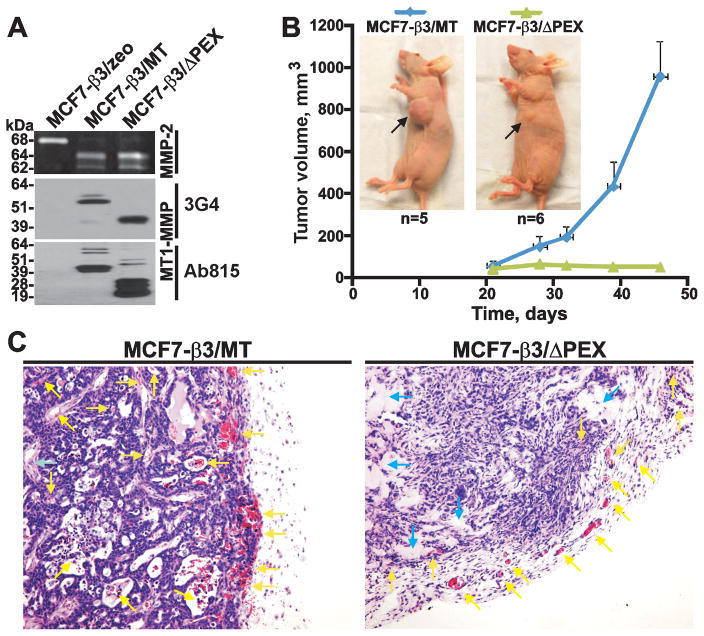
To investigate the tumorigenic role of PEX, we selected MCF7-β3/MT and MCF7-β3/ΔPEX cells. Normally, non-migratory MCF-7 cells do not express MMP-2, MT1-MMP and the β3 integrin. Expression of the β3 integrin subunit reconstitutes the αvβ3 integrin on the MCF-7 cell surface. Compared with the MT1-MMP expression alone, co-expression of MT1-MMP with the β3 integrin subunit confers an invasive phenotype on MCF-7 cells (31, 37, 38). MCF7-β3/MT and MCF7-β3/ΔPEX cells expressed a similar level of the mature MT1-MMP enzyme (as detected by the 3G4 antibody to CAT). Because of MT1-MMP self-proteolysis, degradation species were present in both MCF7-β3/MT and MCF7-β3/ΔPEX cells (as detected by the Ab1815 antibody to the hinge). According to the gelatin zymography analysis, both cell types were similarly efficient in activating the MMP-2 proenzyme. The expression level of MT1-MMP and the ability to activate MMP-2 by MCF7-β3/zeo cells were negligible (39) (Fig. 1A).
Fig. 1. MCF7-β3/MT and MCF7-β3/ΔPEX cells.
(A) Gelatin zymography of MMP-2 (top) and Western blotting with the MT1-MMP 3G4 (middle) and Ab815 (bottom) antibodies. MCF7-β3/zeo cells, control. (B) MCF7-β3/MT and MCF7-β3/ΔPEX tumor growth in mice. Arrows indicate tumors. (C) H&E staining of tumors. Yellow and blue arrows point to blood vessels/T cells and the stroma, respectively.
In our in vivo studies, cells (4×105) were embedded in Matrigel and then xenografted into young immunodeficient female mice (n=5–6). The size of the developing tumors was measured weekly for 46 days. Following a ~20-day lag-period, MCF7-β3/MT xenografts acquired a rapid growth rate. In turn, the size of MCF7-β3/ΔPEX tumors (~25–30 mm3) remained continually low. At day 46, the MCF7-β3/MT tumor volume was ~40-fold larger than that of MCF7-β3/ΔPEX tumors (Fig. 1B).
Microscopic examination revealed extensive vascularization and infiltration of T cells in MCF7-β3/MT xenografts both at the periphery and in the central regions of the tumors. There was a limited development of blood vessels in the central regions of MCF7-β3/ΔPEX tumors. The infiltration of T cells was also reduced in MCF7-β3/ΔPEX tumors compared with MCF7-β3/MT xenografts. There were additional differences between the two tumor types. Thus, MCF7-β3/ΔPEX tumors appeared fibrotic with an increased level of the stroma and connective tissues, but with less tumor cells (Fig. 1C).
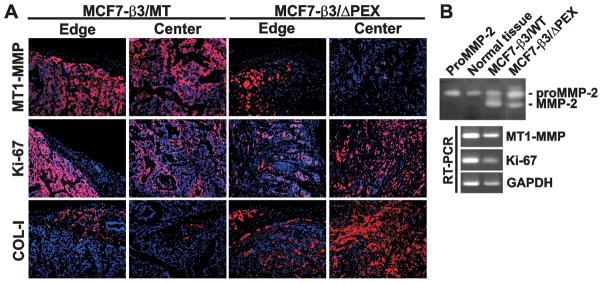
Immunostaining confirmed the presence of high levels of COL-I in MCF7-β3/ΔPEX tumors. In turn, the reduced levels of COL-I were evident in MCF7-β3/MT xenografts. MT1-MMP immunoreactivity was detected in the central and peripheral regions in MCF7-β3/MT tumors. MT1-MMP was mainly observed at the edges of MCF7-β3/ΔPEX xenografts. Ki-67 immunostaining confirmed the presence of proliferating human carcinoma cells in both tumor types (Fig. 2A).
