Abstract
In metazoans, enhancers of gene transcription must often exert their effects over tens of kilobases of DNA. Over the last decade it has become clear that to do this, enhancers come into close proximity with target promoters with the looping away of intervening sequences. In a few cases proteins that are involved in the establishment or maintenance of these loops have been revealed but how the proper gene target is selected remains mysterious. Chromatin insulators had been appreciated as elements that play a role in enhancer fidelity through their enhancer blocking or barrier activity. However, recent work suggests more direct participation of insulators in enhancer-gene interactions. The emerging view begins to incorporate transcription activation by distant enhancers with large scale nuclear architecture and sub-nuclear movement.
Introduction
Enhancers are regulatory elements that increase the transcriptional output of target genes. In metazoans enhancers and the genes they regulate can be as far as 2 or 3 Mbp distant from each other. This geometry produced lively debates on how the distant enhancers could activate their target genes. Models considered included looping and tracking and variations thereof [1]. The advent of new technologies, including 3C [2] confirmed the establishment of close proximity between enhancers and target genes. In the first example, loop formation between the β-globin locus control region enhancer (LCR) and gene was shown to accompany transcriptional activation [3–5]. While this new information did not rule out the possibility that a tracking mechanism contributes to gene activation by distant enhancers [6;7], it did establish a paradigm that was borne out in numerous other loci where developmentally regulated gene clusters and single genes are activated by a distant enhancer. These include the α-globin gene cluster, TH2, IFNG, MHC class II and IgH loci among others [8].
Genomes also contain insulators that modulate enhancer activity. These elements are protein-DNA complexes that prevent an enhancer from activating a gene when positioned between them and can act as barriers to the inappropriate spread of heterochromatin. Chromatin looping underlies their behavior as well (Figure 1). In vertebrates the only known insulator protein is CTCF, which recruits cohesin to many of its functional sites [9]. The cohesin complex forms a ring to embrace chromosomes during sister chromatid exchange and its role at insulator sites could be similar. Although insulators influence enhancer function and gene expression, these elements were thought be distinct; however, the distinctions are blurring. In this review we will discuss new attributes of enhancers and new direct roles for CTCF insulators in enhancer-promoter interactions and in broadly configuring the genome.
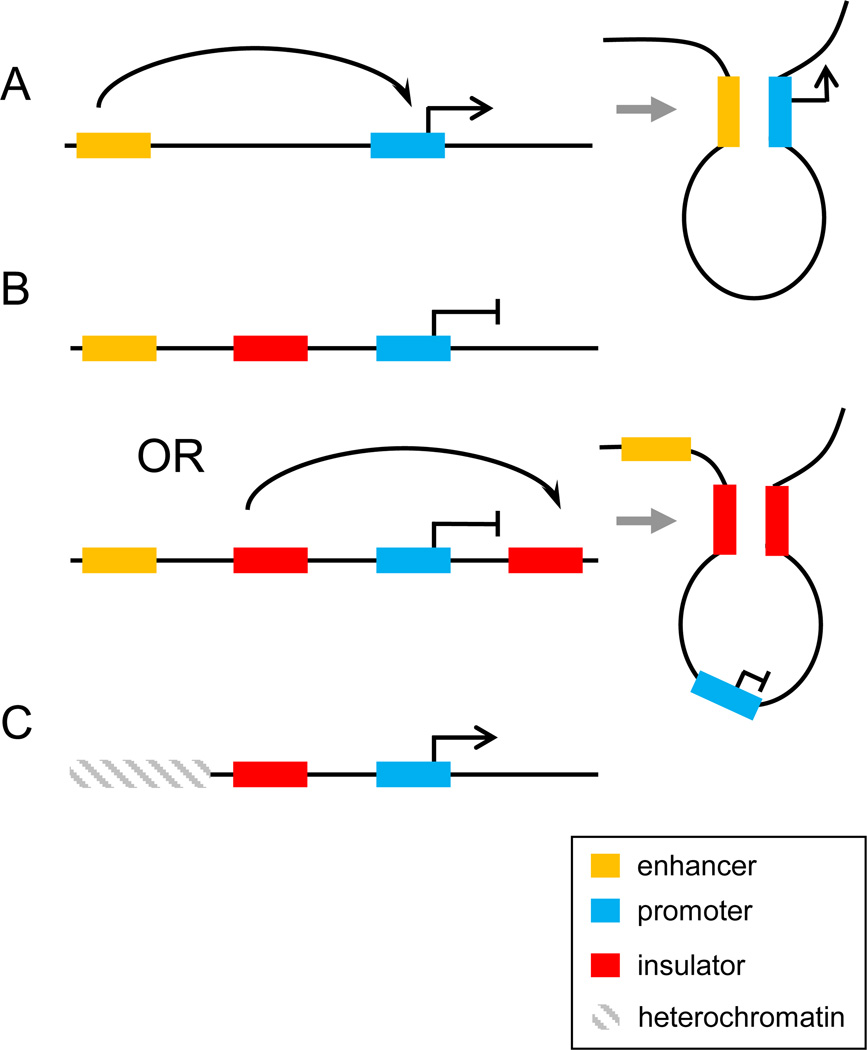
Figure 1. Models of enhancer and insulator function.
