Abstract
Cbln1 is essential for synapse integrity in cerebellum through assembly into complexes that bridge presynaptic β-neurexins (Nrxn) to postsynaptic GluRδ2. However, GluRδ2 is largely cerebellum-specific, yet Cbln1 and its little studied family members, Cbln2 and Cbln4, are expressed throughout brain. Therefore, we investigated whether additional proteins mediate Cbln family actions. Whereas Cbln1 and Cbln2 bound to GluRδ2 and Nrxns1–3, Cbln4 bound weakly or not at all, suggesting it has distinct binding partners. In a candidate receptor-screening assay, Cbln4 (but not Cbln1 or Cbln2) bound selectively to the netrin receptor, DCC (deleted in colorectal cancer) in a netrin-displaceable fashion. To determine whether Cbln4 had a netrin-like function, Cbln4-null mice were generated. Cbln4-null mice did not phenocopy netrin-null mice. Cbln1 and Cbln4 were likely co-localized in neurons thought to be responsible for synaptic changes in striatum of Cbln1-null mice. Furthermore, complexes containing Cbln1 and Cbln4 had greatly reduced affinity to DCC but increased affinity to Nrxns, suggesting a functional interaction. However, Cbln4-null mice lacked the striatal synaptic changes seen in Cbln1-null mice. Thus Cbln family members interact with multiple receptors/signaling pathways in a subunit composition-dependent manner and have independent functions with Cbln4 potentially involved in the less-well characterized role of netrin/DCC in adult brain.
Keywords: Cbln1, Cbln4, DCC, Neurexin, netrin
Introduction
Diverse molecules and signaling pathways have been identified that mediate the critical processes of synapse specification, formation and maintenance in the nervous system. Cerebellin 1 precursor protein (Cbln1) is a secreted glycoprotein belonging to the C1q and tumor necrosis factor superfamily of proteins that is essential for the proper formation and stability of excitatory synapses between cerebellar granule neurons and Purkinje cells (Hirai et al., 2005). The mechanism by which this is achieved is relatively unique in the nervous system. Multi-subunit complexes containing Cbln1 are secreted from granule neurons (Pang et al, 2000; Bao et al., 2006) and serve as a bifunctional ligand that bridges the pre- and post-synaptic membranes by binding to β-neurexins (βNrxns) on granule neurons and δ2 glutamate receptor (GluRδ2) on Purkinje cells, thereby presumptively stabilizing synaptic contacts (Matsuda et, al., 2010; Schmid et al., 2010; Uemura et al., 2010; Joo et al., 2011; Matsuda et, al., 2011). Although this elegant arrangement can explain the roles of Cbln1 and GluRδ2 in the granule cell-Purkinje cell synapse it cannot fully explain the biology of Cbln1 or that of its closely related family members, Cbln2–Cbln4 (Bao et al., 2005).
Unlike βNrxns, expression of GluRδ2 is largely confined to cerebellar Purkinje cells (Araki et al., 1993) yet Cbln1 is expressed outside of cerebellum (Mugnaini and Morgan, 1987; Miura et al., 2006; Wei et al., 2007). For example, Cbln1 is prominently expressed in glutamatergic neurons of the parafascicular nucleus (PF) of the thalamus whose axons synapse onto dendrites of medium spiny neurons (MSN) of the striatum (Wei et al., 2007; Kusnoor et al., 2010). Moreover, loss of Cbln1 results in increased synaptic spine densities on the dendrites of MSNs (Kusnoor et al., 2010). Even within the cerebellum, Cbln1 can undergo transneuronal trafficking at synapses that are not thought to contain GluRδ2 and is accumulated in Bergmann glia that also do not express GluRδ2 (Wei et al., 2009), suggesting it may potentially interact with additional membrane proteins. Finally, Cbln1 is the prototype of a small family of proteins (Cbln1–4) (Bao et al., 2005) that are expressed in neurons with broad, sometimes overlapping distributions in the nervous system (Miura et al., 2006; Wei et al., 2007). Although we have shown that Cbln3 can form heteromeric complexes with Cbln1 (Pang et al., 2000) and influence the turnover of Cbln1 in cerebellum (Bao et al., 2006), nothing is known of the function of the other family members and little is known about their in vivo receptors. Here we have used in vitro and in vivo approaches to identify additional proteins that interact with Cbln family members and determine whether family members have identical functions.
Materials and Methods
Animals and genotyping
Production, characterization and genotyping of Cbln1-null mice have been previously described (Hirai et al., 2005). To generate Cbln4-null mice, a recombineering approach (Liu et al., 2003) was used to generate the Cbln4 knockout constructs. A 13 kb DNA fragment containing the Cbln4 gene was isolated from a BAC DNA clone (Sanger Wellcome Trust) and subcloned into the PL253 vector (NCI-Frederick, MD). The region spaning from the start codon through to the stop codon of Cbln4 was replaced by the neomycin resistant gene cassette as detailed in Fig. 1A. The linearized targeting vector was electroporated into W9.5 embryonic stem cells (kindly provided by P. J. McKinnon, Department of Genetics, St. Jude Children’s Research Hospital) and selected with G418 (Cellgro, Herndon, VA) and fialuridine (Movarek, Biochemicals, Brea, CA). DNA from ES cells was digested with BglII or AseI, and analyzed by Southern blotting using a 0.5 kb 5′ external probe and a 0.36 kb 3′ probe, respectively. One chimera derived from the ES clones underwent germline transmission. Animals were genotyped by Southern blot and PCR (Fig. 1B). PCR primers were: Cbln4 wild type 5′ primer: 5′- ATG AAG CAG GAA GTA CGA AAT A -3′ and Cbln4 wild type 3′ primer: 5′- CTC AAG GGT CTA AAG TGA AAA G -3′, as well as Cbln4 knockout 5′ primer: 5′- TCA AGG GAC AAG TGT GGG TGC C -3′, and Cbln4 knockout 3′ primer: 5′- TGA CGA GTT CTT CTG AGG GGA T -3′. Mice were maintained at St. Jude Children’s Research Hospital and had free access to food and water. Investigational procedures conformed to all applicable federal rules and guidelines and were approved by the Institutional Animal Care and Use Committee.
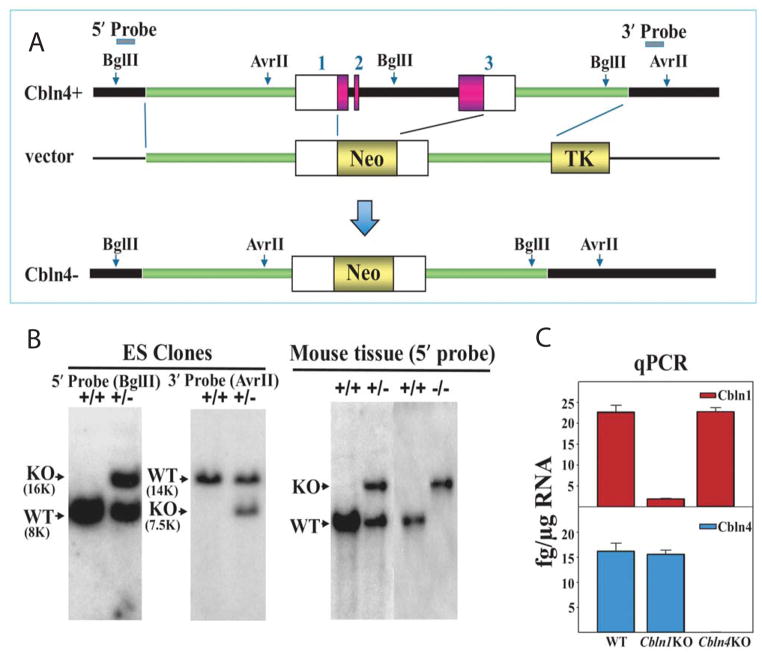
Fig. 1.
Generation of Cbln4-null mice. (A) Schematic representations of Cbln4 genomic DNA (Cbln4+), targeting vector and targeted gene (Cbln4−). Shaded bars indicate the 5′ and 3′ probes for Southern blot analysis. The region spanning from start codon through to stop codon of Cbln4 was deleted by inserting the neomycin resistance gene cassette (Neo). (B) Southern blot analysis of genomic DNA prepared from ES cells (left and middle panels) and tissues of wild-type (+/+), heterozygote (+/−), and knockout (−/−) mice (right panel). DNA from ES cells and mouse tissues was digested with BglII (for 5′ probe) and AseI (for 3′ probe), analyzed using a digoxigenin (DIG)-labeled 0.5 kb 5′ external probe and a 0.36 kb 3′ probe, respectively. Bands with the indicated sizes denote correct targeting in both ES cells and tissues. (C) qPCR showing loss of Cbln1 (red) and Cbln4 mRNA (blue) in Cbln1-null (Cbln1KO) and Cbln4-null (Cbln4KO) mouse brain, respectively. There is no compensatory change of Cbln1 or Cbln4 mRNA levels in Cbln4- or Cbln1-null mice, respectively.