Fig. 2. MCF7-β3/MT and MCF7-β3/ΔPEX tumors.
(A) Immunostaining of xenografts. MT1-MMP, Ki-67 and COL-I were stained with the primary antibody followed by species-specific Alexa 594-conjugated secondary antibody (red). DAPI, blue. (B) Gelatin zymograpy (top) and RT-PCR (bottom) of tumors. Left lane, top – proMMP-2 control. Normal tissue, normal mammary tissue.
Gelatin zymography of MCF7-β3/MT and MCF7-β3/ΔPEX tumor extracts supported these observations. While the normal mammary tissue specimens did not activate the 68 kDa MMP-2 proenzyme, the 62 kDa MMP-2 mature enzyme was readily observed in MCF7-β3/ΔPEX xenografts and, especially, in MCF7-β3/MT tumors. The presence of the human MT1-MMP and Ki-67 transcripts in MCF7-β3/MT and MCF7-β3/ΔPEX xenografts were also confirmed by RT-PCR (Fig. 2B).
We concluded that PEX is essential for both the efficient MT1-MMP proteolysis and the tumor growth. Naturally, if PEX was absent in the MT1-MMP structure, the proteolytically competent MT1-MMP ΔPEX mutant was largely incapable of degrading COL-I and promoting tumor growth. Our results support and extend the findings by others who suggested that PEX was essential for cleaving COL-I fibers at the cell surface (22).
Small-molecule compounds targeting PEX
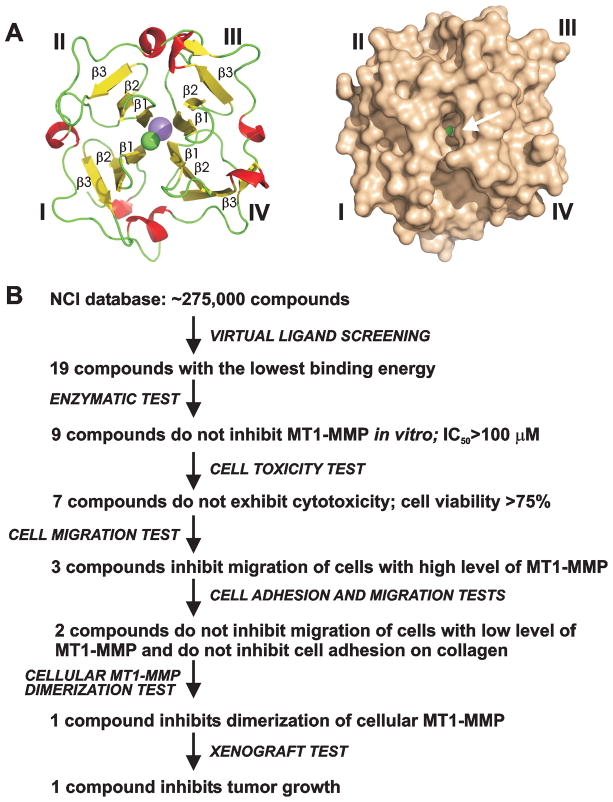
We hypothesized that it would be exceedingly difficult to identify small-molecule inhibitors which would directly bind the PEX dimerization interface and which would interfere with the multi-contact PEX homodimerization interface. Instead, we attempted to identify allosteric rather than competitive inhibitors of homodimerization. For this purpose, we selected a druggable pocket-like site in the center of the PEX structure. This site is largely formed by β-strands 1 of blades I–IV and it is distinct from the PEX dimerization interface (Fig. 3A).
Fig. 3. Selection of PEX-targeting compounds.
(A) Cartoon and surface representation of the four-bladed propeller structure of PEX (left and right, respectively). Blades I–IV and selected β-strands are labeled. Arrow points to the druggable pocket. Sodium atom, pink. Chlorine atom, green. (B) Screening steps and tests we used to select PEX-targeting compounds. Images were created using PyMol.
The diverse 275,000 compound DTP/NCI database was screened using the Q-MOL software against the docking site using the X-ray structure of MT1-MMP (PDB 3C7X). As a result, top-ranking 19 hits were identified, ordered from the DTP/NCI and analyzed further. To select the most efficient compound(s), we used multiple enzyme and cell-based tests which are summarized in Fig. 3B and shown in more detail in Figs. 4 and 5.
