Abstract
Downstream regulatory element antagonist modulator (DREAM) is a Ca2+-binding protein that binds DNA and represses transcription in a Ca2+-dependent manner. Previous work has shown a role for DREAM in cerebellar function regulating the expression of the sodium/calcium exchanger 3 (NCX3) in cerebellar granular neurons to control Ca2+ homeostasis and survival of these neurons. To achieve a global view of the genes regulated by DREAM in the cerebellum, we performed a genome-wide analysis in transgenic cerebellum expressing a Ca2+-insensitive/CREB-independent dominant active mutant DREAM (daDREAM). Here we show that DREAM regulates the expression of the midline 1 (Mid1) gene early after birth. As a consequence, daDREAM mice exhibit a significant shortening of the rostro-caudal axis of the cerebellum and a delay in neuromotor development early after birth. Our results indicate a role for DREAM in cerebellar function.
Keywords: midline 1, cerebellar lobes, nuclear calcium, transcriptomic analysis
Introduction
Downstream regulatory element antagonist modulator (DREAM), also named calsenilin or KChIP-3, is a Ca2+-binding protein of the neuronal calcium sensors family able to repress transcription of specific genes in a Ca2+-dependent manner (Carrion et al., 1999), to interact with presenilins and to modify APP processing (Buxbaum et al., 1998; Lilliehook et al., 2003) and to regulate the membrane expression and gating of Kv4 potassium channels (An et al., 2000; Ruiz-Gomez et al., 2007) and of voltage-dependent calcium channels (Thomsen et al., 2009; Anderson et al., 2010). In addition, DREAM modulates downstream signaling of different membrane receptors including NMDA (Wu et al., 2010; Zhang et al., 2010) and THSR (Rivas et al., 2009).
Transcriptional activity of DREAM is triggered by its sumoylation-dependent nuclear translocation (Palczewska et al., 2011), regulated by redox state (Rivas et al., 2011), and accomplished by its Ca2+-dependent specific binding to DNA and to other nucleoproteins, including CREM and CREB (Ledo et al., 2000, 2002; Rivas et al., 2004; Scsucova et al., 2005; Zaidi et al., 2006). In addition, high affinity binding of DREAM to DRE sequences in the DNA requires Ca2+-dependent DREAM oligomerization (Carrion et al., 1998; Osawa et al., 2001, 2005).
Downstream regulatory element antagonist modulator is widely expressed in the central nervous system as well as in the thyroid gland, testis, and the immune system (Carrion et al., 1999). Tissue-specific target genes for DREAM regulation have been identified first in vitro (Carrion et al., 1999; Link et al., 2004) and more recently in vivo, using DREAM null mice (Cheng et al., 2002) and especially transgenic mice expressing a dominant active DREAM mutant (daDREAM; Gomez-Villafuertes et al., 2005; Savignac et al., 2005, 2010; Rivera-Arconada et al., 2010). Specifically, a significant increase in prodynorphin mRNA was found in spinal cord from DREAM null mice (Cheng et al., 2002) and a decrease in NCX3 and BDNF mRNA and protein levels were reported in hippocampus and cerebellum (Gomez-Villafuertes et al., 2005) and in spinal cord (Rivera-Arconada et al., 2010) respectively, from daDREAM mice. The regulatory effect of daDREAM on the expression of transcriptional targets was shown to be specific, since for instance no change in the expression of other members of the sodium/calcium exchanger family could be observed in hippocampus and cerebellum of daDREAM transgenic mice (Gomez-Villafuertes et al., 2005). As expected, transcriptional regulation by DREAM of various cellular targets has diverse and tissue-specific functional consequences. In the CNS, this includes changes in sensory noxious perception (Cheng et al., 2002; Rivera-Arconada et al., 2010), β-amyloid accumulation (Lilliehook et al., 2003), and learning and memory formation (Alexander et al., 2009), as well as modified T-cell proliferation and Ig production (Savignac et al., 2005, 2010) in the immune system.
Taking the cerebellum as an example, previous work using primary cultures of cerebellar granular neurons has shown the role of DREAM in the regulation of NCX3 expression, which in turn is fundamental for the maintenance of Ca2+ homeostasis and viability of the cerebellar granular neurons in the culture (Gomez-Villafuertes et al., 2005). In the present study, to further investigate the functional involvement of DREAM-mediated transcription in cerebellar function, we have performed a transcriptomic analysis of the cerebellum from daDREAM transgenic mice. We found that expression of the midline 1 gene (Mid 1) is repressed in daDREAM mice. Related to this, daDREAM mice exhibit a significant shortening of the rostro-caudal axis of the cerebellum and a severe delay in neuromotor development early after birth.
Results
Genome-wide analysis in daDREAM cerebellum
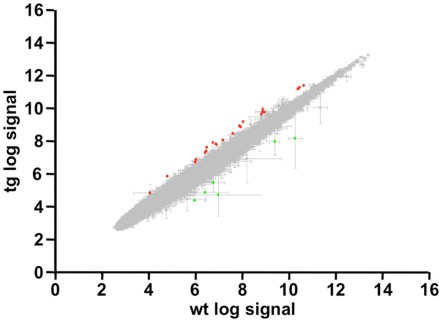
To search for DREAM transcriptional targets that could disclose a role for DREAM in cerebellar function, we performed genome-wide analysis in a line of transgenic mice previously reported to express daDREAM in the cerebellum (Gomez-Villafuertes et al., 2005). Comparison of basal gene expression in wild type and transgenic adult cerebellum, using cDNA microarrays, identified only 11 genes whose expression was significantly altered in daDREAM transgenic cerebellum (Gene Expression Omnibus accession number GSE34765; Figure 1). Up- and down-regulated genes are presented in Table 1 and the bioinformatics analysis is shown in Tables A1 and A2 in Appendix.
Figure 1.
Genome-wide analysis in daDREAM and wild type cerebellum. Statistical values and scatter plot of all probes contained in the genome-wide analysis. Spots with |Fold Change| >1.6 and FDR <0.1 are highlighted in green (repressed genes) and red (induced genes).
Table 1.