Cloning
Nrxn1β and Nrxn2β coding sequence were synthesized by Genscript (Piscataway, NJ) and cloned into p3XFLAG-CMV-14 from Sigma (St Louis, MO, USA) using HindIII and XbaI to obtain Nrxn1β-3XFLAG and Nrxn2β-3XFLAG, respectively. Nrxn3β coding sequence was cloned by PCR from C57Bl/6J mouse cerebellum, using the following primers: Nrxn3β-FLAG-F 5′-CCC AAG CTT ACC ACC ATG CAC CTG AGA ATC CAC CCA AG-3′ and Nrxn3β-FLAG-R 5′-GGG ATC CCA CAT AAT ACT CCT TGT CCT-3′. The PCR product was then cloned into p3XFLAG-CMV-14 using HindIII and XbaI to obtain Nrxn3β-3XFLAG. Nrxn1α-Myc-DKK (MR226542 -NM_020252.3), Nrxn2α-Myc-DKK (MR212442 - NM020253.2) and Nrxn3α-Myc-DKK (MR219440 - NM_172544.3) were purchased from Origene (Rockville, MD). Fcgr1 (OCACo5052A0715D - BC160240), Igdcc5/Prtg (OCACo5052D0818D - BC167223), Igdcc3 (IRAVp968D04145D - BC086481), Igdcc4 (OCACo5052B0414D - BC156227), Ptprg (OCACo5052C0119D - BC168385), Robo1 (OCACo5052H0718D - BC167219), Cntn3 (OCACo5052G122D - BC141426), and Neogenin1 (Neo1) (IRAVp968B08113D -BC054540) were purchased from Imagenes (Berlin, Germany). Unc5b-3XFLAG (Ex-Mm25432-M14 - NM_029770) was purchased from GeneCopoeia (Rockville, MD). Human DCC-pENTR223 (100016116 – BC1522808) was from Open Biosystem (Clone ID 100016116 – BC1522808)]. The numbers in parenthesis refer to the clone ID number and the NCBI sequence contained in each construct. Nrxn1α-Myc-DKK, Nrxn2α-Myc-DKK, Nrxn3α-Myc-DKK and Unc5b-3XFLAG, were used without further modification. Fcgr1, Igdcc5/Prtg, Ptprg and Robo1 ORFs were shuttled into pDEST-pcDNA3-RfA, using Clonase LR II (Invitrogen, Carlsbad, CA) according to manufacturer instructions. Igdcc3 ORF was first shuttled into pDONR221 for Invitrogen using Clonase BP II from Invitrogen. The construct obtained was subsequently used to shuttle Igdcc3 ORF into pDEST-pcDNA3-RfA and pDEST-pCMV14-RfA using Clonase LR II from Invitrogen. Igdcc4 was cloned into pDEST-pCMV14-RfA and pDEST-pCMV14-RfB, to obtain an untagged and a C-terminus 3XFLAG fusion protein, respectively. Cntn3 ORF was shuttled into pDEST-pcDNA3-RfA and pDEST-pCMV14-RfA using Clonase LR II form Invitrogen to obtain an untagged and a C-terminus 3XFLAG fusion protein, respectively. The coding sequence of DCC was shuttled into pDEST-pCMV14-RfA to generate hDCC-3XFLAG Clonase LR II from Invitrogen. pDEST-pcDNA3-RfA, pDEST-pCMV14-RfA and pDEST-pCMV14-RfB were prepared by inserting RfA or RfB from the Gateway® Conversion System (Invitrogen, Carlsbad, CA) into pcDNA3 (Invitrogen, Carlsbad, CA) and p3XFLAG-CMV14 (Sigma, St. Louis, MO), according to manufacturer instructions. Mouse GluRδ1, GluRδ2 and gC1qBP cDNA were cloned by reverse transcriptase-polymerase chain reaction (RT-PCR) from mouse brain total RNA. Cloning of Cbln1, Cbln2 and Cbln4 was as described previously (Bao et al., 2005). The cDNA encoding hemagglutinin (HA) tag was added to the 5′ of Cbln1, Cbln2 and Cbln4, between glutamine (amino acid 22 for Cbln1, Accession: NP_062600; amino acid 52 for Cbln2, Accession: NP_766221; amino acid 25 for Cbln4, Accession: NP_783439) and asparagine (amino acid 23 for Cbln1, 53 for Cbln2 and 26 for Cbln4) amino acids. For Neo1, the coding sequence of Neo1 was amplified by PCR using Platinum Pfx DNA Polymerase (Invitrogen) and the following primers: Neo1-HindIII-F, 5′-CCC AAG CTT GTA GAG ATG GCG GCG GA-3′ and Neo1-NotI-R, 5′-TTT TCC TTT TGC GGC CGC GGC TGT TGT GAT GGC ATT TAG-3′. The coding sequence was then cloned into p3XFLAG-CMV14 using HindIII and NotI sites to obtain a C-terminus flagged fusion protein. GluRδ1, GluRδ2, gC1qBP, HA-Cbln1, HA-Cbln2 and HA-Cbln4 were inserted into pcDNA3.1 (Invitrogen, Carlsbad, CA) according to manufacturer instructions.
In vitro binding assay
To study the binding of HA-Cbln1, HA-Cbln2 and HA-Cbln4 to known or candidate receptors, HEK-293T cells were plated in 6-well plates (300,000 cells/well) and transfected 24 hr later with plasmids containing the receptor of interest or empty vector (Mock). Cells were used 48 hr after transfection. In parallel, cells on separate 6-well plates were transfected with vectors expressing different ligands and medium collected 48 hr after transfection. Medium containing the ligand was added to 6-well plates (2 ml/well) containing receptor- or mock-transfected cells whose media had been removed. Plates were incubated for 4 hr. At the end of the experiment, cells were washed three times with ice-cold medium and lysed with 1X XT-running buffer (BioRad, Hercules, CA) containing 10 μM DTT. Samples were run on Criterion 12% Bis-Tris gels (BioRad, Hercules, CA), and binding of HA-Cbln1, HA-Cbln2, or HA-Cbln4 was detected using mouse anti-HA antibody (Covance, Princeton, NJ). Expression of candidate receptors was detected using rabbit anti-FLAG antibody (Sigma, St Louis, MO).
Netrin-1 competition assays
Recombinant mouse netrin-1 (rmnetrin-1), containing bovine serum albumin (BSA) as carrier (50 μg BSA for every μg of rmnetrin-1), was purchased from R&D Systems (Minneapolis, MN) and dissolved to 100 μg/μl in DMEM-complete (DMEM, containing 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 292 μg/ml glutamine). DMEM was purchased from Lonza (Basel, Switzerland), fetal calf serum from Millipore (Billerica, MA) and penicillin/streptomycin/L-glutamine from Invitrogen (Carlsbad, CA). Medium from HA-Cbln4-transfected HEK293T cells was collected 48 hr after transfection with HA-Cbln4/pcDNA3.1 and diluted as indicated in DMEM-complete from mock-transfected HEK293T cells to obtain a range of HA-Cbln4 concentrations.
For the netrin-1 competition assay, HEK-293T cells were plated in 6-well plates (300,000 cells/well) and transfected 24 hr later with either human DCC/pCMV-3XFLAG or empty vector (Mock). Cells were used for the assay 48 hr after transfection. Cells were pre-incubated with 1 ml DMEM-complete (Mock), DMEM-complete with 1 μg/ml netrin-1 or DMEM-complete with 50 μg/ml BSA for 30 min. BSA was purchased from Sigma (Sigma, St Louis, MO) and dissolved in DMEM-complete 5 μg/μl. After the pre-incubation period, cells were incubated for 3.5 hr with an additional 1 ml of medium containing HA-Cbln4, HA-Cbln4 + netrin-1 1 μg/ml, or HA-Cbln4 + BSA 50 μg/ml respectively.
For the netrin-1 dose response assay, cells were transfected with DCC or empty vector. Cells were subsequently pre-incubated with varying concentrations of netrin-1 (3, 1, 0.5, 0.25 or 0 μg/ml) or BSA carrier for 30 min, and then incubated for an additional 3.5 hr with conditioned medium containing HA-Cbln4 and the appropriate level of netrin-1 or BSA.
Histological methods
One-month-old gene knockout mice and their gender matched wild-type littermates mice were anesthetized, perfused transcardially with buffered 4% paraformaldehyde (PFA), and their brains removed and post-fixed as described previously (Wei et al., 2007). Paraffin sections (5 μm) were stained with hematoxylin and eosin for standard histological analysis. For immunohistochemistry, all sections were subjected to heat-mediated antigen retrieval in 0.01 M sodium citrate buffer (pH 6.0) containing 0.05% Tween 20 (Sigma, St. Louis, MO) as previously described (Wei et al., 2009). Immunostaining for Cbln1 and Cbln4 was performed using rabbit polyclonal antiserum to Cbln1 (E3) (1:1000) (Bao et al., 2005) or rabbit anti-Cbln4 (1:300, Aviva Systems Biology, San Diego, CA) as described previously (Wei et al., 2007). As the anti-Cbln4 antibody used in this experiment cross-reacted with Cbln1 in our hands (data not shown), the brains from Cbln1-null mice were used in the immunohistochemistry studies to localize the expression of Cbln4. After immunostaining, the sections were counterstained with hematoxylin (Vector Labs, Burlingame, CA).
To establish the subcellular localization of Cbln4, heat-mediated antigen retrieval treated paraffin sections were incubated overnight with a mixture of rabbit anti-Cbln4 (1:300) antibody and goat anti-cathepsin D (lysosome marker, 1:300, Santa Cruz Biotechnology, Santa Cruz, CA) (Erickson and Blobel, 1979). Sections were incubated for 1 h with Alexa 488- or Alexa 594- labeled species-specific secondary antibodies (1:200, Invitrogen, Carlsbad, CA) and images acquired using confocal laser microscopy as described previously (Wei et al., 2007).