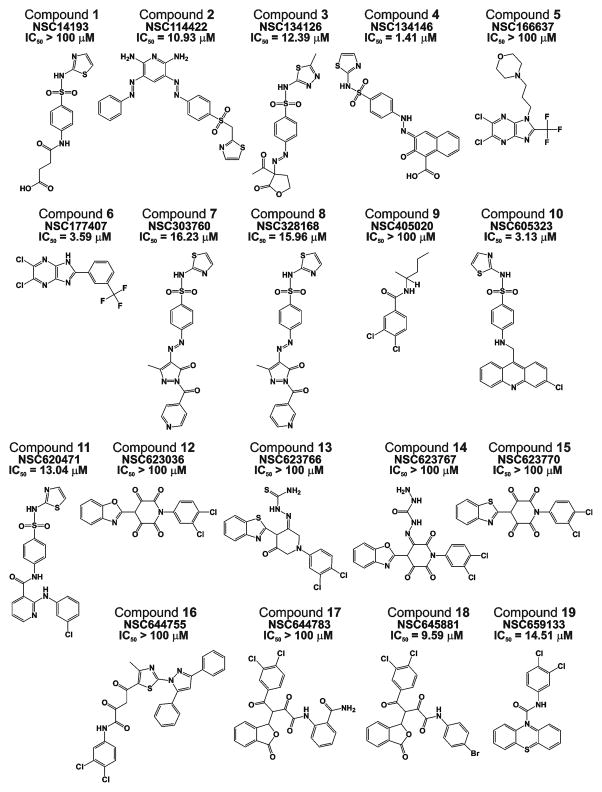
Fig. 4. Top-ranking PEX-targeting compounds.
NSC identifiers and IC50 values against CAT are shown.
Fig. 5. Compound selection steps.
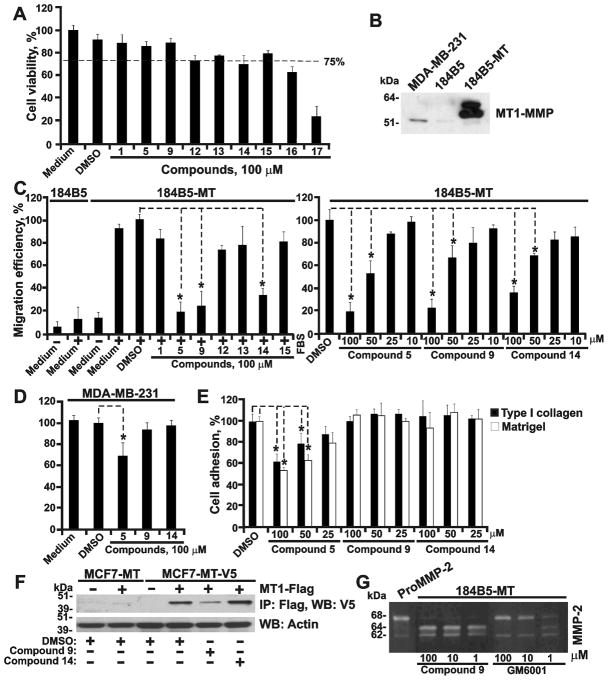
(A) Cell toxicity assay using 184B5-MT cells. (B) Western blotting with the MT1-MMP 3G4 antibody. Cells were co-incubated for 24 h with GM6001. (C) Migration assay on COL-I using 184B5-MT cells. 184B5 cells, non-migratory control. Where indicated, compounds or DMSO (1%) were added to the cells. Left, 100 μM compounds. Right, 10–100 μM compounds 5, 9 and 14. (D) Migration assay on COL-I using MDA-MB-231 cells. Where indicated, compounds (100 μM) or DMSO (1%) were added to the cells. (E) Cell adhesion assay on COL-I and Matrigel using 184B5-MT cells. Compounds 5, 9 and 14 (25–100μM) or DMSO (1%) were added to cells. (F) MT1-MMP dimerization test. MCF7-MT and MCF7-MT-V5 cells transfected with the MT1-MMP-FLAG (MT1-FLAG) construct were co-incubated with compounds 9 and 14 (100 μM) or DMSO. MT1-MMP-FLAG was then immunoprecipitated (IP) using the FLAG antibody-beads and the MT1-MMP-V5 construct was measured in the FLAG-precipitates by Western blotting (WB) with the V5 antibody. Actin, loading control. (G) Gelatin zymography of 184B5-MT cells. Cells were co-incubated with purified proMMP-2, compound 9 (1–100 μM) or GM6001 control (1–100 μM). *, p<0.05.
First, we determined if the compounds were capable of interacting with CAT and inhibiting its catalytic activity. Based on our measurements, we discarded compounds 2, 3, 4, 6–8, 10, 11, 18 and 19 the IC50 values of which were below 20 μM (Fig. 4). Compounds 1, 5, 9 and 12–17 which did not affect the cleavage activity were studied further.
Next, we determined if the compounds affected cell viability. Because normal cells are more sensitive relative to cancer cells, we used normal mammary epithelial 184B5-MT cells in our tests. The original 184B5 cells were obtained from normal augmentation mammoplasty material and immortalized by exposure to benzo(a)pyrene (40). These normal cells are neither malignant nor tumorigenic in immunodeficient mice (30). Compounds 16 and 17 exhibited cell toxicity and were then discarded. Compounds 1, 5, 9 and 12–15 were analyzed further (Fig. 5A).