A. An enhancer is depicted as activating transcription from a target gene promoter through direct interaction over a large distance by creating a chromatin loop. B. An insulator located between an enhancer and a gene can block promoter-enhancer interaction by acting as a road block to a processive signal from the enhancer or by forming a loop with another insulator located distal to the gene. C. An insulator functions as a barrier to block the spreading of repressive chromatin into an inappropriate locus. This function is depicted as a road block but could also be carried out as in panel 1B by the insulator interacting directly with another insulator to form a loop encompassing the active gene. In all panels, the yellow rectangle is the enhancer and the blue rectangle is the gene. The red rectangle is the insulator. The hatched rectangle represents condensed heterochromatin.
Enhancer loops and functions—an update
Genome profiling of enhancers
Two studies localized putative enhancers genome wide by their signature of CBP/p300 binding and H3K4me1 modification [10;11]. However, discovering the targets of these enhancers is a formidable task. A different approach, Hi-C, has allowed investigators to capture long range interactions genome-wide by combining the classical 3C assay with high-throughput sequencing [12]. The resolution of the method was about 2 Mb but was sufficient to show that long range looping interactions underlie the co-localization of chromosomal domains based on functional state. Increased computing power has improved the resolution of Hi-C. Moreover, one could imagine combining this data with enhancer localization by CBP/p300 signature [10;11] to identify novel enhancers that function by long range interaction and their targets. This would allow an assessment of how general this phenomenon is.
In parallel with Hi-C, a different approach called ChIA-PET (chromatin interaction assay with paired end sequencing) was pioneered to investigate chromatin interactions on a genome-wide scale [13]. ChIA-PET is a ChIP-based assay allowing capture of long range chromatin interactions that are established by a specific protein of choice at high resolution. Fullwood et al. used an antibody to estrogen receptor-α (ER-α) to pull down chromatin interactions. Sequencing the resultant ChIA-PET library showed that remote ER-α enhancer-like sites interact with proximal promoters of target genes. It is of great interest to investigate the genome wide looping associations of other enhancer binding proteins.
Mediators of loops between enhancers and genes
Transcription factors or their complexes are thought to mediate enhancer-promoter loop formation but the proteins involved have been functionally identified in only a few cases. In the TH2 cytokine locus, the Il4, Il5 and Il13 genes cluster in close proximity to the TH2 locus control region (LCR) in the poised or active state in T cells but these interactions do not occur in cells lacking the transcription factors STAT6 or GATA-3 [14]. In the β-globin locus, reduction of the erythroid factors GATA-1 and EKLF (KLF1) or the more widely expressed factor Ldb1showed that are required for β-globin activation and for looping between the gene and the β-globin LCR [15–17]. Ren et al now show that OCA-B and general transcription factor TFII-I are required for long range enhancer-promoter communication in the IgH locus [18]. The IgH promoter interacts with 3’ enhancers over 100 kb distant and depletion of OCA-B or TFII-I using RNAi reduced the interactions and IgH transcription. The architectural protein SATB1 also participates in long range enhancer interaction, binding to the BCL2 gene promoter and its distant enhancer to form a loop in cells where the gene is expressed [19]. SATB1 reduction compromised loop formation and transcription. This MAR binding protein engages in many interactions genome wide, making it difficult to rule out the contribution of indirect affects to this outcome [20]. There is a clear need to expand the repertoire of factors that participate in enhancer loop formation and/or maintenance and to decipher specifically what protein-protein contacts suffice for the long range interactions.