List of induced and repressed genes in daDREAM vs. wild type cerebellum.
| Fold change | P-value × 10−5 | FDR | Probe ID | Transcript ID | Gene symbol | Gene description |
|---|---|---|---|---|---|---|
| 1.70 | 0.922 | 0.023 | 1440557_at | Mm.186257.1 | Ipw | Imprinted gene in the Prader–Willi syndrome region |
| 1.75 | 1.792 | 0.038 | 1436733_at | Mm.40013.1 | E130309F12Rik | RIKEN cDNA E130309F12 gene |
| 1.86 | 0.914 | 0.026 | 1460049_s_at | Mm.213028.1 | 1500015O10Rik | RIKEN cDNA 1500015O10 gene |
| 1.95 | 0.319 | 0.0160 | 1417654_at | Mm.3815.1 | Sdc4 | Syndecan 4 |
| 2.10 | 0 | 0 | 1449340_at | Mm.43375.1 | Sostdc1 | Sclerostin domain containing 1 |
| −4.68 | 0.035 | 0.000 | 1436240_at | Mm.216255.1 | B230214O09Rik | RIKEN cDNA B230214O09 gene |
| −4.17 | 0.177 | 0.016 | 1431214_at | Mm.157900.1 | LOC433762 | Hypothetical gene LOC433762 |
| −2.95 | 0.133 | 0.027 | 1440139_at | Mm.174301.1 | gb:BB729836 | Moderately similar to S12207 hypothetical protein |
| −2.83 | 0.231 | 0.030 | 1438239_at | Mm.24820.1 | Mid1 | Midline 1 |
| −2.60 | 0.408 | 0.037 | 1425545_x_at | Mm.33263.2 | H2-D1 | Histocompatibility 2, D region locus 1 |
| −2.38 | 0.639 | 0.048 | 1419327_at | Mm.22635.1 | Pdxdc1 | Pyridoxal-dependent decarboxylase domain containing 1 |
Genes with FDR <0.05 were included in the list of induced or repressed candidates.
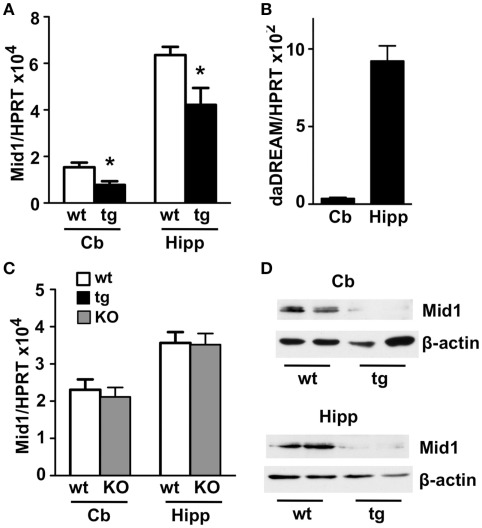
Among the genes with modified expression (Table 1), we focused our attention on the significant down regulation of the Mid 1 gene, an ubiquitin ligase specific for the protein phosphatase 2A (PP2A; Trockenbacher et al., 2001). Loss of function mutations in the Mid1 protein cause the X-linked Opitz BBB/G syndrome, a congenital anomaly disorder characterized by developmental defects of midline structures (Opitz, 1987; Quaderi et al., 1997). Importantly, Mid1 deficient mice mimic the hypoplasia of the anterior portion of the medial cerebellum (Lancioni et al., 2010), a clinical feature present in Opitz patients, suggesting that DREAM, through the regulation of Mid1, might participate in cerebellar development. Real-time qPCR analysis confirmed the reduced level of Mid1 mRNA in the cerebellum of adult transgenic mice as well as in the hippocampus (Figure 2A), areas with significant expression of daDREAM (Figure 2B). As reported for other DREAM target genes like prodynorphin in the hippocampus (Cheng et al., 2002) or CANT1 in the cerebellum (Cali et al., 2012), probably due to functional compensation by other members of the KChIP family, no change in the expression of Mid1 was observed in cerebellum or hippocampus from DREAM knockout mice (Figure 2C). Western blot analysis of cerebellum and hippocampus from daDREAM mice confirmed the reduced levels of Mid1 protein with respect to wild type mice (Figure 2D). Taken together, these results implicate endogenous DREAM/KChIPs in the regulation of Mid1 gene expression in the brain.
Figure 2.
Real-time qPCR and western blot analysis of Mid1 expression in adult brain. Levels of Mid1 (A,C) and daDREAM (B) mRNA in the cerebellum (Cb) and the hippocampus (Hipp) from wild type (wt), transgenic (tg), and DREAM−/− (KO) mice are shown. Values are normalized with respect to HPRT mRNA content. Results are the mean ± SEM of 14–18 mice in three independent experiments. *P < 0.05 (two-tailed, unpaired t-test) relative to wild type mice. (D) Immunoblot analysis of adult brain cerebellum and hippocampal lysates (50 μg) shows that the 70- to 75-kDa expected band for the Mid 1 protein is reduced in transgenic compared to wild type mice. β-Actin was used as protein loading control. The experiment, in duplicates, was repeated twice.
Dream regulates early postnatal Mid1 expression
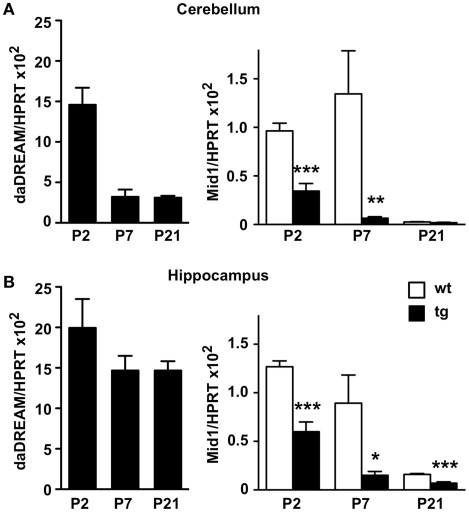
Mid1 is widely expressed at early human embryonic stages, however during organogenesis the expression pattern becomes more restricted to the tissues affected in the Opitz syndrome (Pinson et al., 2004). The development of the cerebellum in mice is mostly postnatal and is complete after the second week of life (reviewed in Millen and Gleeson, 2008). To investigate a role for DREAM in the regulation of Mid1 and cerebellar development, we next analyzed the expression of the daDREAM transgene during cerebellar development and the consequences on Mid1 expression. Real-time qPCR analysis showed that the expression of daDREAM in the cerebellum was very high early after birth (P2) and got down at P7–P21 (Figure 3A) to values slightly higher than adult levels (see Figure 2B). Expression of Mid1 in wild type cerebellum was high early after birth (P2–P7) and reached adult levels at P21 (notice the different scale in Figure 2A). Expression of daDREAM was associated with a significant reduction of Mid1 mRNA levels in transgenic cerebellum (Figure 3A). Early postnatal expression of the transgene was not exclusive to the cerebellum and was observed as well in other brain areas, like the hippocampus (Figure 3B). Expression of daDREAM in the hippocampus was also associated with a significant reduction of Mid1 mRNA levels (Figure 3B).
Figure 3.