We then established if the compounds were capable of affecting cell migration on COL-I. Non-migratory MCF-7 cells could not be rationally used in these experiments. Instead, we used migratory 184B5-MT cells which stably expressed high levels of MT1-MMP (Fig. 5B). Original 184B5 cells do not produce MT1-MMP and do not migrate on COL-I. Following transfection with MT1-MMP, the resulting 184B5-MT cells expressed high levels of the proteinase and acquired an ability to migrate on COL-I. Among the seven compounds we tested, compounds 5, 9 and 14 repressed migration of 184B5-MT cells on COL-I in a dose-dependent manner (Fig. 5C). We next re-tested the inhibitory potency of compounds 5, 9 and 14 using breast carcinoma MDA-MB-231 cells. These cells are naturally invasive regardless of minor levels of MT1-MMP (Fig. 5B). Compounds 9 and 14, as expected, did not show any inhibitory effect in MDA-MB-231 cells. Compound 5, however, inhibited migration of MDA-MB-231 cells, suggesting an off-target effect (Fig. 5D).
We next determined the ability of compounds 5, 9 and 14 to inhibit adhesion of 184B5-MT on the Matrigel and COL-I substrates. While compounds 9 and 14, as expected, did not affect cell adhesion, compound 5, because of its off-target effect, inhibited adhesion of cells on both substrates (Fig. 5E). The data in MDA-MB-231 cells were similar (not shown). Based on these results, we discarded compound 5. Compounds 9 and 14 were studied further.
Compound 9 affects the PEX function
We next determined if compounds 9 and 14 affected homodimerization of MT1-MMP. For this purpose, we employed MCF7-MT-V5 cells which were transiently transfected with the MT1-MMP-FLAG construct. MCF7-MT cells which expressed MT1-MMP without a V5 tag served as a control. In 24 h post-transfection, the cells were co-incubated with compounds 9 and 14 for the next 24 h and then lysed. The MT1-MMP-FLAG construct was pulled-down using anti-FLAG beads. The levels of co-precipitated MT1-MMP-V5 were then measured in the pull-down samples using Western blotting with the V5 antibody. Compound 9, but not compound 14, significantly reduced the levels of co-precipitated MT1-MMP-V5 in the pull-down samples (Fig. 5F). We concluded that compound 9 directly interacted with PEX in the full-length MT1-MMP structure and that these interactions affected PEX homodimerization.
To confirm that the catalytic activity of cellular full-length MT1-MMP was not affected by compound 9, we co-incubated 184B5-MT cells with both the MMP-2 proenzyme (23) and compound 9. The activation level of MMP-2 was then observed using gelatin zymography. This test confirmed that compound 9 did not inhibit the catalytic function of cellular MT1-MMP (Fig. 5G). In addition, compound 9 did not inhibit the catalytic activity of MMP-2 (not shown). As a result of our tests, compound 9 was selected over 14 for our subsequent xenograft studies in mice.
Compound 9 affects pro-tumorigenic MT1-MMP in vivo
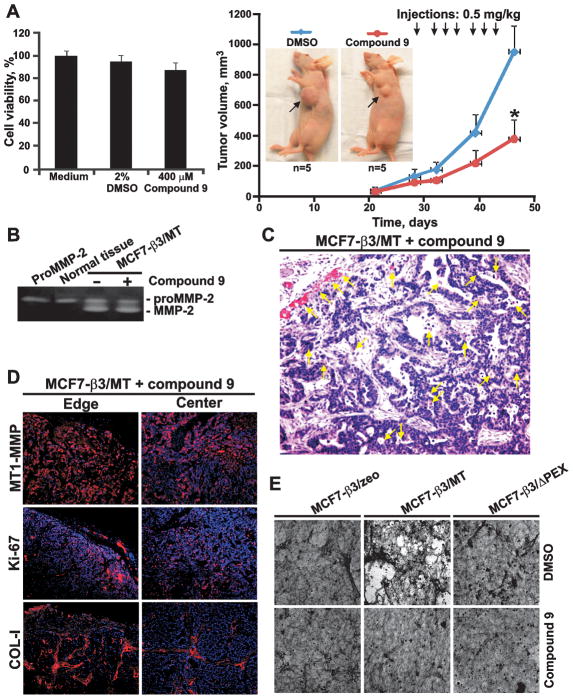
The in vivo effects of compound 9 were assessed using MCF7-β3/MT cells. Cells (4×105) were embedded in Matrigel and xenografted into immunodeficient mice (n=5). On day 29, when the tumors reached a 40–80 mm3 size, a low, 0.5 mg/kg, amount of compound 9 (from a 400 μM stock in PBS) was injected intratumorally. Because of its limited solubility, higher concentrations of compound 9 in aqueous solutions could not be reached. Control mice received vehicle alone. Injections were continued 3 times/week for an additional 2 weeks. A significant reduction in tumor size (366 ± 123 mm3) was observed in the experimental group, while the tumor size reached 933 ± 169 mm3 in the control. We also showed that compound 9 (400 μM) did not demonstrate any cytotoxicity in MCF7-β3/MT cells suggesting that cancer cell death did not affect tumor growth (Fig. 6A). At the end of the experiments, MCF7-β3/MT xenografts were excised and analyzed.