Enhancer-gene looping and transcriptional activation
Numerous enhancers have been described that loop to their target genes and increase transcription but how the transcriptional output is changed is unclear. The loop might increase the local concentration of factors that recruit RNA pol II or pol II might be transferred from an enhancer to a promoter through their proximity [21]. The loop might affect the nuclear localization of the enhancer-promoter pair to a favorable transcriptional compartment (see below). In any case, the enhancer-gene loop appears to be necessary for transcriptional up-regulation as gain or loss of a competing promoter [22;23], reduction of loop-associated proteins [15–18] or interruption of looping by an insulator [24;25] all affect transcriptional outcome. Is the enhancer loop a cause or effect of the onset of transcription? This is a challenging question but a recent study correlated nascent transcripts with proximity of the sonic hedgehog (Shh) promoter and the limb bud enhancer, supporting a causative role [26]. This is one area where careful time course studies might be revealing, perhaps in a system like mouse G1E cells where LCR-β-globin looping and transcriptional activation can be induced by GATA-1 expression [15].
Enhancers and locus migration
Gene re-localization in nuclei upon activation is a well documented finding although how general this phenomenon is remains in question [27–29]. In animal cells, the migration typically involves moving from a peripheral position to a more interior one. Thus, the β-globin locus moves away from the nuclear periphery in maturing fetal liver cell nuclei before becoming highly. Re-localization requires the β-globin LCR but also the protein factor Ldb1, required for looping [30;31]. Recent work shows that GATA-1, its co-factor FOG1, EKLF (KLF1) and chromatin remodeler Mi-2β are also required for migration of the β-globin locus to the nuclear interior [32]. Locus movement occurred before the appearance of key markers of erythroid differentiation, suggesting it is a pre-requisite and not a consequence of high level transcription. Interestingly, inactivation of FOG-1 and Μi-2β after re-localization did not return β-globin loci to the nuclear periphery [32].
Very recently it has been shown that the Eµ enhancer is required for movement of the IgH locus to the nuclear interior [33]. Enhancer dependent long range interactions involved YY1 binding to Eµ and to the sites with which it interacted. Taken together, these results implicate enhancers and factors required for enhancer loops to form in locus movement, raising the question whether looping precedes locus migration. The Eµ enhancer loops and IgH locus migration occur without recombination or transcription of the rearranged gene, arguing that looping and migration happen first (Figure 2A). Alternatively, looping might occur after association with a transcription factory, possibly as a result of transcription (Figure 2B). Further experiments will be necessary to distinguish these models and illuminate this fundamental question.
