Abstract
Mammalian host response to pathogens is associated with fluctuations in high abundant proteins in body fluids as well as in regulation of proteins expressed in relatively low copy numbers like cytokines secreted from immune cells and endothelium. Hence, efficient monitoring of proteins associated with host response to pathogens remains a challenging task. In this paper we present a targeted proteome analysis of a panel of 20 proteins that are widely believed to be key players and indicators of bovine host response to mastitis pathogens. Stable isotope labeled variants of two concordant proteotypic peptides from each of these 20 proteins were obtained through the QconCAT method. We present the quantotypic properties of these 40 proteotypic peptides, and discuss their application to research in host pathogen interactions. Our results clearly demonstrate a robust monitoring of 17 targeted host-response proteins. Twelve of these were readily quantified in a simple extraction of mammary gland tissues, while the expression levels of the remaining proteins were too low for direct and stable quantification; hence their accurate quantification requires further fractionation of mammary gland tissues.
Keywords: SRM, QconCAT assay, quantification, proteomics, quantotypic peptides, mastitis
Introduction
Quantitative and reproducible analyses of specific proteins in complex tissue samples are fundamental to understanding the protein dynamics in cells and tissues and for developing diagnostic methods for a wide range of diseases.1 Antibody based technologies, including sandwich enzyme linked immunosorbent assay (ELISA) and western blotting, as well as isotopic labeled and label-free mass spectrometry based methods have been developed to improve protein detection. However measuring the absolute or relative amounts of specific proteins within complex tissue samples remains a challenging task, particularly for proteins that are present in low concentrations within body fluids or in low copy numbers in cells.2-4 The application of selected reaction monitoring (SRM) mass spectrometry to proteomics has greatly enhanced the selectivity and sensitivity of protein quantification, and the use of precisely quantified stable isotope labeled peptides can support protein quantification in absolute terms. For such quantitative measurements the preparation of accurately quantified stable isotope labeled standards has proven to be a bottleneck. To meet this challenge, the QconCAT method was developed as an affordable alternative to the preparation of stoichiometrically equal, chemically synthesized reference peptides, and is a promising way of providing absolute and relative quantification of specific proteins in complex tissues.5,6
Mastitis caused by bacteria infections in the bovine mammary gland is a major health challenge in the dairy industry, and the detection of improved diagnostic markers for mastitis will benefit early diagnostics and treatment.7,8 Many factors influence the mammalian host response to pathogens, and molecular dissection of host response is complicated by the large number of proteins involved and by the fact that these response proteins are present over a wide concentration range. This range includes highly abundant proteins associated with the acute phase response in body fluids,9 as well as leukocyte derived proteins which are secreted into milk and plasma in relatively low copy numbers.10 Moreover, it is well known that there is considerable individual variation in the timing, extent of response and of the molecular pathways involved, which most likely reflects both complex genetic heterogeneity in the host as well as of the specific pathogens involved.11,12 Therefore, for efficient monitoring of bovine host response to mastitis pathogens, it is important to develop selective and quantitative protein assays that will allow large scale analyses of specific protein fluctuations in complex tissue and milk samples. The ultimate aim is to create better protein quantification assays by increasing sensitivity, precision and speed, all while reducing cost which will allow analysis of the large numbers of individual animal samples that are adequate for dissecting the complex molecular mechanisms of bovine host response to mastitis pathogens.
In this study we have developed a targeted proteome method that may become useful for routine and large scale analysis of 20 proteins that are widely held to be key players and indicators of bovine host response to mastitis pathogens. Each of the 20 proteins was targeted through isotope mass tagged versions of two proteotypic peptides that were obtained through the QconCAT expression system, and we have tested the robustness by which these peptides allow quantification of the 20 targeted proteins in a non-infected and experimentally infected mammary gland. Here we present a thorough analysis of the quantotypic properties of these 40 proteotypic target peptides, and we discuss the potentials of the QconCAT based method in building and optimizing quantitative assays for targeting functional subsets of proteins.
Experimental Methods
Selection of targeted proteins
Proteomics and transcriptomics studies have contributed to biomarker candidates for mastitis and inflammation.9,13-16 From these candidates, 20 target proteins were selected (Table 1), based on previous findings of their relevance to mastitis. A selection of cytokines that are well known to change abundance during mastitis were included, in order to explore quantification of relevant proteins in the low expression range.7
Table 1.