Fig. 6. Compound 9.
(A) Left, Cell toxicity assay using MCF7-β3/MT. Cells were co-incubated for 6 h with compound 9 (400 μM) or DMSO (2%). Right, Effect of compound 9 on MCF7-β3/MT tumor growth. Mice received 7 injections of compound 9 (0.5 mg/kg) starting on day 29. Arrows indicate tumors. (B) Gelatin zymograpy of MCF7-β3/MT tumors. Where indicated, mice received compound 9. Left lane, proMMP-2 control. Normal tissue, normal mammary tissue. (C) H&E staining of MCF7-β3/MT tumors treated with compound 9. Yellow arrows indicate blood vessels. (D) Immunostaining of MCF7-β3/MT tumors treated with compound 9. MT1-MMP, Ki-67 and COL-I were stained with the primary antibodies followed by the Alexa 594-conjugated secondary antibody (red). DAPI, blue. (E) COL-I degradation. Cells were plated on COL-I with compound 9 or DMSO. In 3 days cells were removed. COL-I was stained using Coomassie.
Gelatin zymography of tumor extracts confirmed that compound 9 did not affect the ability of MT1-MMP to activate MMP-2 (Fig. 6B). H&E staining did not reveal any significant effect of compound 9 on xenograft angiogenesis when compared with control mice (Fig. 1C and 6C). There was no physical damage and necrosis in the tumors following the compound 9 injections. Similar levels of the MT1-MMP and Ki-67 immunoreactivity were observed in the experimental and control tumors. In turn, both the immunoreactivity and levels of COL-I were significantly enhanced in the experimental group (Fig. 6D). Notably, the collagen staining pattern in mice treated with compound 9 became comparable with that in MCF7-β3/ΔPEX tumors (Fig. 2A and 6D) suggesting that compound 9 repressed the collagenolytic function of PEX.
To corroborate these results, we employed a COL-I degradation assay and MCF7-β3/MT cells to assess an inhibitory potency of compound 9. As additional controls, we employed MCF7-β3/zeo cells which do not express MT1-MMP and MCF7-β3/ΔPEX cells. Both MCF7-β3/zeo and MCF7-β3/ΔPEX cells were incapable of COL-I degradation. In turn, MCF7-β3/MT cells efficiently degraded the COL-I matrix. Compound 9 dramatically reduced the collagenolytic activity in MCF7-β3/MT cells (Fig. 6E). This result was consistent with our expectations.
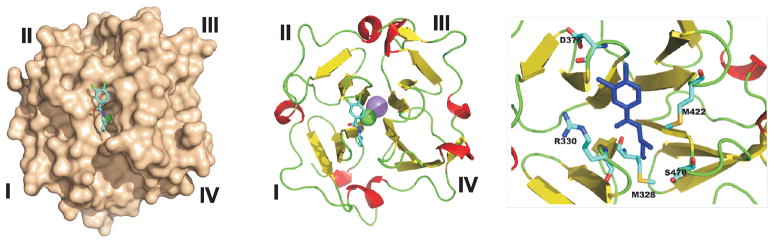
PEX-ligand complex modeling
To visualize the interactions of compound 9 with PEX, we modeled the compund-PEX complex using the PDB 3C7X as a template (Fig. 7). Our results suggested that compound 9 is in a vicinity of Met-328, Arg-330, Asp-376, Met-422 and Ser-470 of the druggable pocket. The latter is formed by the internal portions of the four polypeptide chains the external portions of which shape blades I–IV. Based on our modeling, it is tempting to hypothesize that the binding of compound 9 affected the conformation and flexibility of blades I–IV of the β-propeller and, as a result, decreased the PEX-dependent homodimerization of cellular MT1-MMP.
Fig. 7. Modeled PEX-compound 9 structure.
Left, structure of a PEX-compound 9 complex. PEX is shown as molecular surface. Middle, cartoon representation of PEX with compound 9 (colored by chemical element type). Right, close-up shows residues proximal to compound 9 (blue). Sodium atom, pink. Chlorine atom, green.