Rotarod test
Motor coordination, balance, and motor learning were evaluated with the accelerating rotarod test (Wei et al., 2011). Gender and age matched wild type and knockout mice littermates were tested on an accelerating rotarod (San Diego Instruments, San Diego, CA). The rotarod was programmed to accelerate from 0 to 40 rpm in 4 min and then hold constant speed for a further 1 min. The time elapsed before the mouse fell off was recorded. Animals were given daily sessions consisting of 3 trials, with a 20-minutes inter-trial interval. Animals were repeatedly tested for 5 consecutive days. The latency of the mice to fall from the rod was scored as an index of their motor coordination. Improvement in performance across training days indicates motor learning (Buitrago et al., 2004; Wei et al., 2011).
RNA isolation, cDNA synthesis, and qPCR
Brain tissue samples (about 50 mg) were homogenized in Trizol (Invitrogen, Carlsbad, CA). After homogenization, chloroform was added and the tubes shaken vigorously for 15 seconds. Samples were centrifuged at 12,000 × g for 15 minutes at 4°C. Top aqueous layers were carefully transferred into new nuclease-free tubes and 2-propanol was added.
Samples were briefly vortexed, allowed to stand at room temperature for 10 minutes, then centrifuged at 12,000 × g for 10 minutes at 4°C. RNA pellets were washed with 75% ethanol. After air-drying for 30 minutes, water was added to each RNA pellet. MultiScribeTM Reverse Transcriptase (Applied Biosystems, Foster City, CA) was used for reverse transcription. Reverse transcription master mix containing dNTP, random hexamers, reverse transcriptase, and RNA samples were briefly mixed. Reactions were carried out using thermocycler conditions of: 5 minutes at 25°C, 120 minutes at 37°C, 5 minutes at 85°C, followed by a safety hold at 4°C. For qPCR primer and probe design, the Cbln1, Cbln2, Cbln4, DCC and Ntn1 consensus sequences were entered into Primer Express 3.0 (Applied Biosystems, Foster City, CA). Primers and probes were analyzed for homology to other known sequences using the Basic Local Alignment Search Tool (BLAST). For qPCR reactions, the Applied Biosystems 7900 Fast Real-Time PCR system was used for qPCR amplification and detection. qPCR reactions were prepared in triplicates of 20 μL reaction mixture in MicroAmp optical 96-well reaction plates and sealed with optical adhesive covers. Three replicates of a control sample without DNA template were also included in the runs. The reaction mixture for these assays consisted of 10 μL of 2 × TaqMan Fast Reagents Starter Kit (Applied Biosystems, Foster City, CA), 450 nM each of F and R primers, 450 nM of probe, and 1 μL of cDNA in a final volume of 20 μL. After an incubation of 2 min at 50 °C to allow for uracil-N-glycosylase cleavage, AmpliTaq Gold polymerase was activated by an incubation step for 10 min at 95 °C. All 40 cycles were performed according to the following temperature regime: 95 °C for 15 s and 60 °C for 1 min. Ct determinations were automatically performed by the instrument using default parameters. Seven-point standard curves were constructed using cDNA pooled equally from all brain regions. The curves were constructed from serial 2-fold dilutions and covered a cDNA dilution range of 2 to 0.03125. The concentration of unknown samples was calculated using 7500 Fast System software (Applied Biosystems, Foster City, CA).
Golgi impregnation and dendritic analyses of MSNs
Cbln4-null mice and their gender matched wild-type littermates were perfused with 0.1 M sodium phosphate buffer followed by a solution containing 2% PFA/2.5% glutaraldehyde in 0.1 M sodium phosphate buffer. Coronal sections (150 μm) through the precommissural striatum were cut on a vibrating microtome. The sections were then incubated in 1% osmium tetroxide for 30 minutes and kept in 3.5% potassium dichromate overnight. The next day the sections were developed in 1% silver nitrate for 4–6 hours before being washed extensively, mounted, and coverslipped. Golgi-stained MSNs in the lateral striatum were reconstructed using Neurolucida (MicroBrightField, Colchester, VT). Dendritic spine density was determined on segments of secondary or tertiary dendrites at distances between 70 and 90 μm from the soma, on second or third order dendrites. Segments from three different primary basal dendrites per neuron and 5–6 neurons per animal were analyzed to compute an average dendritic spine density for a given animal. Dendritic spine length was measured in >50 spines/animal of Cbln4 knockout and wild-type mice.
Statistical analysis
For dendritic analyses of MSNs, the spine density/animal values were compared by means of two tailed t-tests. The rotarod data were analyzed for statistical significance using Repeated Measures ANOVA. Significance was set at p<0.05. All statistical analyses were performed using SAS Version 9.2 (SAS Institute Inc., Cary, NC).
Results
To dissect the biology of the Cbln family of proteins we first compared the binding of HA-tagged Cbln family members to HEK293 cells transfected with GluRδ1 or GluRδ2 (Fig. 2A) as well as βNrxns (Fig. 2B) or αNrxns (Fig. 2C) using a western blot assay. HA-Cbln1 bound consistently to GluRδ2 as well as GluRδ1, confirming prior studies (Matsuda et al., 2010; Uemura et al., 2010). HA-Cbln2 bound less well to GluRδ2 and weakly to GluRδ1 whereas HA-Cbln4 did not bind to either receptor (Fig. 2A). HA-Cbln1 and HA-Cbln2 also bound to the S4-containing splice variants of Nrxn1–3β with similar avidity and relative specificities (Nrxn1β=Nrxn2β≫Nrxn3β) (Fig. 2B). HA-Cbln4 bound to Nrxns1β and 2β, albeit much weaker than either HA-Cbln1 or HA-Cbln2, but it had minimal binding to Nrxn3β (Fig. 2B). The three Nrxn genes each utilize two distinct promoters to generate the longer α and shorter β isoforms (Missler et al., 1998). Therefore, we next assessed binding of the Cbln family members to the S4-containing splice variants of the three αNrxns (Fig. 2C). HA-Cbln1 and HA-Cbln2 bound to Nrxn1α and Nrxn2α, but not to Nrxn3α (Fig. 2C). As with the βNrxns, HA-Cbln4 bound to Nrxn1α and 2α, but it did so less avidly than HA-Cbln1 and HA-Cbln2 and like the latter two ligands, it did not bind to Nrxn3α (Fig. 2C). In sum these data confirm and expand prior studies on Cbln family binding to proteins of the Nrxn family (Uemura et al., 2010; Joo et al., 2011) and additionally show that Cbln4 does not exhibit strong interactions with receptors that bind Cbln1 and Cbln2.
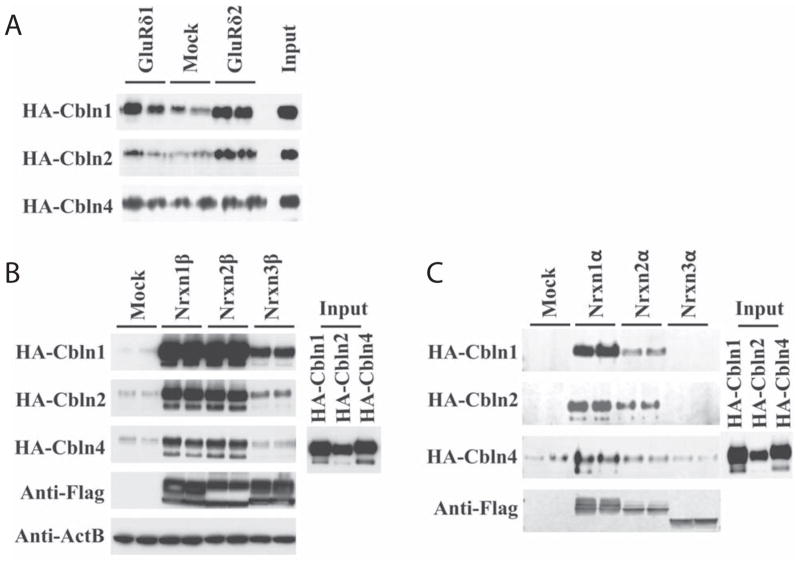
Fig. 2.
Comparison of binding of HA-tagged Cbln family members to GluRδ1, GluRδ2, and α- and β-neurexins. (A) HEK293T cells were transfected with GluRδ1, GluRδ2 or empty vector (Mock). Cells were subsequently exposed to conditioned medium (Input) containing HA-Cbln1, HA-Cbln2, or HA-Cbln4 for 4 h. After washing with medium, cells were lysed with sample buffer and subjected to SDS-PAGE and immunoblotted with anti-hemagglutinin (HA) antiserum. HA-Cbln1 binds both GluRδ1 and GluRδ2. HA-Cbln2 binds to GluRδ2 but weakly or not at all to GluRδ1. HA-Cbln4 does not bind GluRδ1 or GluRδ2. (B) HEK293T cells were transfected with Nrxn1β, Nrxn2β, Nrxn3β or empty vector (Mock). Then cells were incubated with conditioned medium (Input) containing HA-Cbln1, HA-Cbln2, or HA-Cbln4 for 4 h. After washing with medium, cells were lysed with sample buffer and subjected to SDS-PAGE and immunoblotted with anti-hemagglutinin (HA), anti-Flag or anti-β-actin (ActB) antisera. Compared with mock transfection, both Cbln1 and Cbln2 show binding to Nrxn1β, Nrxn2β and Nrxn3β whereas HA-Cbln4 binds to Nrxn1β and Nrxn2β but not Nrxn3β. (C) HEK293T cells were transfected with Nrxn1α, Nrxn2α, Nrxn3α or empty vector (Mock). Then cells were incubated with conditioned medium (Input) containing HA-Cbln1, HA-Cbln2, or HA-Cbln4 for 4 h. After washing with medium, cells were lysed with sample buffer and subjected to SDS-PAGE and immunoblotted with anti-hemagglutinin (HA) or anti-Flag tag antisera. Compared with mock transfection, both HA-Cbln1 and HA-Cbln2 show binding to Nrxn1α and Nrxn2α, but not Nrxn3α, whereas HA-Cbln4 binds only to Nrxn1α.