Real-time qPCR analysis of Mid1 expression during postnatal development. Levels of daDREAM and Mid1 mRNA at the indicated postnatal day from wild type (wt) and transgenic (tg) mice in cerebellum (A) and hippocampus (B) are shown. Values are normalized with respect to HPRT mRNA content. Results are the mean ± SEM of 8–12 mice in two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed, unpaired t-test) relative to wild type mice.
Cerebellar morphological changes in daDREAM mice
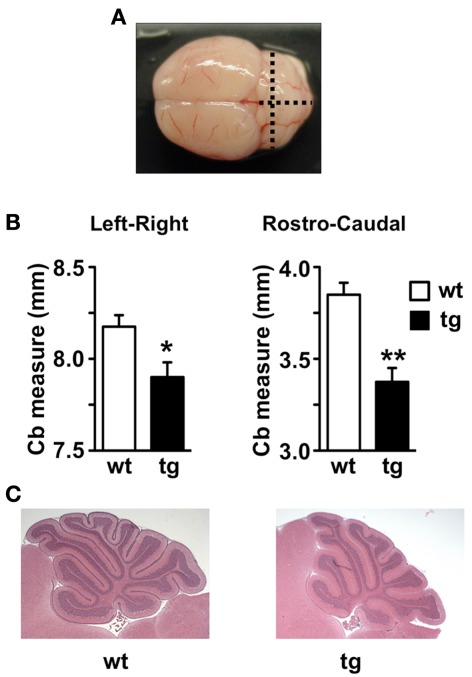
Absence of Mid1 protein in Mid1 null mice results in abnormal formation of the anterobasal cardinal lobe, which is evident already at P0–P2 (Lancioni et al., 2010). In adult Mid1 null mice, although abnormal in shape, even the anterior vermal lobes maintain the correct layer organization and thickness (Lancioni et al., 2010). According to the significant reduction in Mid1 expression, adult daDREAM mice showed a significant shortening in the antero-posterior axis but also in the left–right axis (Figures 4A,B). Sagittal sections from P15 wild type and daDREAM mice confirmed a correct layer organization of transgenic cerebellum and showed the shortening of the rostro-caudal axis (Figure 4C).
Figure 4.
Anatomical analysis of adult and postnatal cerebellum. (A) Image of whole adult mouse brain showing the left–right and rostro-caudal axis used for the measurement. (B) Measurement of left–right and rostro-caudal axis of the cerebellum in wild type and (wt) transgenic (tg) adult mice (n = 6). *P < 0.05, **P < 0.01 (two-tailed, unpaired t-test) relative to wild type mice. (C) For comparison, sagittal sections through the cerebellar vermis of postnatal P15 wild type (wt) and transgenic (tg) mice after staining with hematoxylin/eosin are shown.
Impaired motor development in daDREAM mice
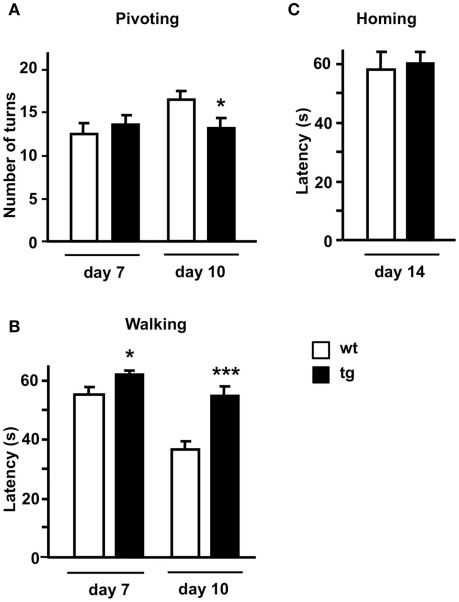
Absence of Mid1 protein in Mid1 null mice has been associated with impaired motor coordination in the adulthood. To assess possible changes during early postnatal development that could be related to Mid1 down regulation in daDREAM mice, we employed a test battery consisting of somatometric, neurologic, and sensorial and motor tests. No differences in somatometric growth, or in the appearance of developmental landmarks, i.e., eye opening, fur appearance, or incisor eruption, were detected in daDREAM mice. However, significant changes were observed in neuromotor development as assessed by the pivoting and the walking tests on postnatal days 7, 10, and 14. In the pivoting locomotion task, wild type, and daDREAM mice showed similar activity at P7. However, the normal age-dependent increase in activity, which is reflecting the adequate maturation of motor systems in wild type, was significantly reduced in daDREAM mice at P10 (Figure 5A). Assessment of walking activity confirmed the hypoactive phenotype in daDREAM mice and, even at P7 the latency to walk in transgenic mice was significantly longer than in wild type mice (Figure 5B). At P10, the reduction in walking latency observed in wild type (33.6% of reduction, P7 vs. P10) was significantly less pronounced in daDREAM mice (10.8% reduction, P7 vs. P10; Figure 5B). This result, along with the affected pivoting activity in daDREAM mice, could suggest a supraspinal deficiency, indicating that the postnatal development of brain structures directly implicated in the motor function might be affected in daDREAM mice. This defect is specific of motor function, since daDREAM mice showed normal latency to reach their nest in the homing test compared to wild types (Figure 5C). This normal latency might suggest a normal development of other functional domains such as sensory perception.
Figure 5.
Neuromotor development in postnatal mice. Behavioral analysis (A) pivoting test, (B) walking test, and (C) homing test were performed at the indicated postnatal days in wild type and (wt) transgenic (tg) mice (n = 16–18). *P < 0.05, ***P < 0.001 (two-tailed, unpaired t-test) relative to wild type mice.
Discussion
Genome-wide analysis of different tissues or brain areas from daDREAM transgenic mice has rendered discrete lists of potential DREAM regulated genes with some overlapping as well as tissue-specific targets (Savignac et al., 2010; Jose R. Naranjo, unpublished observations). In all cases, including the present data, these lists include both repressed and induced genes suggesting that besides the direct repressor action of DREAM, interactions between DREAM and other transcriptional regulators (Ledo et al., 2002; Rivas et al., 2004, 2011; Scsucova et al., 2005; Zaidi et al., 2006) or indirect/secondary transcriptional effects might contribute to gene induction in daDREAM transgenic tissues. In the case of the daDREAM cerebellum, the list of genes with modified expression is especially short. This is not related to technical problems with the hybridization of the array since the bioinformatics analysis of the hybridization signals (see Figure 1) shows a perfectly normal distribution. Instead, the low number of hits might reflect the limitation of the technique, showing for instance a minor and not significant down regulation (−1.15-fold; P-value 0.06) of the NCX3 gene (also named Slc8a3: solute carrier family 8) previously validated as a direct DREAM target and shown to be repressed by more than 50% in daDREAM cerebellum (Gomez-Villafuertes et al., 2005). Alternatively, the low number of targets may simply reflect the limited but specific functionality of DREAM in the cerebellum, a brain region where DREAM expression shows the lowest level among the different areas in the CNS (Carrion et al., 1999; Rhodes et al., 2004).