Discussion
MT1-MMP is essential to pericellular proteolysis and cell invasion (1). MT1-MMP is a multi-domain enzyme that exhibits several structural domains. These domains are additional to and distinct from CAT. The role of these domains in the net pro-invasive, pro-tumorigenic function of MT1-MMP remained unclear, especially for PEX. To record the function of PEX, we designed the MT1-MMP construct lacking PEX and then, for the first time, characterize the ΔPEX mutant in the MT1-MMP-dependent model of tumor growth. In contrast with malignant cells, which expressed the wild-type MT1-MMP and which generated the rapidly growing xenografts, the ΔPEX tumors ceased growing after reaching a metastasis-like, 25–30 mm3, size. Additional distinctions were observed in the fibrotic phenotype, the abundance of COL-I and the reduced angiogenesis in the ΔPEX tumors compared with the wild-type MT1-MMP xenografts. These observation combined with the results by others (19, 20, 22, 25, 28) led us to suggest that the ΔPEX mutant was largely incapable of COL-I degradation and, hence, that efficient MT1-MMP proteolysis of COL-I was essential for tumor growth in vivo.
It is established that homodimerization of MT1-MMP takes place via PEX (22). The structure of PEX has been solved recently (20). It became apparent that homodimerization involves multiple residue interactions between the PEX monomers. Furthermore, homodimerization occurs in the course of exocytosis of the de novo synthesized MT1-MMP rather than at the cell surface (22). For this reason, we focused on the druggable pocket that was distinct from the PEX dimerization interface.
For this purpose, we employed virtual ligand screening using the 275,000 compound DTP/NCI library as a ligand source and the X-ray structure of PEX as a target. Compounds were docked in silico to the druggable pocket-like site. This pocket is distinct from the PEX dimerization interface and localized in the center of PEX at a roughly equal distance from blades I–IV. As a result of our screens, we selected the top-ranking compounds, ordered them from DTP/NCI and tested using multiple in vitro and cell-based assays. Consequently, we identified a novel inhibitory compound that selectively targeted PEX, reduced PEX homodimerization and repressed MT1-MMP-mediated cell migration and degradation of COL-I. This compound, however, did not exhibit any observable off-target effects and did not inhibit the cleavage activity of CAT, the efficiency of MMP-2 activation by MT1-MMP and cell adhesion to the COL-I substratum.
The anti-tumor efficacy of this PEX-targeting compound 9 (NSC405020) was directly demonstrated in the in vivo tests. The low, 0.5 mg/kg, intratumoral injections of the compound significantly repressed tumor growth. Compound 9 injections also caused a fibrotic tumor phenotype and increased the level of COL-I. The fibrotic phenotype caused by the compound was similar to that we observed in the ΔPEX tumors. These findings suggested that the compound affected PEX and the ability of MT1-MMP to function properly in degrading COL-I to support tumor cell invasion. We have already identified additional, potent compounds of the same scaffold (Supplemental Table S1). Based on our ADME studies, we are confident that compound derivatives rather than the original compound 9 will be most suitable for the subsequent systemic administration and the in vivo PK studies.
We believe that our study, especially when combined with the results of others (25, 41, 42), provides proof of concept for a design of those non-catalytic domain inhibitors which will selectively target PEX and which will affect pro-tumorigenic MT1-MMP in a clinically beneficial manner. In addition, as shown by the pioneering work by others (18, 25, 41, 42), targeting PEX appears to be a novel and promising approach to modulating the function of other MMPs, including MMP-9, rather than MT1-MMP alone. Overall, our proof-of-principle work provides conceptual support to probe MT1-MMP PEX with small molecule ligands and a rationale for design of selective PEX-targeting drugs.
Supplementary Material
Footnotes
The work reported here was supported by NIH Grants CA83017 and CA77470 (to AYS).
References
- 1.Zucker S, Pei D, Cao J, Lopez-Otin C. Membrane type-matrix metalloproteinases (MT-MMP) Curr Top Dev Biol. 2003;54:1–74. doi: 10.1016/s0070-2153(03)54004-2. [DOI] [PubMed] [Google Scholar]
- 2.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–92. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 3.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–74. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 4.Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J Cell Biol. 2000;149:1309–23. doi: 10.1083/jcb.149.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hotary KB, Yana I, Sabeh F, Li XY, Holmbeck K, Birkedal-Hansen H, et al. Matrix metalloproteinases (MMPs) regulate fibrin-invasive activity via MT1-MMP-dependent and -independent processes. J Exp Med. 2002;195:295–308. doi: 10.1084/jem.20010815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itoh Y, Seiki M. MT1-MMP: a potent modifier of pericellular microenvironment. J Cell Physiol. 2006;206:1–8. doi: 10.1002/jcp.20431. [DOI] [PubMed] [Google Scholar]