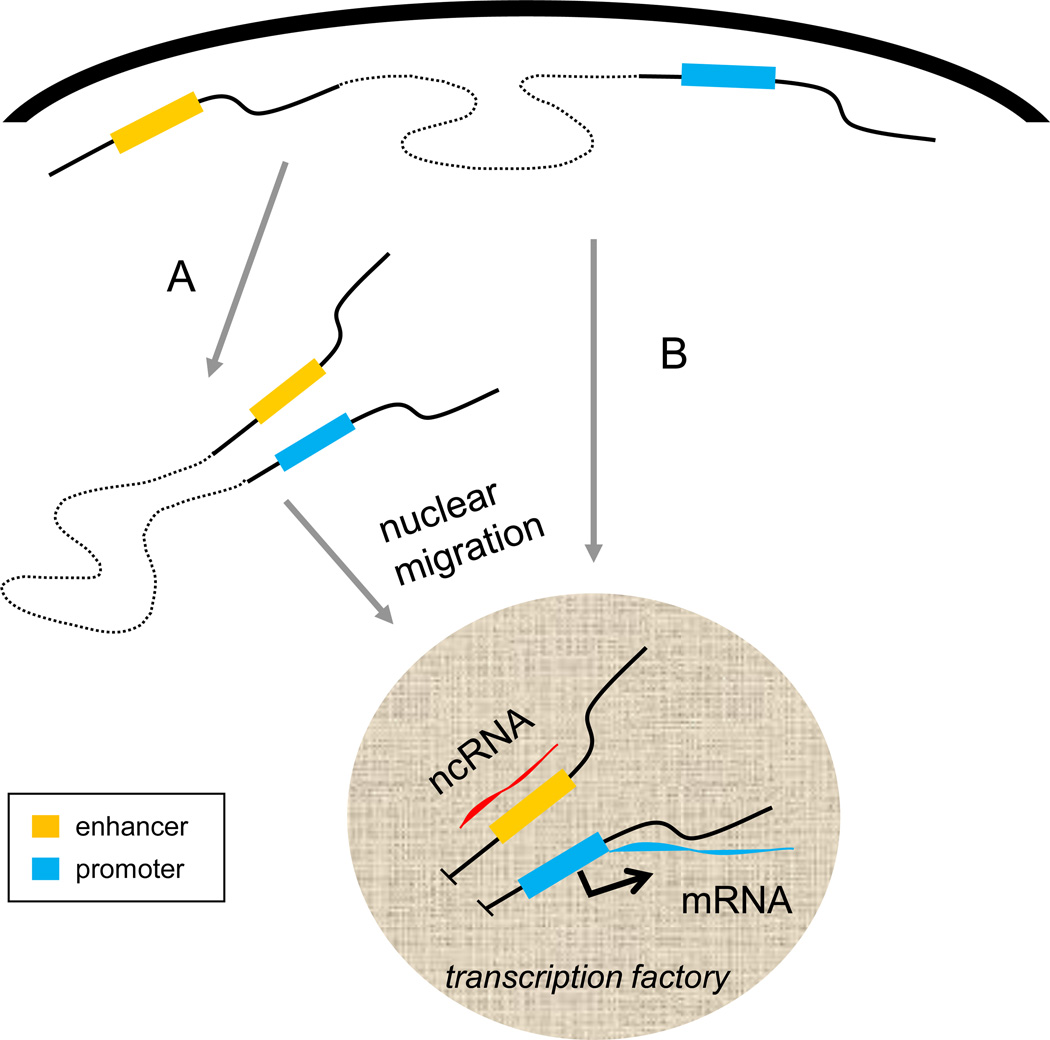
Figure 2. Influence of enhancer-promoter interaction on intra-nuclear migration to a transcription factory.
Before activation a locus occupies a position near the nuclear periphery and the enhancer and promoter do not yet interact with each other. A. In one view, after activation the enhancer interacts with the promoter which in turn leads to locus migration into the nuclear interior and localization in an RNA pol II transcription factory. Subsequently, transcription at the enhancer and promoter produces ncRNA that stabilize the interaction and mRNA respectively. B. Alternatively, the locus migrates to an interior position without communication between the enhancer and the target gene promoter and their interaction is established in the RNA polII factory, possibly with the participation of the ncRNA transcript. Designations are the same as in Figure 1.
Intra-nuclear migration during transcription activation correlates with entry into a transcription factory [34]. These entities are independent nuclear sub-compartments that are repositories of hyper-phosphorylated RNA pol II [35]. The β-globin locus enters a transcription factory after migration away from the nuclear periphery [30]. If transcription is interrupted the LCR- β-globin loop is retained, arguing that at least the maintenance of this proximity does not require ongoing transcription or even residence in a transcription factory [35;36]. Furthermore, the choice of transcription factory is non-random [37]. EKLF-dependent genes, including β-globin, co-localize at a subset of “specialized” factories that are enriched for EKLF. The underlying mechanisms are unclear. Do EKLF regulated genes seek out the proper factory, or are genes regulated by EKLF first bound by the factor and then co-migrate to a factory making it “specialized”. The precise sequence of events and interrelationships among factor binding, enhancer looping and intra-nuclear migration remain to be established.
Enhancers and long, non-coding RNAs
Genome profiling revealed that it is not unusual for RNA pol II to localize at enhancers [10]. It has also been known for many years that RNA pol II localizes at LCRs and that sense and antisense transcripts arise from these regions, although the function of such transcripts is unknown [38–41]. Using the pol II hallmark and p300 and H3K4me1 localization, Kim et al used ChIP-seq to identify thousands of neuronal activity-regulated putative enhancers genome wide [42]. A subset of the enhancers was transcribed bi-directionally by pol II into enhancer RNAs (eRNAs) which were not poly-adenylated and were generally short (<2 kb). The eRNA levels correlated with mRNA synthesis at nearby genes, suggesting that eRNA transcription might be necessary for activation of the genes. Interestingly, eRNA transcription, although not pol II occupancy, required the presence of an intact target promoter, at least in the case of the Arc gene [42].