Among induced genes, syndecan 4 is a transmembrane proteoglycan for which there are no reports relating a function in brain or cerebellum. Sclerostin domain containing 1 (Sostdc1) is a secreted inhibitor of the Wnt and Bmp pathways, that antagonizes BMP signaling in the mesenchymal induction of teeth and orofacial morphogenesis, regulating also the spatial patterning of teeth and hair. Deletion of Sostdc1 leads to the full development of single extra incisors adjacent to the main incisors (Munne et al., 2009). On the other hand, induced expression of sclerostin domain containing 1 (Sostdc1) has been reported in the brain of mice deficient in phenylalanine hydroxylase (PAH), an animal model for the autosomal recessive disorder phenylketonuria (Park et al., 2009), though the functional meaning of this increased expression is not known. In daDREAM mice, however, induced levels of Sostdc1 do not have an effect on dentation and hair appearance since both are normal.
Among repressed genes, the Mid1 protein is biochemically well characterized and a role in brain development has been reported. Thus, early studies of Mid1 function recognized its ubiquitin ligase activity and have identified its specific role in the degradation of protein phosphatase 2A by the proteosome (Trockenbacher et al., 2001). Importantly, PP2A is the major serine–threonine–tau phosphatase in the brain and dephosphorylates also other microtubule-associated proteins like 4EBP1 and p70S6K, two targets of the mTOR kinase (Nojima et al., 2003). Loss of function mutations of the Mid1 protein are associated with developmental midline malformations present in Opitz syndrome patients (Opitz, 1987; Quaderi et al., 1997). Furthermore, cells derived from Opitz syndrome patients show decreased formation of mTORC1, the complex between mTOR and its interacting proteins Raptor and mLST8, as well as S6K1 phosphorylation, cell size, and cap-dependent translation. Expression of wild type Mid1 or activated mTOR protein rescue the phenotype, suggesting that mTORC1 plays a key role in Opitz syndrome pathogenesis (Liu et al., 2011). In addition, a recent study has identified the binding of the cytoskeletal-associated Mid1/α4/PP2A complex to polyribosomes and mRNAs via a purine-rich sequence motif called MIDAS (MID1 association sequence) increasing the stability and translational efficiency of these mRNAs (Aranda-Orgilles et al., 2011). Notably, the MIDAS motif is frequent in mRNAs related to development and energy metabolism and mutated Mid1 does not interact with MIDAS-containing mRNAs suggesting that defects associated with the Opitz syndrome could be produced by altered protein translation of some of these mRNAs (Aranda-Orgilles et al., 2011). About the mechanism by which Mid1 could affect brain development it has been hypothesized that by controlling PP2A activity, Mid1 is regulating the activity of Gli3 (Krauss et al., 2008) a Wnt target gene related to the development of the hippocampus (Hasenpusch-Theil et al., 2012), which has been implicated in three human pathological conditions sharing common features with Opitz syndrome (Biesecker, 2006). Lack of the murine Mid1 gene results in motor coordination defects as well as motor, non-associative, and procedural learning impairments that may correlate with the developmental delays of Opitz syndrome patients (Lancioni et al., 2010). The behavioral analysis in this study was done in adult mice with no specific focus on postnatal development. Our data report an early down regulation of Mid1 expression in daDREAM mice and suggest a delayed neuromotor development in these mice. Down regulation of Mid1 mRNA levels is not restricted to the cerebellum and is also observed in the hippocampus, a brain area with strong expression of the daDREAM mutant. Whether hippocampal morphology is altered in daDREAM mice and to what extent the reduction in other genes like NCX3 contribute to the morphological and neuromotor phenotypes remain to be investigated.
As expected for a protein with developmental functions, expression of Mid1 in adult brain is reduced compared to postnatal levels. Interestingly, however, expression in the adult hippocampus is higher than in the adult cerebellum. This may suggest a specific function for Mid1 in adult hippocampus and the existence of strong trans-activating mechanisms to compensate a presumable stronger repression due to higher levels of DREAM protein in hippocampus compared to cerebellum.
Analysis of the post-transcriptional regulation of the Mid1 mRNA has shown that alternative use of different transcription start sites, alternative splicing, and the existence of different polyadenylation signal results in a wide variety of Mid1 isoforms (Landry and Mager, 2002; Winter et al., 2004). Less is known, however, about the transcriptional mechanisms that regulate Mid1 expression. Like in the case of the BDNF gene, another example of a gene with multiple transcription start sites, specific regulatory regions have been assigned to each transcription start site. Interestingly, in silico analysis of these different promoters, both in the human and the mouse genes, has identified putative DRE sites (Table 2), mostly located in the complementary strand, a circumstance previously reported for DRE sites in the ICER (Link et al., 2004) and the interferon γ (Savignac et al., 2005) genes. Nevertheless, in vitro analysis of the ability of these sites to bind recombinant DREAM and especially chromatin immunoprecipitation studies should confirm the in silico predictions.
Table 2.
Downstream response elements, DREs, present in the 5′ untranslated region of different Mid1 isoforms.
| Species | Isoform | Position in mRNA | Sequencea,b | Orientation |
|---|---|---|---|---|
| Mouse | Var 1c | 155–164 | tgatgacccc | ← |
| Var 2 | 68–78 | cttgacattgt | ← | |
| Var 2 | 261–270 | gggtcatgga | ← | |
| Human | Var 1 | 317–328 | gggtcatggga | → |
| Var 2d | 161–173 | ctgatgacaca | ← | |
| Var 4 | 158–168 | taatgactcca | ← | |
| Var 4 | 259–268 | aatgacgtaa | ← |
aThe core sequence is underlined.
bFor comparison the mouse interferon γ DRE site in the complementary strand (←) is atcggctgacctagag and the human dynorphin forward element (→) is agccggagtcaaggag.
cThe same element is present in mouse variant 2 at position 316–327.
dThe same element is present in variants 1, 3, 6, 7, 8, and 9.
In conclusion, the results from the genome-wide analysis of the cerebellum from daDREAM mice indicate that endogenous DREAM, in addition to regulate NCX3, might also contribute to cerebellar function through the regulation of Mid1 gene expression.