- 7.Friedl P, Wolf K. Proteolytic interstitial cell migration: a five-step process. Cancer Metastasis Rev. 2009;28:129–35. doi: 10.1007/s10555-008-9174-3. [DOI] [PubMed] [Google Scholar]
- 8.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 9.Strongin AY. Mislocalization and unconventional functions of cellular MMPs in cancer. Cancer Metastasis Rev. 2006;25:87–98. doi: 10.1007/s10555-006-7892-y. [DOI] [PubMed] [Google Scholar]
- 10.Strongin AY. Proteolytic and non-proteolytic roles of membrane type-1 matrix metalloproteinase in malignancy. Biochim Biophys Acta. 2010;1803:133–41. doi: 10.1016/j.bbamcr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagase H, Woessner JF., Jr Matrix metalloproteinases. J Biol Chem. 1999;274:21491–4. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 12.Golubkov VS, Chernov AV, Strongin AY. Intradomain cleavage of inhibitory prodomain is essential to protumorigenic function of membrane type-1 matrix metalloproteinase (MT1-MMP) in vivo. J Biol Chem. 2011;286:34215–23. doi: 10.1074/jbc.M111.264036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei D, Weiss SJ. Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature. 1995;375:244–7. doi: 10.1038/375244a0. [DOI] [PubMed] [Google Scholar]
- 14.Ota I, Li XY, Hu Y, Weiss SJ. Induction of a MT1-MMP and MT2-MMP-dependent basement membrane transmigration program in cancer cells by Snail1. Proc Natl Acad Sci U S A. 2009;106:20318–23. doi: 10.1073/pnas.0910962106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabeh F, Li XY, Saunders TL, Rowe RG, Weiss SJ. Secreted versus membrane-anchored collagenases: relative roles in fibroblast-dependent collagenolysis and invasion. J Biol Chem. 2009;284:23001–11. doi: 10.1074/jbc.M109.002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun TH, Hotary KB, Sabeh F, Saltiel AR, Allen ED, Weiss SJ. A pericellular collagenase directs the 3-dimensional development of white adipose tissue. Cell. 2006;125:577–91. doi: 10.1016/j.cell.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 17.Cao J, Kozarekar P, Pavlaki M, Chiarelli C, Bahou WF, Zucker S. Distinct roles for the catalytic and hemopexin domains of membrane type 1-matrix metalloproteinase in substrate degradation and cell migration. J Biol Chem. 2004;279:14129–39. doi: 10.1074/jbc.M312120200. [DOI] [PubMed] [Google Scholar]
- 18.Dufour A, Sampson NS, Zucker S, Cao J. Role of the hemopexin domain of matrix metalloproteinases in cell migration. J Cell Physiol. 2008;217:643–51. doi: 10.1002/jcp.21535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collier IE, Legant W, Marmer B, Lubman O, Saffarian S, Wakatsuki T, et al. Diffusion of MMPs on the surface of collagen fibrils: the mobile cell surface-collagen substratum interface. PLoS ONE. 2011;6:e24029. doi: 10.1371/journal.pone.0024029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tochowicz A, Goettig P, Evans R, Visse R, Shitomi Y, Palmisano R, et al. The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain: crystal structure and biological functions. J Biol Chem. 2011;286:7587–600. doi: 10.1074/jbc.M110.178434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tam EM, Wu YI, Butler GS, Stack MS, Overall CM. Collagen binding properties of the membrane type-1 matrix metalloproteinase (MT1-MMP) hemopexin C domain. The ectodomain of the 44-kDa autocatalytic product of MT1-MMP inhibits cell invasion by disrupting native type I collagen cleavage. J Biol Chem. 2002;277:39005–14. doi: 10.1074/jbc.M206874200. [DOI] [PubMed] [Google Scholar]
- 22.Itoh Y, Ito N, Nagase H, Evans RD, Bird SA, Seiki M. Cell surface collagenolysis requires homodimerization of the membrane-bound collagenase MT1-MMP. Mol Biol Cell. 2006;17:5390–9. doi: 10.1091/mbc.E06-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem. 1995;270:5331–8. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- 24.Mori H, Tomari T, Koshikawa N, Kajita M, Itoh Y, Sato H, et al. CD44 directs membrane-type 1 matrix metalloproteinase to lamellipodia by associating with its hemopexin-like domain. Embo J. 2002;21:3949–59. doi: 10.1093/emboj/cdf411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarrabi K, Dufour A, Li J, Kuscu C, Pulkoski-Gross A, Zhi J, et al. Inhibition of matrix metalloproteinase 14 (MMP-14)-mediated cancer cell migration. J Biol Chem. 2011;286:33167–77. doi: 10.1074/jbc.M111.256644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P, Nie J, Pei D. The hemopexin domain of MT1-MMP is not required for its activation of proMMP2 on cell surface, but essential for MT1-MMP mediated invasion in 3-D type I collagen. J Biol Chem. 2004;279:51148–55. doi: 10.1074/jbc.M409074200. [DOI] [PubMed] [Google Scholar]