Characteristics of the quantotypic peptides used in the mastitis QconCAT. For each target protein, peptides were selected by a combination of prediction algorithms and ad hoc rules. For each protein, the table shows the selected peptides, the respective peptide molecular weights (MW), 5-6 best transitions (Q3 ions) for each peptide, CV of total and filtered transition data of standard QconCAT peptides in control (−) and challenged (+) mammary gland analyses. The Q3 ions: ions that passed the manual validation step and were further used and analyzed in the study are emphasized in bold text. Q3 ions are named according to charge state and position in the y ion series; hence 2y6 refers to a doubly charged y6 ion. Peptide Type classification: Type A (A/S): both good analyte and standard detection was obtained, Type B (−/S): good standard detection but weak analyte detection and Type C (−/−): weak detection of both standard and analyte.
| Protein | Peptide | Peptide Sequence | MW | Q3 ions | Peptide quality | CV(%) Control (−) |
Challenged (+) | ||
|---|---|---|---|---|---|---|---|---|---|
| Total | Filtered | Total | Filtered | ||||||
| Alpha-1 antitrypsin | A1AT-1 | ADLSGITK | 1167.6 | 2y5;2y6;2y7;3y3;3y4 | Type A (A+/S+) | 4.48 | 4.52 | 7.01 | 7.04 |
| A1AT-2 | LSISETYDLK | 803.44 | 2y5;2y6;2y7;2y8;2y9 | Type A (A+/S+) | 5.09 | 5.09 | 4.36 | 4.36 | |
| Alpha 2 macroglobulin |
A2M-1 | IQHHTLLASPVR | 1370.78 | 2y8;2y9;3y5;3y6;3y7;3y8 | Type A (A+/S+) | 3.54 | 3.54 | 8.53 | 8.53 |
| A2M-2 | LPPNVVEESAR | 1209.64 | 2y6;2y7;2y8;2y9;3y4 | Type A (A+/S+) | 8.52 | 8.52 | 6.11 | 6.11 | |
| Calgranulin B (S100A9) |
CALGB-1 | ELPNFLK | 859.48 | 2y4;2y5;2y6;3y3 | Type A (A+/S+) | 1.54 | 1.53 | 9.61 | 9.91 |
| CALGB-2 | LGHYDTLIQK | 1186.63 | 2y6;2y7;3y4;3y5;3y6 | Type A (A+/S+) | 8.99 | 8.99 | 7.46 | 7.46 | |
| Calgranulin C (S100A12) |
CALGC-1 | DQPTIDK | 815.4 | 2y4;2y5;2y6;3y3;3y4 | Type C (A−/S−) | 143.29 | 149.58 | 30.96 | 7.15 |
| CALGC-2 | VGHFDTLNK | 1029.52 | 2y5;2y6;3y3;3y4;3y5 | Type A (A+/S+) | 11.88 | 11.88 | 14.53 | 14.53 | |
| Cathepsin C | CATC-1 | NVHGINFVTPVR | 1351.74 | 2y7;2y9;3y4;3y5;3y6 | Type A (A+/S+) | 7.11 | 7.11 | 25.99 | 25.99 |
| CATC-2 | TGNTSENVNVNTAR | 1475.7 | 2y10;2y11;2y7;2y8;2y9 | Type A (A+/S+) | 2.03 | 2.03 | 2.06 | 2.06 | |
| CD14 | CD14-1 | LGAAQVPAQLLVAVLR | 1617.99 | 2y10;2y8;2y9;3y6;3y7;3y8 | Type A (A+/S+) | 5.55 | 5.74 | 10.86 | 11.1 |
| CD14-2 | VQPQSLDLSHNSLR | 1592.83 | 2y10;3y5;3y6;3y7;3y8;3y9 | Type A (A+/S+) | 5.85 | 5.82 | 4.54 | 4.41 | |