Materials and Methods
Transgenic mice
A cDNA encoding human DREAM with two amino acid substitutions at EF-hands 2, 3, and 4 and at the N-terminal LCD (daDREAM) was cloned downstream of the human CaMK-IIα promoter (Mayford et al., 1995) in a bicistronic expression vector containing an IRES and the LacZ reporter gene. DREAM transgenic mice were prepared by pronuclear microinjection of this cassette in the C57BL/6xCBA hybrid background. Transgenic progeny were identified by Southern blotting and qualitative PCR of tail DNA using specific primers; forward 5′-TTGCAGTGCACGGCAGATACACTTGCTGA-3′ and reverse 5′-CCACTGGTGTGGG CCATAATTCAATTCGC-3′. An amplified fragment of 326 bp indicated the presence of the transgene. Founder males were backcrossed to C57BL/6 females to generate lines that were maintained as heterozygous and age- and sex-matched littermates were used as controls. Of the different transgenic lines generated, in this study we used transgenic line 33 that shows expression of the transgene in different brain areas, including the cerebellum (Gomez-Villafuertes et al., 2005; Wu et al., 2010).
Microarray
RNA from whole cerebellum from wild type and transgenic mice was prepared using TRIzol (Invitrogen) and the RNAeasy Mini Kit (Qiagen). RNA was quantified and the quality was assessed with a 2100 Bioanalyzer (Agilent technologies). cDNA was synthesized from 4 μg of total RNA using one-cycle target labeling and control reagents (Affymetrix) to produce biotin labeled cRNA. The cRNA preparation (15 μg) was fragmented at 94°C for 35 min into 35–200 bases in length. Labeled cRNAs were hybridized to Affymetrix chips (GeneChip Mouse Genome 430 2.0 Array). Each sample was added to a hybridization solution containing 100 mM 2-(N-morpholino) ethanesulfonic acid, 1 M Na+, and 20 mM of EDTA in the presence of 0.01% of Tween-20 to a final cRNA concentration of 0.05 μg/ml. Hybridization was performed for 16 h at 45°C. Each microarray was washed and stained with streptavidin–phycoerythrin in a Fluidics station 450 (Affymetrix) and scanned at 1.56 μm resolution in a GeneChip® Scanner 3000 7G System (Affymetrix).
Microarray data analysis
Three biological replicates were independently hybridized for each cell type. GeneChip intensities were background-corrected, normalized and summarized by the RMA method (Irizarry et al., 2003) using the “Affy” package (Gautier et al., 2004) from Bioconductor. Rank Products method (Breitling et al., 2004) was applied to identify differentially expressed genes as implemented in the “RankProd” package (Hong et al., 2006) from Bioconductor. Genes with FDR <0.05 were included in the list of induced or repressed candidates. FIESTA viewer was used to facilitate the application of these numerical filters and the selection of candidate genes (Oliveros, 2007). For the functional annotation of the results GeneCodis 3.0 was used (Nogales-Cadenas et al., 2009) applying a hypergeometric distribution and a FDR threshold of 0.05 for detecting over-represented Gene Ontology terms in candidate genes.
Real-time quantitative PCR
RNA was isolated from whole tissues using TRIzol (Invitrogen), treated with DNAse (Ambion), and reverse transcribed using hexamer primer and Moloney murine leukemia virus reverse transcriptase. To confirm the absence of genomic DNA, each sample was processed in parallel without reverse transcriptase. Real-time quantitative PCR (qPCR) for endogenous DREAM, and daDREAM was performed as described (Savignac et al., 2005). Validation of microarray up- or down-regulated genes was with specific primers and TaqMan MGB probes (Applied Biosystems). The results were normalized as indicated by parallel amplification of HPRT.
Western blot analysis
Fifty micrograms of total protein from cerebellum or hippocampus were resolved in SDS-PAGE and transferred to PVDF membranes (Millipore). Antibodies against Mid1 (Ab 70770, Abcam) and β-actin (Sigma) were used.
Anatomical analysis
For observation of adult cerebellum, 2-months-old wild type and transgenic mice were anesthetized and whole brains were carefully removed from the skull. Coronal and sagittal sections were taken and measurements of left–right and rostro-caudal axis were performed. For histological analysis, brains from 15-days-old pups (P15) were harvested and fixed in 4% paraformaldehyde for 48 h at 4°C. Brains were processed for paraffin embedding and microtome sectioning. Brain sagittal sections of 10 μm were stained with hematoxylin/eosin using standard procedures.
Behavioral analysis
Experiments were performed in newborn mice homozygous for the transgene and wild type littermates. Mice were initially housed in a temperature (21 ± 1°C) and humidity (65 ± 10%) controlled room with a 12/12-h light/dark cycle (lights on from 0800 to 2000 hours) with ad libitum food and water. Breeding pairs were formed and females visibly close to parturition were isolated. To avoid the effects of parity on behavioral ontogeny (Crusio and Schmitt, 1996) the first litter not used for the experiments. Testing involved approximately equal amounts of males and females. Experiments were carried out during the dark phase of the light–dark cycle. Behavioral tests and animal care were conducted according the EU and local ethical guidelines (EU directive 86/609 and Appendix A of the Council of Europe Convention ETS123, EU decree 2001-486 Decree 214/97 and Spanish law 32/2007) and approved by the local ethical committee (CEEA-PRBB). All behavioral experiments were carried out with the experimenter blind to the genotype. Developmental landmarks analyzed include: (i) assessment of body growth and body length were recorded from P1, the day of birth; (ii) fur appearance: beginning on P2 appearance of immature fur was observed and fur was defined as the mature hair being raised; (iii) incisor eruption: beginning on P7 pups were inspected daily for the emergence of both lower and upper incisor from the gingival; and (iv) eye opening, beginning on P9 pups were inspected daily for the complete opening of both eyelids. Neuromotor development was assessed on P7 and P10 by means of the pivoting and walking tests and general psychomotor development on P14 by means of the homing test.
In the pivoting locomotion test, the total number of degrees turned by the pup during a 60-s period was recorded. The test was performed on a flat surface covered with a paper on which lines had been drawn to delineate four 90° quadrants. The number of degrees was scored only in completed 90° segments.
In the walking test, the latency for a mouse to lift up on all four legs and walk a distance exceeding its body length was measured on a flat surface covered with a green paper.
For the homing test, individual pups on postnatal day 14 were transferred to a cage containing new sawdust in 3/4 and 1/4 of sawdust of the home litter (“goal arena”). The pups were placed in the opposite side of the goal arena, near to the wall. The time taken to reach the home litter sawdust was recorded. A cut-off time of 180 s was applied.
Significance of the effects was assessed by two-tailed, unpaired Student’s t-test was used for comparisons between groups. Analysis was processed using the SPSS program.