Orom et al took a different approach and looked directly at the function of long non-coding RNAs (ncRNAs) [43]. They began with the GENCODE annotation of the human genome and filtered out transcripts over-lapping protein coding genes or belonging to known classes of non-coding RNAs. The resulting collection of ncRNA loci displayed CBP/p300 and RNA pol II occupancy but the ncRNAs were polyadenylated and less than 1% of them were bi-directional, in contrast to eRNAs described by Kim et al. Knock down of a subset of the ncRNAs (termed ncRNA-a, for activating) resulted in decreased transcription of neighboring protein coding genes and, in one case, a gene 150 kb distant. The ncRNA-a itself was required for the enhancer effect and not just transcription of the ncRNA gene. A direct role for a broader class of large non-coding RNAs (lincRNAs) in gene regulation was suggested by isolation of a subset as part of chromatin regulatory protein complexes in ES cells [44]. However, a comprehensive survey of lincRNAs in ES cells revealed that 15% of them overlapped enhancers but only 1% did so in neuronal cells [45]. At this point, the enhancer function of eRNAa and nc-RNA-a requires considerable further study and validation.
Interestingly, for nc-RNA-a there were often genes intervening between the putative enhancer and the activated target that were unaffected [43]. Viewed from an enhancer-centric perspective, the strong suggestion is that these enhancers loop to their target genes to activate their transcription, skipping over intervening genes, although the study did not include 3C experiments. Possibly, eRNAs and ncRNAs-a have a structural role in establishing or stabilizing enhancer-promoter loops although this remains unclear (Figure 2).
Enhancer and insulator functions converge
CTCF insulators protect enhancer-promoter interactions in vertebrates and insects. The β-globin LCR and genes are encompassed within a CTCF-mediated loop [46]. While reduction of CTCF in precursor cells not yet transcribing the globin genes does not appear to affect the locus, reduction of CTCF in cells actively transcribing γ-globin results in decreased transcription and incursion of repressive histone modifications, consistent with insulator function for these CTCF sites [46;47]. Likewise, disruption of interaction among surrounding insulators by CTCF depletion negatively impacts interaction of an enhancer with the APO gene promoters [48]. These examples illustrate positive indirect effects of CTCF insulator loops on enhancer-mediated gene expression (Figure 3A).
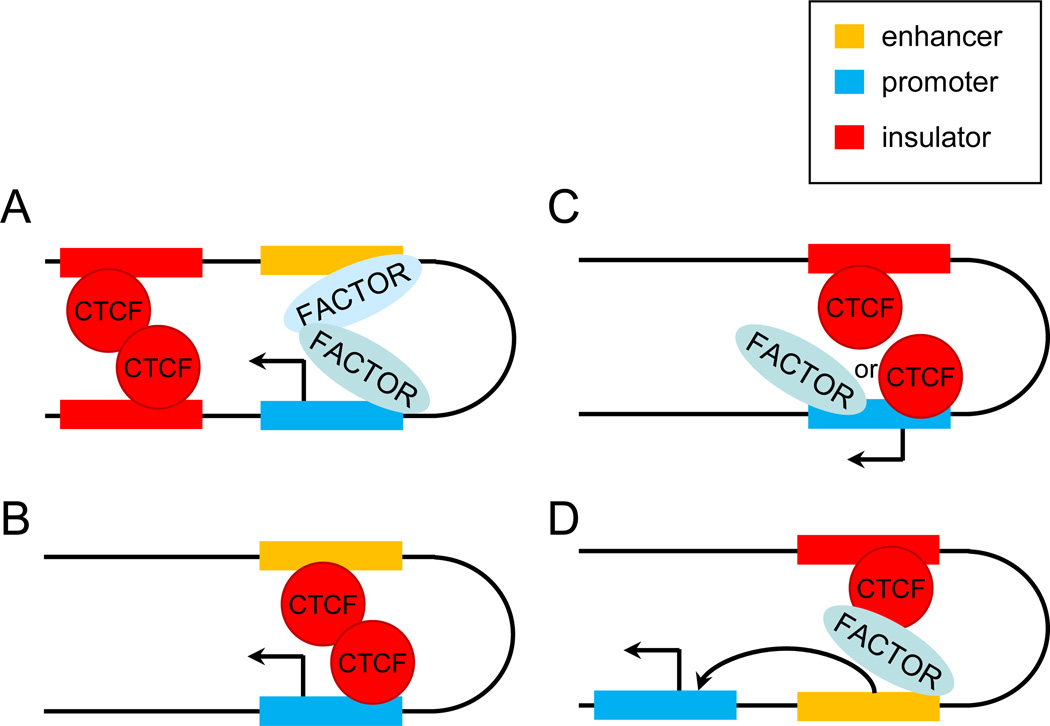
Figure 3. CTCF role in facilitating long range interaction between an enhancer and promoter.