The lack of interaction of Cbln4 with GluRδ2 and its weaker interactions with α and βNrxns suggested it has other binding partners. Therefore, we undertook a search for additional membrane proteins that interacted with the Cbln family members using a candidate approach in the binding assay described above. Several criteria were used in selecting candidate membrane proteins. First they should be expressed in neurons. Second, as the Cbln family is characterized by the presence of a complement C1q domain that is also found in some cytokines, we included proteins known to bind this type of motif or that contained immunoglobulin-like domains. The proteins surveyed included (amongst others), the complement C1q binding glycoprotein, gC1qBP, the immunoglobulin gamma Fc binding protein, Fcgr1, the immunoglobulin domain-containing proteins, contactin-3 (Cntn3), DCC and Robo1 and the cytokine receptor motif-containing tyrosine phosphatase, Ptprg. With one exception, all of the membrane proteins tested did not show significant interactions with any of the Cbln family members (Fig. 3A and data not shown). However, Cbln4 but not Cbln1 or Cbln2 bound robustly to the netrin receptor, DCC (Fig. 3A). The binding of Cbln4 to DCC was proportionately much higher than the binding of Cbln1 to GluRδ2 and more akin to binding with Nrxns. DCC is the prototype of a family of receptors that have been implicated in diverse developmental processes (Fazeli et al., 1997; Mehlen et al., 1998; Jiang et al., 2003; Cole et al., 2007). As neogenin also binds netrin and has the greatest sequence homology to DCC we assessed whether it bound to Cbln4 (Fig. 3B). Neither Cbln4 (Fig. 3B) nor Cbln1 or Cbln2 (data not shown) bound to Neo1. We next determined the binding specificities of HA-Cbln1, 2 and 4 to other DCC family members, to establish not only whether Cbln4 bound to additional DCC-like proteins but also if Cbln1 and Cbln2 bound to other DCC-related receptors. As with Neo1, none of the Cbln family members bound to Igdcc3, Igdcc4 or protogenin (prtg, a.k.a. Igdcc5) (Fig. 3C). Thus, Cbln4 uniquely bound to the netrin receptor, DCC. The Slit receptor, Robo-1, was amongst the other proteins tested. Robo-1 is reported to bind DCC (Stein and Tessier-Lavigne, 2001), but it did not bind Cbln4 (Fig. 3C). Netrin can also interact with Unc5 family members (Geisbrecht et al., 2003; Krauss, 2010; Leonardo et al., 1997). However, HA-Cbln4 did not bind to Unc5B (Fig. 3D). These data further underscore the specificity of the interaction of Cbln4 with DCC.
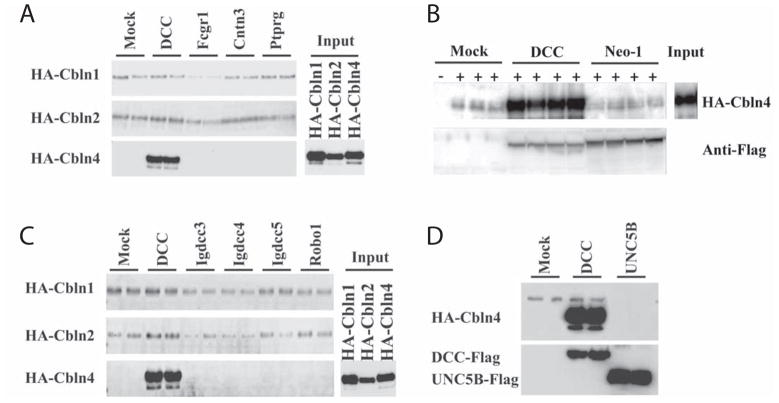
Fig. 3.
HA-Cbln4 but not HA-Cbln1 or HA-Cbln2 binds to DCC. (A) HEK293T cells were transfected with DCC, Fcgr1, Cntn3, Ptprg or empty vector (Mock). Then cells were incubated with conditioned medium (Input) containing HA-Cbln1, HA-Cbln2, or HA-Cbln4 for 4 h. After washing with medium, cells were lysed with sample buffer and subjected to SDS-PAGE and immunoblotted with anti-hemagglutinin (HA) antiserum. HA-Cbln4 bound to DCC but not other membrane proteins. Neither HA-Cbln1 nor HA-Cbln2 bound to any of the membrane proteins tested. (B) Neogenin (Neo-1) is not a receptor for Cbln4. HEK-293 cells were transfected with empty vector (Mock), DCC or Neo-1 containing a C-terminal FLAG tag. Cells were subsequently exposed to conditioned medium from mock transfected (−) or HA-Cbln4-transfected (+) cells (Input) for 4 hours, washed with medium, lysed in loading buffer, subjected to SDS-PAGE and immunoblotted with anti-hemagglutinin (HA) or anti-Flag tag antisera. Whereas HA-Cbln4 bound to DCC it did not bind to cells transfected with Neo-1. (C) Cbln family members do not bind to other DCC family members. HEK293T cells were transfected with DCC, Igdcc3, Igdcc4, Igdcc5, Robo1 or empty vector (Mock). Cells were subsequently exposed to conditioned medium (Input) containing HA-Cbln1, HA-Cbln2, or HA-Cbln4 for 4 h. After washing with medium, cells were lysed with sample buffer and subjected to SDS-PAGE and immunoblotted with anti-hemagglutinin (HA) antisera. HA-Cbln4 bound to DCC but no other proteins whereas HA-Cbln1 and HA-Cbln2 did not bind to any of the membrane proteins tested. (D) UNC5B is not a receptor for Cbln4. HEK-293 cells were transfected with empty vector (Mock), DCC or UNC5B and subsequently exposed to conditioned medium containing HA-Cbln4 (Input) for 4 hours, washed with medium, lysed in loading buffer, subjected to SDS-PAGE and immunoblotted with anti-hemagglutinin (HA) or anti-Flag tag antisera. Whereas HA-Cbln4 bound to DCC it did not bind to cells transfected with UNC5B.
To establish whether netrin-1 and Cbln4 compete for binding to DCC we performed a competition-binding assay using HA-Cbln4 and purified recombinant netrin-1. DCC-expressing cells were pre-treated for 30 minutes with 1ml of tissue culture medium containing either netrin-1 (1μg/ml with 50μg/ml BSA carrier) or carrier BSA (50μg/ml) alone. Subsequently an additional 1ml of medium was added that contained increasing dilutions of HA-Cbln4 and either netrin-1 at a final concentration of 1μg/ml or BSA carrier and the cells incubated for an additional 3.5 hours. Subsequently cell-bound HA-Cbln4 was measured by western blotting as in prior assays. HA-Cbln4 binding to DCC was observed at all dilutions of HA-Cbln4 tested when compared with binding to mock transfected cells (Fig. 4A). The presence of 1μg/ml netrin-1 consistently reduced HA-Cbln4 binding to DCC when compared to its binding in the presence of BSA carrier alone (Fig. 4A). To extend this finding we performed a binding assay in which HA-Cbln4 input was kept constant but the concentration of netrin-1 was varied (Fig. 4B). Netrin-1 showed a dose-dependent inhibition of HA-Cbln4 binding. Indeed, at 3μg/ml netrin-1, Cbln4 binding was completely blocked.
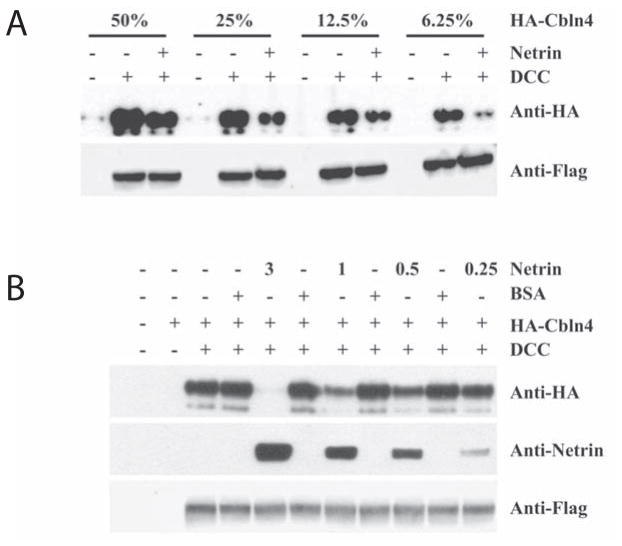
Fig. 4.
Netrin-1 competes with HA-Cbln4 for DCC binding. (A) HEK293T cells were transfected with DCC (+) or empty vector (−). Cells were subsequently exposed to varying dilutions (as a %) of conditioned medium containing HA-Cbln4 with (+) or without (−) netrin-1 (1μg/ml). Subsequently cells were harvested and processed for immunoblotting as in Figure 2. The presence of netrin-1 reduced HA-Cbln4 binding to DCC at all dilutions of HA-Cbln4 tested. (B) Cells were transfected with DCC (+) or empty vector (−). Cells were subsequently pre-incubated with varying concentrations of netrin-1 (3, 1, 0.5, 0.25 or 0 μg/ml) or BSA carrier for 30 min, and then incubated for an additional 3.5 hours with conditioned medium containing HA-Cbln4 and the appropriate level of netrin-1 or BSA. Netrin-1 competed with HA-Cbln4 for DCC binding in a dose-dependent manner.