Author Contributions
Conceived and designed the experiments: Mara Dierssen, Britt Mellström, Marisa Brini, Ernesto Carafoli, and Jose R. Naranjo. Performed the experiments: Laura Fedrizzi, Rosa Gomez-Villafuertes, María Martinez de Lagran, Alfonso Gutierrez-Adan, Ignasi Sahún, Belen Pintado, Xose M. Dopazo, and Paz Gonzalez. Analyzed the data: Britt Mellström, Juan C. Oliveros, Mara Dierssen, and Jose R. Naranjo. Wrote the paper: Mara Dierssen, Britt Mellström, and Jose R. Naranjo.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work was supported by grants from DGICYT (SAF2005-04682; SAF2007: /-62449/-64062/-31093E/-60827; SAF2008-03469; SAF2010-16427; SAF2010-21784), Spanish Ministry of Health (PI 082038), DURSI (2009SGR1313), Fundación Reina Sofía (PI 006/09), La Marató (2007-062231), La Caixa (BM04-167-0), and Areces, EU sixth Framework Program (PROMEMORIA; PHECOMP_037669; LSH-037627; NeuroNE, LSHG-CT-2006-037627; PS09/02673; and CureFXS, ERA-NET E-Rare, EU/FIS PS09102673).
Appendix
Table A1.
Functional annotation of over-represented candidate genes according to Gene Ontology (Cellular components).
| GO ID | Term | Probe ID | P-value | FDR |
|---|---|---|---|---|
| GO:0005925 | Focal adhesion | 1417654_at | 0.0134524 | 0.045738 |
| GO:0005615 | Etracellular space | 1449340_at, 1460049_s_at | 0.0050619 | 0.0286841 |
| GO:0015630 | Microtubule cytoskeleton | 1438239_at | 0.0116782 | 0.0496325 |
| GO:0043034 | Costamere | 1417654_at | 0.0024424 | 0.0207604 |
| GO:0042612 | MHC class I protein complex | 1425545_x_at | 0.00211703 | 0.0359896 |
Table A2.
Functional annotation of over-represented candidate genes according to Gene Ontology (Biological process).
| GO ID | Term | Probe ID | P-value | FDR |
|---|---|---|---|---|
| GO:0007389 | Pattern specification process | 1449340_at | 0.0142579 | 0.0161589 |
| GO:0016055 | Wnt receptor signaling pathway | 1449340_at | 0.0262754 | 0.0262754 |
| GO:0042475 | Odontogenesis of dentin-containing tooth | 1449340_at | 0.00731229 | 0.0088792 |
| GO:0006955 | Immune response | 1425545_x_at | 0.0243609 | 0.0258834 |
| GO:0032874 | Positive regulation of stress-activated MAPK cascade | 1438239_at | 0.00211703 | 0.00514137 |
| GO:0030514 | Negative regulation of BMP signaling pathway | 1449340_at | 0.00471748 | 0.0066831 |
| GO:0051894 | Positive regulation of focal adhesion assembly | 1417654_at | 0.000977557 | 0.00415462 |
| GO:0090398 | Cellular senescence | 1460049_s_at | 0.00130323 | 0.00443099 |
| GO:0051496 | Positive regulation of stress fiber assembly | 1417654_at | 0.00227973 | 0.00484442 |
| GO:0070314 | G1 to G0 transition | 1460049_s_at | 0.000488878 | 0.00415546 |
| GO:0045860 | Positive regulation of protein kinase activity | 1417654_at | 0.00504214 | 0.00659357 |
| GO:0007026 | Negative regulation of microtubule depolymerization | 1438239_at | 0.00260505 | 0.00492065 |
| GO:0019882 | Antigen processing and presentation | 1425545_x_at | 0.00471748 | 0.0066831 |
| GO:0008054 | Cyclin catabolic process | 1460049_s_at | 0.000651793 | 0.00369349 |
| GO:0002474 | Antigen processing and presentation of peptide antigen via MHC class I | 1425545_x_at | 0.00195432 | 0.00553723 |
| GO:0001916 | Positive regulation of T-cell mediated cytotoxicity | 1425545_x_at | 0.00276768 | 0.00470505 |
| GO:0002485 | Antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent | 1425545_x_at | 0.000488878 | 0.00415546 |
References
- Alexander J. C., Mcdermott C. M., Tunur T., Rands V., Stelly C., Karhson D., Bowlby M. R., An W. F., Sweatt J. D., Schrader L. A. (2009). The role of calsenilin/DREAM/KChIP3 in contextual fear conditioning. Learn. Mem. 16, 167–177 10.1101/lm.1261709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W. F., Bowlby M. R., Betty M., Cao J., Ling H. P., Mendoza G., Hinson J. W., Mattsson K. I., Strassle B. W., Trimmer J. S., Rhodes K. J. (2000). Modulation of A-type potassium channels by a family of calcium sensors. Nature 403, 553–556 10.1038/35000592 [DOI] [PubMed] [Google Scholar]
- Anderson D., Mehaffey W. H., Iftinca M., Rehak R., Engbers J. D., Hameed S., Zamponi G. W., Turner R. W. (2010). Regulation of neuronal activity by Cav3-Kv4 channel signaling complexes. Nat. Neurosci. 13, 333–337 10.1038/nn.2493 [DOI] [PubMed] [Google Scholar]
- Aranda-Orgilles B., Rutschow D., Zeller R., Karagiannidis A. I., Kohler A., Chen C., Wilson T., Krause S., Roepcke S., Lilley D., Schneider R., Schweiger S. (2011). Protein phosphatase 2A (PP2A)-specific ubiquitin ligase MID1 is a sequence-dependent regulator of translation efficiency controlling 3-phosphoinositide-dependent protein kinase-1 (PDPK-1). J. Biol. Chem. 286, 39945–39957 10.1074/jbc.M111.224451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker L. G. (2006). What you can learn from one gene: GLI3. J. Med. Genet. 43, 465–469 10.1136/jmg.2004.029181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004). Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83–92 10.1016/j.febslet.2004.07.055 [DOI] [PubMed] [Google Scholar]
- Buxbaum J. D., Choi E. K., Luo Y., Lilliehook C., Crowley A. C., Merriam D. E., Wasco W. (1998). Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat. Med. 4, 1177–1181 10.1038/2673 [DOI] [PubMed] [Google Scholar]
- Cali T., Fedrizzi L., Ottolini D., Gomez-Villafuertes R., Mellstrom B., Naranjo J. R., Carafoli E., Brini M. (2012). Ca2+ -activated nucleotidase 1, a novel target gene for the transcriptional repressor DREAM (downstream regulatory element antagonist modulator), is involved in protein folding and degradation. J. Biol. Chem. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion A. M., Link W. A., Ledo F., Mellstrom B., Naranjo J. R. (1999). DREAM is a Ca2+ -regulated transcriptional repressor. Nature 398, 80–84 10.1038/18044 [DOI] [PubMed] [Google Scholar]