A. CTCF mediates interaction between two insulators which positively influences enhancer-promoter interaction. This model reflects the arrangement in the β-globin and APO loci [4;48]. B. CTCF interacts with an enhancer and target promoter and participates directly in long range interaction between them that leads to transcription activation. Examples of this arrangement have been recently been described [49]. C. CTCF interacts with an insulator and provides interaction with a promoter which in turn activates transcription. The INFG and MHC class II loci provide examples of this mechanism [51;53]. D. Insulator bound CTCF provides interaction with an enhancer which in turn interacts with a target promoter. This mechanism is utilized in the IgH locus [54;55]. Variants A, C and D can be incorporated into the concept of the active chromatin hub which provides an environment conducive to transcription activation. Designations are as in Figure 1.
Genome wide role of insulator loops
Genome profiling by Hi-C revealed that chromatin loops are central to the organization of active and silent chromatin into separate functional domains [12]. The emerging picture is that insulators are key contributors to this organization. Using ChIA-PET, new data show that CTCF mediates interaction between thousands of loci and organizes the genome into different functional compartments [49]. Handoko et al observed that many of the CTCF-mediated long range interacting sites coincided with an enhancer and promoter. Depletion of CTCF using RNAi reduced looping between the elements at select loci and reduced transcription of the gene involved. The suggestion is that CTCF facilitates enhancer-promoter interaction directly (Figure 3B). Thus, in addition to the classical view of domain separation by insulators to topologically isolate an enhancer and a proper gene target, we can envision other arrangements more directly involving insulators in this communication. Of note, insulator interaction with an enhancer and promoter can also be associated with a negative influence on transcription activation by the enhancer [50].
Direct interaction of enhancers and genes with insulators
Recent work documents CTCF occupied sites at insulators, within genes and in enhancers that participate in looping interactions and play a role in transcription activation. For example, the INFG gene has a CTCF site in the first intron that contacts two distant CTCF sites in the locus to form loops that are required for activation (Figure 3C) [51]. The INFG CTCF site interactions are modulated by recruitment of cohesin. In the MHC-II locus, a CTCF bound insulator site interacts with HLA-DRB1 and HLA-DQA1 promoters to form a chromatin loop [52]. These long range interactions depend on promoter-bound transcription factors CIITA and RFX and on CTCF whose reduction compromises gene transcription (Figure 3C). In addition, other insulator sites in the locus form long range associations among themselves that are dependent on CTCF but not on CIITA and RFX. Cohesin is also important for contact between the CTCF insulator site and MHC-II gene promoters [53].
In addition to promoters, enhancers can directly interact with insulators (Figure 3D). The Eμ recombination enhancer in the IgH locus of pro-B cells interacts with upstream and downstream CTCF insulators forming loops that are thought to contract the locus and facilitate recombination [54;55]. These interactions are CTCF and cohesin dependent and necessary for IgH locus functioning. At least one of the sites downstream from Eµ has enhancer blocking activity. Interestingly, one of the CTCF insulators downstream of Eμ (HS4) was found in the work of Ren et al (see above) to interact with the VDJ promoter in an OCA-B and TFII-I dependent fashion adding complexity to the overall locus structure. Very recent work using 4C details two sets of loops at the IgH locus; those dependent on CTCF but not on Eµ and those dependent on Eµ along with YY1 [33]. Several of the YY1 sites involved in Eµ looping bind CTCF and thus represent heterotypic interactions of the two factors [33;55].
In another example, T cell receptor rearrangement in the Tcra locus in mouse thymocytes involves CTCF and cohesin occupancy and long range interactions among the TEA gene, the Eα enhancer and the Tcra LCR. Deletion of the cohesin subunit Rad21 disrupted the long range interactions, accompanied by down-regulation of TEA, H3K4me3 depletion and aberrant TCRα rearrangement [56]. A regulatory element that engages in a similar mechanism is revealed in studies of the general transcription factor TAF3 [57]. Some distant TAF3 binding sites were found to colocalize with CTCF and cohesin and CTCF tethers these sites to TAF3 dependent promoters. However, the distant TAF3 sites were not otherwise tested for enhancer activity. The data discussed above collectively indicate that enhancer/insulator interplay is more complex that it was thought to be. In some cases, insulator protein CTCF and/or cohesin can facilitate enhancer-promoter interaction directly by interacting with these elements to bring them together.