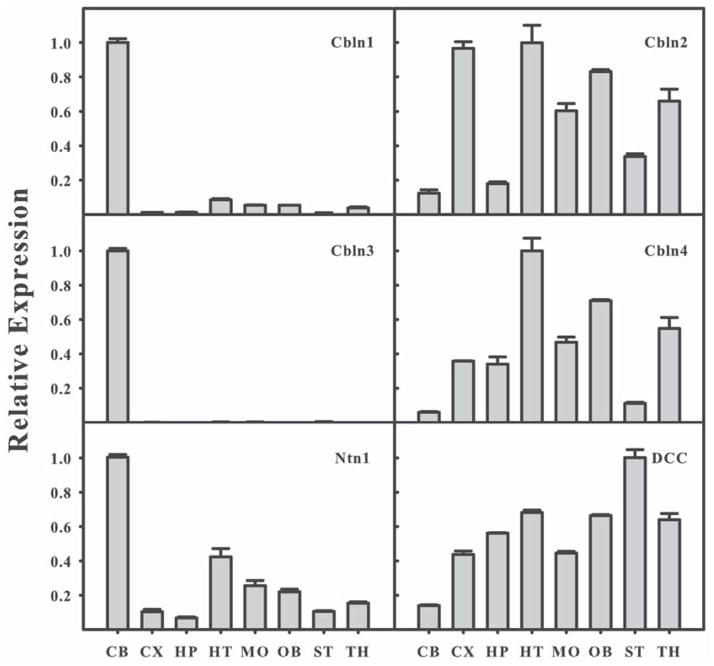
Although Cbln4 and netrin bind competitively to DCC it is possible that they do not coexist in the same location in brain and do not compete functionally. Therefore we assessed the levels of mRNA of all 4 Cbln family members as well as netrin-1 and DCC in various regions of adult brain using qPCR. In brain regions critical to this investigation, DCC and netrin had inverse expression patterns with cerebellum (CB) showing the highest levels of netrin and the lowest levels of DCC mRNA whereas striatum (ST) had the highest levels of DCC and low levels of netrin mRNA (Fig. 5). Cbln4 was expressed at low levels in both cerebellum and striatum but in agreement with a prior study (Miura et al., 2006), it is expressed at relatively high levels in thalamus (TH, Fig. 5). This opens the possibility that Cbln4 may be expressed in the thalamic neurons that project to striatum, the location of the highest levels of DCC.
Fig. 5.
Relative expression levels of mRNAs of Cbln1, Cbln2, Cbln3, Cbln4, Ntn1 and DCC in different adult mouse brain regions. Levels of mRNAs were determined by qPCR. The abundance of mRNA was normalized to β-actin and is expressed as the magnitude of the change in abundance relative to the brain region with the highest level for that transcript. CB, cerebellum; CX, cerebral cortex; HP, hippocampus; HT, hypothalamus; MO, medulla oblongata; OB, olfactory bulb; ST, striatum; TH, thalamus. Results expressed as mean ±SE of measurements of 3 samples from independent mice.
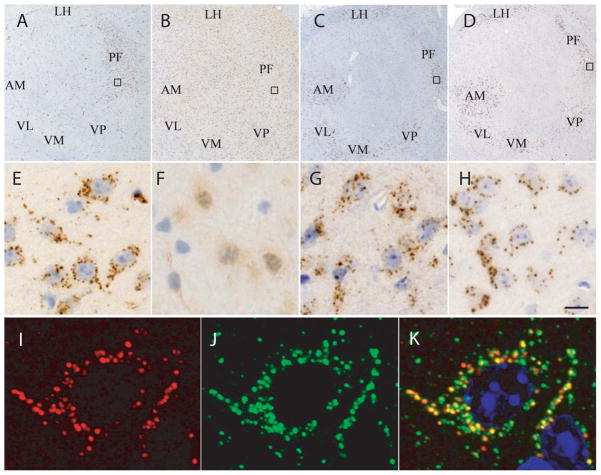
To compare the location of Cbln1 and Cbln4 in thalamus we performed immunohistochemistry. As the commercial anti-Cbln4 antiserum cross-reacted with Cbln1 but not other Cbln family members in our hands (data not shown) we used Cbln1-null mice when localizing Cbln4-ir. Prominent punctate Cbln4-ir (panels A, E) and Cbln1-ir (Panels C, G) was detected in neurons within the parafascicular nucleus (PF) of the thalamus (Fig. 6). As with Cbln1-ir (Kusnoor et al., 2010) Cbln4-ir is present in the majority of PF-neurons. However, whereas Cbln4-ir was confined to PF-neurons Cbln1-ir was also observed in other regions of thalamus (Fig. 6C). No Cbln4-ir was seen in thalamus of Cbln1/Cbln4-double null mice (panels B, F of Fig. 6) establishing that the anti-Cbln4 antiserum does not cross-react with additional unidentified proteins. As with Cbln1-ir (Wei et al., 2007) Cbln4-ir was coincident with staining for the lysosome marker, cathepsin D (Fig. 6I–K) suggesting that both are trafficked through the endolysosomal compartment (Wei et al., 2009). Notably, loss of Cbln4 did not markedly influence the level of Cbln1-ir the way loss of Cbln3 does in the cerebellum (Bao et al., 2006; Wei et al., 2007) (Fig. 6D, 6H) neither did it appear to influence the subcellular localization of Cbln1 in these neurons (Fig. 6H). Thus Cbln1 and Cbln4 are present in the same subcellular compartments of the thalamic neurons that project axons to medium spiny neurons in striatum.
Fig. 6.
Localization of Cbln4-like and Cbln1-like immunoreactivities in thalamus. As the anti-Cbln4 antibody used in this experiment cross-reacted with Cbln1 in our hands, the brains from Cbln1-null mice were used in the immunohistochemistry studies to localize the expression of Cbln4. (A) In sagittal sections of adult Cbln1-null moue brains, Cbln4-like immunoreactivity is observed in a punctate pattern (E is enlargement of box in A) in the cytoplasm of thalamic neurons. The most prominent staining is seen in neurons of the parafascicular nucleus of thalamus (PF). (B) Specificity of the anti-Cbln4 antiserum is shown by the lack of Cbln4-like immunoreactivity in thalamic neurons from Cbln1-/Cbln4-double null mice (F is enlargement of box in B). (C) Compared with the localization of Cbln4, Cbln1-like immunoreactivity in a wild type mouse is more widespread in the thalamus with staining observed in not only PF neurons but also neurons in the ventral (VL, VM, VP) and anterior thalamus (AM). Like Cbln4, Cbln1-like immunoreactivity is punctate (G is enlargement of box in C) and of roughly the same incidence as for Cbln4. (D) In Cbln4-null mice there is no overt change in the level or distribution of Cbln1-like immunoreactivity (H is enlargement of box in D). (I–K) Double immunofluorescence labeling was performed on sections from thalamus of Cbln1-null mice using the anti-Cbln4 antiserum (red, I and K) and an antiserum to the lysosome marker cathepsin D (green, J and K). Merged confocal images (K) revealed that the Cbln4 vesicle-like staining almost always co-localizes with cathepsin D (yellow staining). Scale bar: A–D, 360 μm; E–H, 20 μm; I–K, 4 μm. AM, anteromedial thalamic nucleus; LH, lateral habenular nucleus; PF, parafascicular nucleus; VL, ventrolateral thalamic nucleus; VM, ventromedial thalamic nucleus; VP, ventral posterior tier thalamic nuclei.
As Cbln1 and Cbln4 are likely co-expressed in PF neurons of the thalamus we assessed what the consequences of co-expression of these two family members had on binding specificity. As HA-Cbln4 binds to DCC but not Nrxn3β, whereas HA-Cbln1 has the opposite specificity we focused on these two receptors. Co-expression of Cbln1 with HA-Cbln4 reduced, but did not eliminate HA-Cbln4 binding to DCC but HA-Cbln4 was now recruited to Nrxn3β (Fig. 7). Thus, heteromeric complexes of Cbln1 and Cbln4 have distinct binding preferences compared to the homomeric complexes.
Fig. 7.
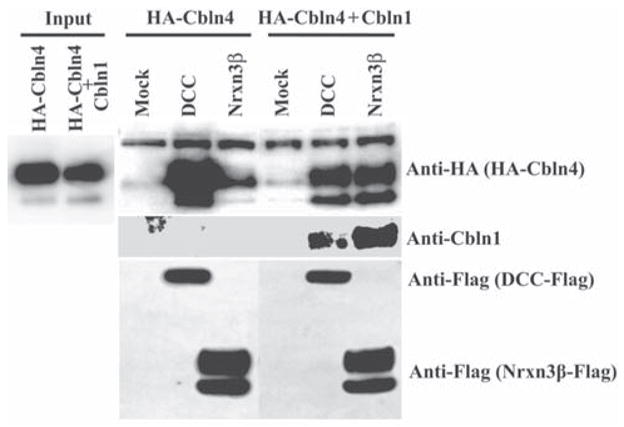
Co-expression of Cbln1 with Cbln4 alters the receptor binding specificity of HA-Cbln4. HEK293T cells were transfected with DCC, Nrxn3β or empty vector (Mock). Cells were subsequently exposed to conditioned medium (Input) derived from HEK293 cells co-transfected with either HA-Cbln4 and mock plasmid or HA-Cbln4 and Cbln1 for 4 h. After washing with medium, cells were lysed with sample buffer and subjected to SDS-PAGE and immunoblotted with anti-hemagglutinin (HA), anti-Cbln1 or anti-Flag tag antisera. HA-Cbln4 shows strong binding to DCC and minimal binding to Nrxn3β. Co-expression of Cbln1 with HA-Cbln4 alters the binding specificity of HA-Cbln4 by reducing its association with DCC and promoting its association with Nrxn3β.