- Carrion A. M., Mellstrom B., Naranjo J. R. (1998). Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element. Mol. Cell. Biol. 18, 6921–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. Y., Pitcher G. M., Laviolette S. R., Whishaw I. Q., Tong K. I., Kockeritz L. K., Wada T., Joza N. A., Crackower M., Goncalves J., Sarosi I., Woodgett J. R., Oliveira-Dos-Santos A. J., Ikura M., Van Der Kooy D., Salter M. W., Penninger J. M. (2002). DREAM is a critical transcriptional repressor for pain modulation. Cell 108, 31–43 10.1016/S0092-8674(01)00629-8 [DOI] [PubMed] [Google Scholar]
- Crusio W. E., Schmitt A. (1996). Prenatal effects of parity on behavioral ontogeny in mice. Physiol. Behav. 59, 1171–1174 10.1016/0031-9384(95)02251-1 [DOI] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B. M., Irizarry R. A. (2004). Affy – analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- Gomez-Villafuertes R., Torres B., Barrio J., Savignac M., Gabellini N., Rizzato F., Pintado B., Gutierrez-Adan A., Mellstrom B., Carafoli E., Naranjo J. R. (2005). Downstream regulatory element antagonist modulator regulates Ca2+ homeostasis and viability in cerebellar neurons. J. Neurosci. 25, 10822–10830 10.1523/JNEUROSCI.3912-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenpusch-Theil K., Magnani D., Amaniti E. M., Han L., Armstrong D., Theil T. (2012). Transcriptional analysis of Gli3 mutants identifies Wnt target genes in the developing hippocampus. Cereb. Cortex[Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F., Breitling R., Mcentee C. W., Wittner B. S., Nemhauser J. L., Chory J. (2006). RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22, 2825–2827 10.1093/bioinformatics/btl331 [DOI] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Krauss S., Foerster J., Schneider R., Schweiger S. (2008). Protein phosphatase 2A and rapamycin regulate the nuclear localization and activity of the transcription factor GLI3. Cancer Res. 68, 4658–4665 10.1158/0008-5472.CAN-07-6174 [DOI] [PubMed] [Google Scholar]
- Lancioni A., Pizzo M., Fontanella B., Ferrentino R., Napolitano L. M., De Leonibus E., Meroni G. (2010). Lack of Mid1, the mouse ortholog of the Opitz syndrome gene, causes abnormal development of the anterior cerebellar vermis. J. Neurosci. 30, 2880–2887 10.1523/JNEUROSCI.4196-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J. R., Mager D. L. (2002). Widely spaced alternative promoters, conserved between human and rodent, control expression of the Opitz syndrome gene MID1. Genomics 80, 499–508 10.1006/geno.2002.6863 [DOI] [PubMed] [Google Scholar]
- Ledo F., Kremer L., Mellstrom B., Naranjo J. R. (2002). Ca2+ -dependent block of CREB-CBP transcription by repressor DREAM. EMBO J. 21, 4583–4592 10.1093/emboj/cdf440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledo F., Link W. A., Carrion A. M., Echeverria V., Mellstrom B., Naranjo J. R. (2000). The DREAM-DRE interaction: key nucleotides and dominant negative mutants. Biochim. Biophys. Acta 1498, 162–168 10.1016/S0167-4889(00)00092-6 [DOI] [PubMed] [Google Scholar]
- Lilliehook C., Bozdagi O., Yao J., Gomez-Ramirez M., Zaidi N. F., Wasco W., Gandy S., Santucci A. C., Haroutunian V., Huntley G. W., Buxbaum J. D. (2003). Altered Abeta formation and long-term potentiation in a calsenilin knock-out. J. Neurosci. 23, 9097–9106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link W. A., Ledo F., Torres B., Palczewska M., Madsen T. M., Savignac M., Albar J. P., Mellstrom B., Naranjo J. R. (2004). Day-night changes in downstream regulatory element antagonist modulator/potassium channel interacting protein activity contribute to circadian gene expression in pineal gland. J. Neurosci. 24, 5346–5355 10.1523/JNEUROSCI.1460-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E., Knutzen C. A., Krauss S., Schweiger S., Chiang G. G. (2011). Control of mTORC1 signaling by the Opitz syndrome protein MID1. Proc. Natl. Acad. Sci. U.S.A. 108, 8680–8685 10.1073/pnas.1100131108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M., Wang J., Kandel E. R., O’Dell T. J. (1995). CaMKII regulates the frequency-response function of hippocampal synapses for the production of both LTD and LTP. Cell 81, 891–904 10.1016/0092-8674(95)90009-8 [DOI] [PubMed] [Google Scholar]
- Millen K. J., Gleeson J. G. (2008). Cerebellar development and disease. Curr. Opin. Neurobiol. 18, 12–19 10.1016/j.conb.2008.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munne P. M., Tummers M., Jarvinen E., Thesleff I., Jernvall J. (2009). Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development 136, 393–402 10.1242/dev.025064 [DOI] [PubMed] [Google Scholar]
- Nogales-Cadenas R., Carmona-Saez P., Vazquez M., Vicente C., Yang X., Tirado F., Carazo J. M., Pascual-Montano A. (2009). GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 37, W317–W322 10.1093/nar/gkp416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., Yoshino K., Hara K., Tanaka N., Avruch J., Yonezawa K. (2003). The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278, 15461–15464 10.1074/jbc.C200665200 [DOI] [PubMed] [Google Scholar]
- Oliveros J. C. (2007). “FIESTA at BioinfoGP” in An Interactive Server for Analyzing DNA Microarray Experiments with Replicates. Available at: http://bioinfogp.cnb.csic.es/tools/FIESTA
- Opitz J. M. (1987). G syndrome (hypertelorism with esophageal abnormality and hypospadias, or hypospadias-dysphagia, or “Opitz-Frias” or “Opitz-G” syndrome) – perspective in 1987 and bibliography. Am. J. Med. Genet. 28, 275–285 10.1002/ajmg.1320280129 [DOI] [PubMed] [Google Scholar]
- Osawa M., Dace A., Tong K. I., Valiveti A., Ikura M., Ames J. B. (2005). Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J. Biol. Chem. 280, 18008–18014 10.1074/jbc.M503002200 [DOI] [PubMed] [Google Scholar]
- Osawa M., Tong K. I., Lilliehook C., Wasco W., Buxbaum J. D., Cheng H. Y., Penninger J. M., Ikura M., Ames J. B. (2001). Calcium-regulated DNA binding and oligomerization of the neuronal calcium-sensing protein, calsenilin/DREAM/KChIP3. J. Biol. Chem. 276, 41005–41013 10.1074/jbc.M105842200 [DOI] [PubMed] [Google Scholar]