Influence of cohesion on enhancer-promoter interactions
Cohesin is recruited to CTCF sites genome wide but a subset of sites is unique to cohesin [58;59]. New data has shed light on the function of these sites. ER-α binding sites colocalize with cohesin sites that lack CTCF [60]. It was proposed that ER-α together with cohesin provides long range interactions, although this was not tested directly by 3C. In another example, cohesin and the Mediator complex are implicated in enhancer-promoter interaction [61]. Kagey et al found that Mediator subunits colocalize with cohesin at enhancers and promoters and are necessary for loop formation between them. The data illustrate the potential role of cohesin as a physical tether mediating enhancer-gene proximity in combination with mediator, which may coordinate signals between enhancers and the general transcription machinery. What recruits cohesin to these non-CTCF sites and whether they have insulator function are unknown.
Conclusions
The very recent data summarized here strikingly illustrate that enhancer/insulator interplay in regulation of gene transcription is more complex than was previously appreciated. In some cases, enhancer-promoter interaction appears to be directly facilitated by CTCF/cohesin occupancy. In other cases, the promoter or the enhancer is occupied by CTCF/cohesin and additional factors cooperate to support interaction between them. It will be important to determine the exact geometry of the CTCF/cohesin binding sites vis-à-vis transcription factors when they co-occupy enhancers or promoters. In view of its interaction with potential enhancers [49], RNA pol II transcription factories [62] and the general transcription machinery [57], does CTCF have a direct role in transcription activation? Are all CTCF and cohesin sites insulator sites in the classic view? The genome wide architectural role of CTCF loops may provide a clue to how enhancer-promoter ensembles migrate within the nucleus. Almost nothing is known about this process although it seems to involve actin-myosin motors [63;64]. The novel convergence of enhancer-promoter pairs and CTCF insulators impacts important unresolved questions such as how enhancers and promoters establish a connection, how they move within the nucleus and how transcriptional output is increased by these events.
Acknowledgements
We would like to thank Dr. Gerd Blobel for helpful comments on the manuscript and members of our laboratory for their suggestions. Work in our laboratory is supported by the Intramural Program of NIDDK, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Dean A. On a chromosome far, far away: LCRs and gene regulation. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 3.Carter D, Chakalova L, Osborne CS, Dai Y, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 4.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 5.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The β-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 6.Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 7.Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–4919. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim. Biophys Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wendt KS, Peters JM. How cohesin and CTCF cooperate in regulating gene expression. Chromosome. Res. 2009;17:201–214. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- 10.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 11.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. This is the first paper that uses the Hi-C, a method for identifying long range interactions genome wide based on the 3C assay followed by massively parallel sequencing. The resolution of this technique is 2 Mb. The authors demonstrated that chromatin interacts non-randomly and the interactions reflect the functional state of interacting regions.
- 13. Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. This is the first study that uses the ChIA-PET assay for identifying all long range interactions provided by a single protein of interest genome wide. Interacting chromatin was pulled down by antibodies to ERα, tagged and sequenced. As a result, it was shown that distant ERα binding sites interact with ERα binding promoters. This new approach was validated by FISH and 3C.