To pursue the possibility of biological interactions amongst Cbln4, netrin and DCC we generated Cbln4-null mice using standard techniques and the strategy summarized in Fig. 1A. The elimination of the gene and the absence of the cognate mRNA were confirmed by Southern blotting (Fig. 1B) and qPCR (Fig. 1C), respectively. Using qPCR we also showed no transcriptional compensation amongst family members in the brains of Cbln1-or Cbln4-null mice (Fig. 1C), precluding this potential confounder and supporting the immunohistochemistry data (Fig. 6D, H).
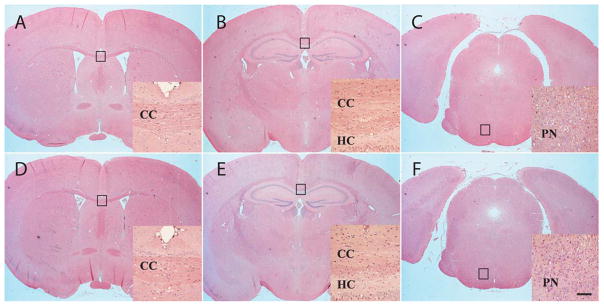
Cbln4 knockout mice are fertile, have normal life spans and have no overt anatomical abnormalities. In marked contrast, netrin-1-null mice and DCC-null mice exhibit perinatal mortality and have various neuroanatomical deficits, notably the absence of a corpus callosum, hippocampal commisure and pontine nucleus (Serafini et al., 1996; Fazeli et al., 1997). When compared to wild type littermates (Fig. 8, panels A–C) Cbln4 knockout mice (Fig. 8, panels D–F) have a normal corpus callosum (CC), hippocampal commisure (HC) and pontine nucleus (PN). In addition, we see no other gross neuroanatomical abnormalities in Cbln4-null mice (Fig. 8).
Fig. 8.
Cbln4-null mice have no obvious neuroanatomical abnormalities. Representative matched coronal sections from wild type (A–C) or Cbln4-null (D–E) mice were stained with hematoxylin and eosin. The sections were selected to highlight regions of brain (boxes in A–F) that have anomalies in netrin- and DCC-null mice. Compared with wild-type (A–C) mice, Cbln4-null mice (D–F) show no overt neuroanatomical defects. Furthermore, specific structures such as corpus callosum (CC in D, E), hippocampal commissure (HC, in E), and pontine nuclei (PN in F) that are lacking in netrin- and DCC-null mice are present and indistinguishable from wild type littermates in Cbln4-null mice. Scale bar, 750 μm for A–F and 100 μm for the insets.
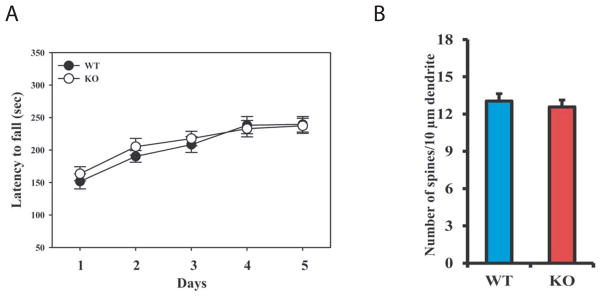
Netrin-1- and DCC-null mice have abnormal body posture and limb flexion (Serafini et al., 1996; Fazeli et al., 1997). However, even when tested on the accelerating rotarod assay Cbln4-null mice are indistinguishable from wild type littermates (P>0.5, n=9) (Fig. 9A). Cbln1 is expressed in PF neurons and its deficiency results in increased synaptic spine density on dendrites of MSN neurons of striatum (Kusnoor et al., 2010). As Cbln4-ir is also detected in the same population of PF neurons and Cbln1 and Cbln4 can form heteromeric complexes in vitro and change receptor-binding specificity, we assessed whether loss of Cbln4 mimicked loss of Cbln1 in the thalamo-striatal pathway. As seen in Fig. 9B, there is no difference in MSN dendritic spine densities of Cbln4-null mice and wild type littermates. Thus, loss of Cbln4 does not mimic loss of Cbln1 in the same neuronal population.
Fig. 9.
Cbln4-null mice do not exhibit the same deficits as Cbln1-null mice. (A) Cbln4-null mice (open circles, KO) are indistinguishable from wild type (filled circles, WT) gender-matched littermates (p>0.5, n=9/genotype) on the accelerating rotarod assay. Data is mean ± SEM. (B) There is no difference in dendritic spine densities on striatal medium spiny neurons of Cbln4-null (KO) compared to wild type (WT) littermates (p>0.1, n=4/genotype).
Discussion
The data presented here support the hypothesis that Cbln family members serve distinct functions in vivo through their differential interactions with various classes of membrane receptors and signaling pathways. Cbln1 and Cbln2 bind avidly to the S4-containing splice variants of α and βNrxns as well as to GluRδ1 and GluRδ2 whereas Cbln4 binds weakly to α-and βNrxns 1 and 2 and not at all to α-and βNrxn3, GluRδ1 and GluRδ2. In contrast, Cbln4 binds avidly to the netrin receptor, DCC whereas Cbln1 and Cbln2 do not bind at all to DCC. Therefore, Cbln1 and Cbln2 may predominantly contribute to the biology of Nrxns whereas Cbln4 may participate in DCC-dependent processes. Furthermore, our binding data imply that these two apparently distinct signaling pathways have crosstalk that is controlled by the subunit composition of the Cbln-containing complexes.
Recombinant Cbln1 complexes are composed of a dimer of trimers, where the trimers are assembled through interactions between the C1q domains in the C-terminus of the proteins and subsequently two trimers are covalently linked together via conserved cysteine residues in the N-terminal region (Bao et al., 2005). We showed previously that Cbln family proteins also associate with one another and form heteromeric complexes in vitro (Pang et al., 2000; Bao et al., 2005) and in the case of Cbln1 and Cbln3 they exist in heteromeric complexes in vivo (Bao et al., 2006). Therefore, our present finding that Cbln1–Cbln4 heteromeric complexes bind, albeit weaker than the respective homomeric complexes, to both Nrxn3β and DCC has profound implications as it establishes for the first time that the subunit composition of the complex determines binding specificity to receptors in distinct signaling pathways. In essence, Cbln1 has a dominant function over Cbln4 by decoying it to Nrxns and nullifying its interaction with DCC. As Cbln1 and Cbln4 are likely co-expressed in PF neurons of wild type mice they are expected to exist in both homomeric and heteromeric complexes with different binding affinities for Nrxns and DCC. Presumably, the relative concentrations of Cbln1 and Cbln4 in the PF neurons determine the ratio of Cbln1 and Cbln4 homomeric complexes to Cbln1–Cbln4 heteromeric complexes. By extension, disruption of this stoichiometry, as occurs in Cbln1- or Cbln4-null mice, is predicted to alter the ratio of the complexes and thereby influence multiple downstream signaling pathways. In Cbln1-null mice signaling through Nrxns should decrease while signaling through DCC should increase as the proportion of Cbln4 homomeric complexes would increase. That is, the Cbln1-null mouse is functionally a Cbln4 hypermorphic animal in the thalamo-striatal tract.
Although Cbln4 binds to DCC in vitro and is competed by netrin-1, the Cbln4-null mouse does not recapitulate the major features of DCC-null or netrin-1-null mice, which exhibit perinatal mortality, abnormal body posture and limb flexion and several substantial neuroanatomical defects such as absence of a corpus callosum and pontine nucleus (Serafini et al., 1996; Fazeli et al., 1997). That Cbln4 deficiency does not phenocopy the netrin-1-null mouse may be explained in a number of ways. First, an obvious distinction between the two ligands is that whereas netrin-1 binds to a number of receptor proteins that include DCC, neogenin, and Unc5 family members (Serafini et al., 1994; Keino-Masu et al., 1996; Ackerman et al., 1997; Bennett et al., 1997; Leonardo et al., 1997; Corset et al., 2000; Stein and Tessier-Lavigne, 2001; Nikolopoulos and Giancotti; 2005; Shipp and Hsieh-Wilson, 2007; Andrews, et al., 2008; Ly et al., 2008; Liu et al., 2009), Cbln4 exclusively binds to DCC. Thus, loss of netrin is likely to affect signaling through diverse receptors and have a more severe phenotype. Second, Cbln4 may be a netrin antagonist, and therefore its elimination may produce a netrin gain-of-function rather than loss-of-function phenotype that is limited to DCC signaling. Third, Cbln4 is predominantly expressed in the adult nervous system whereas netrin and DCC are expressed more prominently in the developing organism. Therefore, Cbln4 and netrin may not be present in the same location, as suggested by our qPCR data (Fig. 5) and might exert quite distinct DCC-dependent responses. In addition, the principal focus of netrin/DCC biology has focused upon axon guidance and cell migration during development and although they have been implicated in axon branching, synaptogenesis, oligodendroglia development and maturation (Lai Wing Sun et al., 2011, for review), their role in the adult nervous system is less clear and so more subtle shared phenotypes may be present in the adult brains of the various knockout strains of mice.