- 27.Remacle AG, Rozanov DV, Baciu PC, Chekanov AV, Golubkov VS, Strongin AY. The transmembrane domain is essential for the microtubular trafficking of membrane type-1 matrix metalloproteinase (MT1-MMP) J Cell Sci. 2005;118:4975–84. doi: 10.1242/jcs.02610. [DOI] [PubMed] [Google Scholar]
- 28.Itoh Y, Takamura A, Ito N, Maru Y, Sato H, Suenaga N, et al. Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. Embo J. 2001;20:4782–93. doi: 10.1093/emboj/20.17.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh Y, Ito N, Nagase H, Seiki M. The second dimer interface of MT1-MMP, the transmembrane domain, is essential for ProMMP-2 activation on the cell surface. J Biol Chem. 2008;283:13053–62. doi: 10.1074/jbc.M709327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Golubkov VS, Chekanov AV, Savinov AY, Rozanov DV, Golubkova NV, Strongin AY. Membrane type-1 matrix metalloproteinase confers aneuploidy and tumorigenicity on mammary epithelial cells. Cancer Res. 2006;66:10460–5. doi: 10.1158/0008-5472.CAN-06-2997. [DOI] [PubMed] [Google Scholar]
- 31.Deryugina EI, Bourdon MA, Jungwirth K, Smith JW, Strongin AY. Functional activation of integrin alpha V beta 3 in tumor cells expressing membrane-type 1 matrix metalloproteinase. Int J Cancer. 2000;86:15–23. doi: 10.1002/(sici)1097-0215(20000401)86:1<15::aid-ijc3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 32.Golubkov VS, Cieplak P, Chekanov AV, Ratnikov BI, Aleshin AE, Golubkova NV, et al. Internal cleavages of the autoinhibitory prodomain are required for membrane type 1 matrix metalloproteinase activation, although furin cleavage alone generates inactive proteinase. J Biol Chem. 2010;285:27726–36. doi: 10.1074/jbc.M110.135442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratnikov B, Deryugina E, Leng J, Marchenko G, Dembrow D, Strongin A. Determination of matrix metalloproteinase activity using biotinylated gelatin. Anal Biochem. 2000;286:149–55. doi: 10.1006/abio.2000.4798. [DOI] [PubMed] [Google Scholar]
- 34.Shiryaev SA, Remacle AG, Savinov AY, Chernov AV, Cieplak P, Radichev IA, et al. Inflammatory proprotein convertase-matrix metalloproteinase proteolytic pathway in antigen-presenting cells as a step to autoimmune multiple sclerosis. J Biol Chem. 2009;284:30615–26. doi: 10.1074/jbc.M109.041244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiryaev SA, Cheltsov AV, Gawlik K, Ratnikov BI, Strongin AY. Virtual ligand screening of the National Cancer Institute (NCI) compound library leads to the allosteric inhibitory scaffolds of the West Nile Virus NS3 proteinase. Assay Drug Dev Technol. 2011;9:69–78. doi: 10.1089/adt.2010.0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J Am Chem Soc. 1996;118:11225–36. [Google Scholar]
- 37.Deryugina EI, Ratnikov B, Monosov E, Postnova TI, DiScipio R, Smith JW, et al. MT1-MMP initiates activation of pro-MMP-2 and integrin alphavbeta3 promotes maturation of MMP-2 in breast carcinoma cells. Exp Cell Res. 2001;263:209–23. doi: 10.1006/excr.2000.5118. [DOI] [PubMed] [Google Scholar]
- 38.Deryugina EI, Ratnikov BI, Postnova TI, Rozanov DV, Strongin AY. Processing of integrin alpha(v) subunit by membrane type 1 matrix metalloproteinase stimulates migration of breast carcinoma cells on vitronectin and enhances tyrosine phosphorylation of focal adhesion kinase. J Biol Chem. 2002;277:9749–56. doi: 10.1074/jbc.M110269200. [DOI] [PubMed] [Google Scholar]
- 39.Rozanov DV, Deryugina EI, Ratnikov BI, Monosov EZ, Marchenko GN, Quigley JP, et al. Mutation analysis of membrane type-1 matrix metalloproteinase (MT1-MMP). The role of the cytoplasmic tail Cys(574), the active site Glu(240), and furin cleavage motifs in oligomerization, processing, and self-proteolysis of MT1-MMP expressed in breast carcinoma cells. J Biol Chem. 2001;276:25705–14. doi: 10.1074/jbc.M007921200. [DOI] [PubMed] [Google Scholar]
- 40.Walen KH, Stampfer MR. Chromosome analyses of human mammary epithelial cells at stages of chemical-induced transformation progression to immortality. Cancer Genet Cytogenet. 1989;37:249–61. doi: 10.1016/0165-4608(89)90056-3. [DOI] [PubMed] [Google Scholar]
- 41.Dufour A, Sampson NS, Li J, Kuscu C, Rizzo RC, Deleon JL, et al. Small-molecule anticancer compounds selectively target the hemopexin domain of matrix metalloproteinase-9. Cancer Res. 2011;71:4977–88. doi: 10.1158/0008-5472.CAN-10-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dufour A, Zucker S, Sampson NS, Kuscu C, Cao J. Role of matrix metalloproteinase-9 dimers in cell migration: design of inhibitory peptides. J Biol Chem. 2010;285:35944–56. doi: 10.1074/jbc.M109.091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.