- Palczewska M., Casafont I., Ghimire K., Rojas A. M., Valencia A., Lafarga M., Mellstrom B., Naranjo J. R. (2011). Sumoylation regulates nuclear localization of repressor DREAM. Biochim. Biophys. Acta 1813, 1050–1058 10.1016/j.bbamcr.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Park J. W., Park E. S., Choi E. N., Park H. Y., Jung S. C. (2009). Altered brain gene expression profiles associated with the pathogenesis of phenylketonuria in a mouse model. Clin. Chim. Acta 401, 90–99 10.1016/j.cca.2008.11.019 [DOI] [PubMed] [Google Scholar]
- Pinson L., Auge J., Audollent S., Mattei G., Etchevers H., Gigarel N., Razavi F., Lacombe D., Odent S., Le Merrer M., Amiel J., Munnich A., Meroni G., Lyonnet S., Vekemans M., Attie-Bitach T. (2004). Embryonic expression of the human MID1 gene and its mutations in Opitz syndrome. J. Med. Genet. 41, 381–386 10.1136/jmg.2003.014829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaderi N. A., Schweiger S., Gaudenz K., Franco B., Rugarli E. I., Berger W., Feldman G. J., Volta M., Andolfi G., Gilgenkrantz S., Marion R. W., Hennekam R. C., Opitz J. M., Muenke M., Ropers H. H., Ballabio A. (1997). Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat. Genet. 17, 285–291 10.1038/ng1197-285 [DOI] [PubMed] [Google Scholar]
- Rhodes K. J., Carroll K. I., Sung M. A., Doliveira L. C., Monaghan M. M., Burke S. L., Strassle B. W., Buchwalder L., Menegola M., Cao J., An W. F., Trimmer J. S. (2004). KChIPs and Kv4 alpha subunits as integral components of A-type potassium channels in mammalian brain. J. Neurosci. 24, 7903–7915 10.1523/JNEUROSCI.0776-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M., Aurrekoetxea K., Mellstrom B., Naranjo J. R. (2011). Redox signaling regulates transcriptional activity of the Ca(2+)-dependent repressor DREAM. Antioxid. Redox Signal. 14, 1237–1243 10.1089/ars.2010.3385 [DOI] [PubMed] [Google Scholar]
- Rivas M., Mellstrom B., Naranjo J. R., Santisteban P. (2004). Transcriptional repressor DREAM interacts with thyroid transcription factor-1 and regulates thyroglobulin gene expression. J. Biol. Chem. 279, 33114–33122 10.1074/jbc.M403526200 [DOI] [PubMed] [Google Scholar]
- Rivas M., Mellstrom B., Torres B., Cali G., Ferrara A. M., Terracciano D., Zannini M., Morreale De Escobar G., Naranjo J. R. (2009). The DREAM protein is associated with thyroid enlargement and nodular development. Mol. Endocrinol. 23, 862–870 10.1210/me.2008-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Arconada I., Benedet T., Roza C., Torres B., Barrio J., Krzyzanowska A., Avendano C., Mellstrom B., Lopez-Garcia J. A., Naranjo J. R. (2010). DREAM regulates BDNF-dependent spinal sensitization. Mol. Pain 6, 95. 10.1186/1744-8069-6-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gomez A., Mellstrom B., Tornero D., Morato E., Savignac M., Holguin H., Aurrekoetxea K., Gonzalez P., Gonzalez-Garcia C., Cena V., Mayor F., Jr., Naranjo J. R. (2007). G protein-coupled receptor kinase 2-mediated phosphorylation of downstream regulatory element antagonist modulator regulates membrane trafficking of Kv4.2 potassium channel. J. Biol. Chem. 282, 1205–1215 10.1074/jbc.M607166200 [DOI] [PubMed] [Google Scholar]
- Savignac M., Mellstrom B., Bebin A. G., Oliveros J. C., Delpy L., Pinaud E., Naranjo J. R. (2010). Increased B cell proliferation and reduced Ig production in DREAM transgenic mice. J. Immunol. 185, 7527–7536 10.4049/jimmunol.1000152 [DOI] [PubMed] [Google Scholar]
- Savignac M., Pintado B., Gutierrez-Adan A., Palczewska M., Mellstrom B., Naranjo J. R. (2005). Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. EMBO J. 24, 3555–3564 10.1038/sj.emboj.7600810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scsucova S., Palacios D., Savignac M., Mellstrom B., Naranjo J. R., Aranda A. (2005). The repressor DREAM acts as a transcriptional activator on Vitamin D and retinoic acid response elements. Nucleic Acids Res. 33, 2269–2279 10.1093/nar/gki503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen M. B., Wang C., Ozgen N., Wang H. G., Rosen M. R., Pitt G. S. (2009). Accessory subunit KChIP2 modulates the cardiac L-type calcium current. Circ. Res. 104, 1382–1389 10.1161/CIRCRESAHA.109.196972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trockenbacher A., Suckow V., Foerster J., Winter J., Krauss S., Ropers H. H., Schneider R., Schweiger S. (2001). MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat. Genet. 29, 287–294 10.1038/ng762 [DOI] [PubMed] [Google Scholar]
- Winter J., Lehmann T., Krauss S., Trockenbacher A., Kijas Z., Foerster J., Suckow V., Yaspo M. L., Kulozik A., Kalscheuer V., Schneider R., Schweiger S. (2004). Regulation of the MID1 protein function is fine-tuned by a complex pattern of alternative splicing. Hum. Genet. 114, 541–552 10.1007/s00439-004-1114-x [DOI] [PubMed] [Google Scholar]
- Wu L. J., Mellstrom B., Wang H., Ren M., Domingo S., Kim S. S., Li X. Y., Chen T., Naranjo J. R., Zhuo M. (2010). DREAM (Downstream Regulatory Element Antagonist Modulator) contributes to synaptic depression and contextual fear memory. Mol. Brain 3, 3. 10.1186/1756-6606-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N. F., Kuplast K. G., Washicosky K. J., Kajiwara Y., Buxbaum J. D., Wasco W. (2006). Calsenilin interacts with transcriptional co-repressor C-terminal binding protein(s). J. Neurochem. 98, 1290–1301 10.1111/j.1471-4159.2006.03972.x [DOI] [PubMed] [Google Scholar]
- Zhang Y., Su P., Liang P., Liu T., Liu X., Liu X. Y., Zhang B., Han T., Zhu Y. B., Yin D. M., Li J., Zhou Z., Wang K. W., Wang Y. (2010). The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. J. Neurosci. 30, 7575–7586 10.1523/JNEUROSCI.5168-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]