As Cbln4 has its highest level of expression in PF-thalamic neurons that project to the striatum, if DCC is involved in its biological function in the adult brain it should either be expressed in these neurons (analogous to presynaptic Nrxns) or their targets, striatal MSNs (analogous to postsynaptic GluRδ2). Using in situ hybridization, Livesey and Hunt (Livesey and Hunt, 1997) localized netrin to sporadic large (presumptively cholinergic) neurons whereas DCC was expressed in numerous smaller neurons throughout the adult striatum that likely include MSNs (Livesey and Hunt, 1997). Our qPCR data also show that striatum is the site of the highest levels of DCC mRNA. Therefore, DCC is expressed in the appropriate target location in the adult brain. However, the function of DCC and netrin in this brain region in the adult is less clear although they have been implicated in the migration of striatal neurons during development (Hamasaki et al., 2001). In the adult nervous system netrin is expressed at its highest level in the substantia nigra pars compacta (Livesey and Hunt, 1997) and netrin and DCC have been implicated in the control of dopaminergic neuron number and arborization during development (Xu et al., 2010). Axons from dopaminergic neurons in the substantia nigra pars compacta synapse onto the same MSNs that are innervated by the PF neurons (Dubé et al., 1988; Parent and Hazrai, 1995). Therefore, it is conceivable that Cbln4 released from PF neurons and netrin released from dopaminergic neurons in the substantia nigra pars compacta interact with DCC on MSNs and mediate some synaptic function in the adult. This circuit is critical for the control of movement and its perturbation, such as occurs in Parkinson’s disease where dopaminergic neurons of the substantia nigra pars compacta selectively degenerate (Fahn, 2003), cause myriad locomotor symptoms (Fahn, 2003).
Interestingly, netrin has also been implicated in development of the thalamo-cortical tract (Braisted et al., 2000). Besides their profuse connections to the striatum/basal ganglia, PF neurons also project to the frontal and cingulate cortices (Marini et al., 1996; Vercelli et al., 2003). Furthermore Cbln2 is also prominently expressed in both the frontal and cingulate cortices (Miura et al., 2006), implying that Cbln family members may have both pre- and postsynaptic locations in the thalamo-cortical pathway. It also implies that Cbln4, and other Cbln family members may play a role in additional aspects of PF function, such as sensory and motor functions (Uhlrich et al., 1995; Schlag-ray and Schlag, 1984; Marini et al., 1999; Vercelli et al., 2003). The Cbln4-null mice reported here provide a new potential tool to investigate the molecular mechanisms that control synaptic function in these regions of the nervous system and to dissect the role of DCC/netrin signaling in adult brain.
Table 1.
Primer and probe sequence information for qPCR
| Gene name | Primer & probe sequences (5′ to 3′) | Reference sequence | |
|---|---|---|---|
| Cbln1 | F-primer | GGTGAAAGTCTACAACAGACAGACCAT | NM_019626 |
| R-primer | CGGCGAAGGCTGAAATCA | ||
| probe | CAGGTGAGCCTCATGTTGAACGGGT | ||
| Cbln2 | F-primer | GTGGTCAAAGTGTACAACAGACAAACT | NM_0172633 |
| R-primer | CCGGCAAATGCAGAGATCA | ||
| probe | TCCAGGTCAGCTTAATGCAGAATGGCTACC | ||
| Cbln3 | F-primer | GGTACCAGTGGTGCCATCTACTT | NM_019820 |
| R-primer | GCCCGAGGTCCGATCAA | ||
| probe | CCAGGTACTGGTCAATGAGGGCGAGG | ||
| Cbln4 | F-primer | CCAACCACGAGCCATCTGA | NM_175631 |
| R-primer | AGATTCCTTTCCTCGGTGCC | ||
| probe | CATCATTTACTTTGATCAGATCCTGGTTAACGTGGGT | ||
| Dcc | F-primer | AAAGTAGTGCCCAGCTGATTGTC | NM_007831 |
| R-primer | AAGGGAGGATGCTGGAGCTT | ||
| probe | CCAAGCCTGCCATC | ||
| Ntn1 | F-primer | CGAGACATGGGCAAGCCTAT | NM_008744 |
| R-primer | GGCAATCACAGGCTTTGCA | ||
| probe | CCCACCGGAAGGC |
Acknowledgments
This work was supported in part by the NCI Cancer Center Support Grant CA 21765, National Institutes of Health grants NS040361 and NS042828, and ALSAC (American Lebanese Syrian Associated Charities) to J. I. M. and NS44282 and MH-077298 and National Parkinson Foundation Center of Excellence at Vanderbilt to A. Y. D. We thank R. J. Smeyne, Department of Developmental Neurobiology, St. Jude Children’s Research Hospital, for advice with neuroanatomy. We thank the Hartwell Center for Bioinformatics and Biotechnology at St Jude Children’s Research Hospital for all DNA sequencing and synthesis.
Abbreviations
- ANOVA
one-way analysis of variance
- BAC
Bacterial artificial chromosome
- BSA
bovine serum albumin
- Cbln
Cerebellin precursor protein
- Cntn
contactin
- DCC
deleted in colorectal cancer
- PF
parafascicular nucleus
- MSN
medium spiny neurons
- ES cells
Embryonic stem cells
- Fcgr
Fc fragment of IgG receptor
- GluR
glutamate receptor
- HA
hemagglutinin
- Igdcc
immunoglobulin superfamily, DCC subclass
- Neo1
Neogenin1
- Nrxn
neurexin
- PFA
paraformaldehyde
- Prtg
protogenin homolog
- Ptprg
protein tyrosine phosphatase receptor type G
- qPCR
quantitative real time PCR
- Robo
roundabout
- RT-PCR
reverse transcriptase-polymerase chain reaction
- TH
thalamus
- Unc5b
unc-5 homolog B
- WT
wild type
Footnotes
None of the authors have any financial interests or conflicts of interest in the data presented in the manuscript.
References
- Ackerman SL, Kozak LP, Przyborski SA, Rund LA, Boyer BB, Knowles BB. The mouse rostral cerebellar malformation gene encodes an UNC-5-like protein. Nature. 1997;386:838–842. doi: 10.1038/386838a0. [DOI] [PubMed] [Google Scholar]
- Andrews GL, Tanglao S, Farmer WT, Morin S, Brotman S, Berberoglu MA, Price H, Fernandez GC, Mastick GS, Charron F, Kidd T. Dscam guides embryonic axons by Netrin-dependent and independent functions. Development. 2008;135:3839–3848. doi: 10.1242/dev.023739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Meguro H, Kushiya E, Takayama C, Inoue Y, Mishina M. Selective expression of the glutamate receptor channel delta 2 subunit in cerebellar Purkinje cells. Biochem Biophys Res Commun. 1993;197:1267–1276. doi: 10.1006/bbrc.1993.2614. [DOI] [PubMed] [Google Scholar]
- Bao D, Pang Z, Morgan MA, Parris J, Rong Y, Li L, Morgan JI. Cbln1 is essential for interaction-dependent secretion of Cbln3. Mol Cell Biol. 2006;26:9327–9337. doi: 10.1128/MCB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao D, Pang Z, Morgan JI. The structure and proteolytic processing of Cbln1 complexes. J Neurochem. 2005;95:618–629. doi: 10.1111/j.1471-4159.2005.03385.x. [DOI] [PubMed] [Google Scholar]
- Bennett KL, Bradshaw J, Youngman T, Rodgers J, Greenfield B, Aruffo A, Linsley PS. Deleted in colorectal carcinoma (DCC) binds heparin via its fifth fibronectin type III domain. J Biol Chem. 1997;272:26940–26946. doi: 10.1074/jbc.272.43.26940. [DOI] [PubMed] [Google Scholar]
- Braisted JE, Catalano SM, Stimac R, Kennedy TE, Tessier-Lavigne M, Shatz CJ, O’Leary DD. Netrin-1 promotes thalamic axon growth and is required for proper development of the thalamocortical projection. J Neurosci. 2000;20:5792–5801. doi: 10.1523/JNEUROSCI.20-15-05792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitrago MM, Schulz JB, Dichgans J, Luft AR. Short and long-term motor skill learning in an accelerated rotarod training paradigm. Neurobiol Learn Mem. 2004;81:211–216. doi: 10.1016/j.nlm.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Cole SJ, Bradford D, Cooper HM. Neogenin: A multi-functional receptor regulating diverse developmental processes. Int J Biochem Cell Biol. 2007;39:1569–1575. doi: 10.1016/j.biocel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Corset V, Nguyen-Ba-Charvet KT, Forcet C, Moyse E, Chédotal A, Mehlen P. Netrin-1-mediated axon outgrowth and cAMP production requires interaction with adenosine A2b receptor. Nature. 2000;407:747–750. doi: 10.1038/35037600. [DOI] [PubMed] [Google Scholar]
- Dubé L, Smith AD, Bolam JP. Identification of synaptic terminals of thalamic or cortical origin in contact with distinct medium-size spiny neurons in the rat neostriatum. J Comp Neurol. 1988;267:455–471. doi: 10.1002/cne.902670402. [DOI] [PubMed] [Google Scholar]
- Erickson AH, Blobel G. Early events in the biosynthesis of the lysosomal enzyme cathepsin D. J Biol Chem. 1979;254:11771–11774. [PubMed] [Google Scholar]
- Fahn S. Description of Parkinson’s disease as a clinical syndrome. Ann N Y Acad Sci. 2003;991:1–14. doi: 10.1111/j.1749-6632.2003.tb07458.x. [DOI] [PubMed] [Google Scholar]
- Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, Stoeckli ET, Keino-Masu K, Masu M, Rayburn H, Simons J, Bronson RT, Gordon JI, Tessier-Lavigne M, Weinberg RA. Phenotype of mice lacking functional Deleted in colorectal cancer (DCC) gene. Nature. 1997;386:796–804. doi: 10.1038/386796a0. [DOI] [PubMed] [Google Scholar]
- Geisbrecht BV, Dowd KA, Barfield RW, Longo PA, Leahy DJ. Netrin binds discrete subdomains of DCC and UNC5 and mediates interactions between DCC and heparin. J Biol Chem. 2003;278:32561–32568. doi: 10.1074/jbc.M302943200. [DOI] [PubMed] [Google Scholar]
- Hamasaki T, Goto S, Nishikawa S, Ushio Y. A role of netrin-1 in the formation of the subcortical structure striatum: repulsive action on the migration of late-born striatal neurons. J Neurosci. 2001;21:4272–4280. doi: 10.1523/JNEUROSCI.21-12-04272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Pang Z, Bao D, Miyazaki T, Li L, Miura E, Parris J, Rong Y, Watanabe M, Yuzaki M, Morgan JI. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat Neurosci. 2005;8:1534–1541. doi: 10.1038/nn1576. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Liu MT, Gershon MD. Netrins and DCC in the guidance of migrating neural crest-derived cells in the developing bowel and pancreas. Dev Biol. 2003;258:364–384. doi: 10.1016/s0012-1606(03)00136-2. [DOI] [PubMed] [Google Scholar]
- Joo JY, Lee SJ, Uemura T, Yoshida T, Yasumura M, Watanabe M, Mishina M. Differential interactions of cerebellin precursor protein (Cbln) subtypes and neurexin variants for synapse formation of cortical neurons. Biochem Biophys Res Commun. 2011;406:627–632. doi: 10.1016/j.bbrc.2011.02.108. [DOI] [PubMed] [Google Scholar]
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- Krauss RS. Regulation of promyogenic signal transduction by cell-cell contact and adhesion. Exp Cell Res. 2010;316:3042–3049. doi: 10.1016/j.yexcr.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnoor SV, Parris J, Muly EC, Morgan JI, Deutch AY. Extracerebellar role for Cerebellin1: modulation of dendritic spine density and synapses in striatal medium spiny neurons. J Comp Neurol. 2010;518:2525–2537. doi: 10.1002/cne.22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Wing Sun K, Correia JP, Kennedy TE. Netrins: versatile extracellular cues with diverse functions. Development. 2011;138:2153–2169. doi: 10.1242/dev.044529. [DOI] [PubMed] [Google Scholar]
- Leonardo ED, Hinck L, Masu M, Keino-Masu K, Ackerman SL, Tessier-Lavigne M. Vertebrate homologues of C. elegans UNC-5 are candidate netrin receptors. Nature. 1997;386:833–838. doi: 10.1038/386833a0. [DOI] [PubMed] [Google Scholar]
- Liu G, Li W, Wang L, Kar A, Guan KL, Rao Y, Wu JY. DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc Natl Acad Sci USA. 2009;106:2951–2956. doi: 10.1073/pnas.0811083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Hunt SP. Netrin and netrin receptor expression in the embryonic mammalian nervous system suggests roles in retinal, striatal, nigral, and cerebellar development. Mol Cell Neurosci. 1997;8:417–429. doi: 10.1006/mcne.1997.0598. [DOI] [PubMed] [Google Scholar]
- Ly A, Nikolaev A, Suresh G, Zheng Y, Tessier-Lavigne M, Stein E. DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin-1. Cell. 2008;133:1241–1254. doi: 10.1016/j.cell.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini G, Pianca L, Tredici G. Thalamocortical projection from the parafascicular nucleus to layer V pyramidal cells in frontal and cingulate areas of the rat. Neurosci Lett. 1996;203:81–84. doi: 10.1016/0304-3940(95)12266-4. [DOI] [PubMed] [Google Scholar]
- Marini G, Pianca L, Tredici G. Descending projections arising from the parafascicular nucleus in rats: trajectory of fibers, projection pattern and mapping of terminations. Somatosens Mot Res. 1999;16:207–222. doi: 10.1080/08990229970465. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33:1447–1461. doi: 10.1111/j.1460-9568.2011.07638.x. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, Watanabe M, Yuzaki M. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- Mehlen P, Rabizadeh S, Snipas SJ, Assa-Munt N, Salvesen GS, Bredesen DE. The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature. 1998;395:801–804. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- Missler M, Fernandez-Chacon R, Südhof TC. The making of neurexins. J Neurochem. 1998;71:1339–1347. doi: 10.1046/j.1471-4159.1998.71041339.x. [DOI] [PubMed] [Google Scholar]
- Miura E, Iijima T, Yuzaki M, Watanabe M. Distinct expression of Cbln family mRNAs in developing and adult mouse brains. Eur J Neurosci. 2006;24:750–756. doi: 10.1111/j.1460-9568.2006.04950.x. [DOI] [PubMed] [Google Scholar]
- Mugnaini E, Morgan JI. The neuropeptide cerebellin is a marker for two similar neuronal circuits in rat brain. Proc Natl Acad Sci U S A. 1987;84:8692–8696. doi: 10.1073/pnas.84.23.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulos SN, Giancotti FG. Netrin-integrin signaling in epithelial morphogenesis, axon guidance and vascular patterning. Cell Cycle. 2005;4:e131–e135. [PubMed] [Google Scholar]
- Pang Z, Zuo J, Morgan JI. Cbln3, a novel member of the precerebellin family that binds specifically to Cbln1. J Neurosci. 2000;20:6333–6339. doi: 10.1523/JNEUROSCI.20-17-06333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent A, Hazrati LN. Functional anatomy of the basal ganglia. I. The cortico-basal ganglia-thalamo-cortical loop. Brain Res Brain Res Rev. 1995;20:91–127. doi: 10.1016/0165-0173(94)00007-c. [DOI] [PubMed] [Google Scholar]
- Schlag-Rey M, Schlag J. Visuomotor functions of central thalamus in monkey. I. Unit activity related to spontaneous eye movements. J Neurophysiol. 1984;51:1149–1174. doi: 10.1152/jn.1984.51.6.1149. [DOI] [PubMed] [Google Scholar]
- Schmid SM, Hollmann M. Bridging the synaptic cleft: lessons from orphan glutamate receptors. Sci Signal. 2010;3:pe28. doi: 10.1126/scisignal.3136pe28. [DOI] [PubMed] [Google Scholar]
- Serafini T, Kennedy TE, Galko MJ, Mirzayan C, Jessell TM, Tessier-Lavigne M. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell. 1994;78:409–424. doi: 10.1016/0092-8674(94)90420-0. [DOI] [PubMed] [Google Scholar]
- Serafini T, Colamarino SA, Leonardo ED, Wang H, Beddington R, Skarnes WC, Tessier-Lavigne M. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell. 1996;87:1001–1014. doi: 10.1016/s0092-8674(00)81795-x. [DOI] [PubMed] [Google Scholar]
- Shipp EL, Hsieh-Wilson LC. Profiling the sulfation specificities of glycosaminoglycan interactions with growth factors and chemotactic proteins using microarrays. Chem Biol. 2007;14:195–208. doi: 10.1016/j.chembiol.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Stein E, Tessier-Lavigne M. Hierarchical organization of guidance receptors: silencing of netrin attraction by slit through a Robo/DCC receptor complex. Science. 2001;291:1928–1938. doi: 10.1126/science.1058445. [DOI] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Uhlrich DJ, Tamamaki N, Murphy PC, Sherman SM. Effects of brain stem parabrachial activation on receptive field properties of cells in the cat’s lateral geniculate nucleus. J Neurophysiol. 1995;73:2428–2447. doi: 10.1152/jn.1995.73.6.2428. [DOI] [PubMed] [Google Scholar]
- Vercelli A, Marini G, Tredici G. Anatomical organization of the telencephalic connections of the parafascicular nucleus in adult and developing rats. Eur J Neurosci. 2003;18:275–289. doi: 10.1046/j.1460-9568.2003.02743.x. [DOI] [PubMed] [Google Scholar]
- Wei P, Blundon JA, Rong Y, Zakharenko SS, Morgan JI. Impaired Locomotor Learning and Altered Cerebellar Synaptic Plasticity in pep-19/pcp4-Null Mice. Mol Cell Biol. 2011;31:2838–2844. doi: 10.1128/MCB.05208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Smeyne RJ, Bao D, Parris J, Morgan JI. Mapping of Cbln1-like immunoreactivity in adult and developing mouse brain and its localization to the endolysosomal compartment of neurons. Eur J Neurosci. 2007;26:2962–2978. doi: 10.1111/j.1460-9568.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- Wei P, Rong Y, Li L, Bao D, Morgan JI. Characterization of trans-neuronal trafficking of Cbln1. Mol Cell Neurosci. 2009;41:258–273. doi: 10.1016/j.mcn.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Goldman JS, Rymar VV, Forget C, Lo PS, Bull SJ, Vereker E, Barker PA, Trudeau LE, Sadikot AF, Kennedy TE. Critical roles for the netrin receptor deleted in colorectal cancer in dopaminergic neuronal precursor migration, axon guidance, and axon arborization. Neuroscience. 2010;169:932–949. doi: 10.1016/j.neuroscience.2010.05.025. [DOI] [PubMed] [Google Scholar]