- 14.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 15.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 16.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song S-H, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range β-globin locus control region function. Mol. Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren X, Siegel R, Kim U, Roeder RG. Direct interactions of OCA-B and TFII-I regulate immunoglobulin heavy-chain gene transcription by facilitating enhancer-promoter communication. Mol Cell. 2011;42:342–355. doi: 10.1016/j.molcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong F, Sun L, Wang Z, Shi J, Li W, Wang S, Han X, Sun Y. The BCL2 gene is regulated by a special AT-rich sequence binding protein 1-mediated long range chromosomal interaction between the promoter and the distal element located within the 3'-UTR. Nucleic Acids Res. 2011;39:4640–4652. doi: 10.1093/nar/gkr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 21.Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue- specific locus control region and a downstream promoter. Mol. Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 22.De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de JP, Cheng JF, Rubin EM, Wood WG, Bowden D, Higgs DR. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 23.Lower KM, Hughes JR, De GM, Henderson S, Viprakasit V, Fisher C, Goriely A, Ayyub H, Sloane-Stanley J, Vernimmen D, Langford C, Garrick D, Gibbons RJ, Higgs DR. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci U. S. A. 2009;106:21771–21776. doi: 10.1073/pnas.0909331106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 25.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci U. S. A. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, Trowsdale J, Sheer D. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- 28.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 29.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song SH, Kim A, Ragoczy T, Bender MA, Groudine M, Dean A. Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation. Blood. 2010;116:2356–2364. doi: 10.1182/blood-2010-03-272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee HY, Johnson KD, Boyer ME, Bresnick EH. Relocalizing genetic loci into specific subnuclear neighborhoods. J Biol Chem. 2011;286:18834–18844. doi: 10.1074/jbc.M111.221481. These two studies show that several proteins that are necessary for long range interaction between the β-globin promoter and LCR are also necessary for nuclear migration of globin loci. Lee et al show that the relocalization of β-globin loci after activation is an irreversible process. The data clearly show a connection between enhancer-promoter interaction and nuclear relocalization.
- 33. Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. Two Forms of Loops Generate the Chromatin Conformation of the Immunoglobulin Heavy-Chain Gene Locus. Cell. 2011 doi: 10.1016/j.cell.2011.08.049. This work uses 4C to comprehensively determine looping interactions in the IgH locus. One set of loops are enhancer dependent and involve YY1 and CTCF. A second set of loops are CTCF dependent and not dependent on the Eµ enhancer. The Eµ enhancer is shown to be required for migration of the locus away from the nuclear periphery.
- 34.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de LW. Maintenance of Long-Range DNA Interactions after Inhibition of Ongoing RNA Polymerase II Transcription. PLoS. ONE. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. This study shows that co-regulated genes tend to occupy the same RNA pol II factory. Furthermore, evidence is presented for the existence of specialized transcription factories. Only a subset of transcription factories in erythroid cells are enriched for the transcription factor KLF1 but it is in these factories that KLF1 regulated erythroid genes congregate when activated.
- 38.Johnson KD, Grass JA, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, Bresnick EH. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad.Sci. U.S.A. 2002;99:11760–11765. doi: 10.1073/pnas.192285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 40.Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 2003;4:132–137. doi: 10.1038/ni883. [DOI] [PubMed] [Google Scholar]
- 41.Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, Fraser PJ, Corcoran AE. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 42. Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. This work shows that transcription is common at enhancers genome wide. Enhancer specific RNA (eRNA) levels correlated with the level of mRNA synthesis at nearby genes required the promoter of the gene suggesting the eRNA transcription was directly linked to mRNA transcription.
- 43. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. This study describes long non-coding RNAs (ncRNAs) that are necessary for activation of genes near the ncRNA coding sequence. Selective knock down of ncRNAs caused gene repression indicating that the transcript per se was required for the activation.
- 44.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U.S.A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. In this paper the ChIA-PET assay was used to identify the CTCF interactome in mouse ES cells. It was shown that CTCF partitions chromatin into different functional domains and in some cases may participate directly in long range interaction between potential enhancers and promoters.
- 50.Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K. Analysis of the H19ICR insulator. Mol. Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Majumder P, Boss JM. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol. Cell Biol. 2010;30:4211–4223. doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majumder P, Boss JM. Cohesin Regulates MHC Class II Genes through Interactions with MHC Class II Insulators. J Immunol. 2011;187:4236–4244. doi: 10.4049/jimmunol.1100688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, Bates JG, Richards N, Myers D, Patel H, Gallagher M, Schlissel MS, Murre C, Busslinger M, Giallourakis CC, Alt FW. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, Murre CS, Birshtein BK, Schork NJ, Schlissel MS, Riblet R, Murre C, Feeney AJ. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. These two studies, together with reference 31 provide an in depth view of looping interactions in the IgH locus that are thought to be required to condense the locus and facilitate V/D/J recombination. Guo et al show the locus contraction is Eµ enhancer dependent using FISH. Degner et al also show that cohesin participates along with CTCF in mediating loops in the locus.
- 56.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of Embryonic Stem Cell Lineage Commitment by Core Promoter Factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 59.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 60.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR. The proteomes of transcription factories containing RNA polymerases I, II or III. Nat. Methods. 2011 doi: 10.1038/nmeth.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U.S.A. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]