| CXCL1 (GRO1) | CXCL1-1 | VTTPGPHCDQTEVIASLK | 1894.95 | 3y10;3y7;3y8;3y9 | Type C (A−/S−) | 104.3 | n/a | 13.06 | n/a |
| CXCL1-2 | VTTPGPHCDQTEVIATLK | 1908.96 | 3y10;3y7;3y8;3y9 | Type C (A−/S−) | 84.98 | n/a | 32.96 | n/a | |
| Chemokine (C-X-C motif) ligand 3 |
CXCL3-1 | AATAAAPR | 1846.96 | 2y4;2y5;2y6;2y7;3y2 | Type B (A−/S+) | 27.29 | 3.06 | 16.17 | 32.61 |
| CXCL3-2 | VTPPGPHCGQTEVIATLK | 727.4 | 3y10;3y7;3y8;3y9 | Type B (A−/S+) | 3.63 | 3.63 | 7.06 | 7.06 | |
| Haptoglobin | HP-1 | NQLVEVEK | 979.49 | 2y4;2y5;2y6;2y7;3y3 | Type A (A+/S+) | 3.76 | 4.15 | 10.37 | 10.92 |
| HP-2 | VGYVSGWGR | 957.51 | 2y5;2y6;2y7;2y8;3y3 | Type A (A+/S+) | 4.78 | 4.56 | 6.35 | 6.47 | |
| Interleukin 1β | IL1B-1 | GGQDITDFR | 1658.74 | 2y4;2y5;2y6;2y7;3y3;3y4 | Type A (A+/S+) | 5.31 | 5.02 | 4.73 | 6.21 |
| IL1B-2 | NSAYAHVFHDDDLR | 1007.47 | 2y7;2y8;3y5;3y6;3y7;3y8 | Type B (A−/S+) | 11.12 | 11.12 | 18.65 | 18.65 | |
| IL 1 receptor antagonist |
IL1RA-1 | NNQLVAGYLQGPNTK | 1041.49 | 2y10;2y11;2y8;2y9;3y6;3y7 | Type A (A+/S+) | 14.25 | 14.25 | 19.37 | 19.37 |
| IL1RA-2 | SETACHPLGK | 1615.83 | 2y6;2y7;2y8;3y4;3y5;3y6 | Type C (A−/S−) | 15.17 | n/a | 33.52 | n/a | |
| Interleukin 6 | IL6-1 | IADLITTPATNTDLLEK | 1379.72 | 2y9;3y6;3y7;3y8;3y9 | Type B (A−/S+) | 96.68 | 3.05 | 22.69 | 8.71 |
| IL6-2 | NLENFLQFSLR | 1827.98 | 2y6;2y7;3y4;3y5;3y6 | Type B (A−/S+) | 3.56 | 3.58 | 12.44 | 4.47 | |
| IL-6 receptor | IL6R-1 | EDHPAGSVR | 966.45 | 2y6;2y7;3y3;3y4;3y5;3y6 | Type B (A−/S+) | 8.01 | 8.01 | 9.7 | 12.51 |
| IL6R-2 | QVFQEPCQYSPESQR | 1824.81 | 2y8;2y9;3y6;3y7;3y8 | Type B (A−/S+) | 3.79 | 3.79 | 10.68 | 10.68 | |
| Interleukin 8 | IL8-1 | VIESGPHCENSEIIVK | 1752.87 | 2y10;3y6;3y7;3y8;3y9 | Type C (A−/S−) | 51.64 | n/a | 16.8 | n/a |
| IL8-2 | VVQVFVK | 817.51 | 2y4;2y5;2y6;3y3;3y4 | Type B (A−/S+) | 4.7 | 4.7 | 2.25 | 2.25 | |
| Lectin, galactoside- binding, soluble, 1 |
LGALS1-1 | DAGAWGAEQR | 1059.47 | 2y5;2y6;2y7;2y8;3y3;3y4 | Type A (A+/S+) | 3.26 | 3.03 | 4.8 | 4.42 |
| LGALS1-2 | DDNNLCLHFNPR | 1456.65 | 2y6;2y7;3y4;3y5;3y6;3y7 | Type A (A+/S+) | 8.86 | 8.86 | 13.67 | 13.67 | |
| Lectin, galactoside- binding, soluble, 3 |
LGALS3-1 | GNDVAFHFNPR | 1323.72 | 2y5;2y6;2y7;3y4;3y5;3y6 | Type A (A+/S+) | 12.17 | 12.17 | 84 | 53.83 |
| LGALS3-2 | IQVLVEPDHFK | 1272.6 | 2y8;2y9;3y10;3y5;3y6;3y7 | Type A (A+/S+) | 1.87 | 1.87 | 15.19 | 7.98 | |
| Lactotransferrin | LTF-1 | GEADALNLDGGYIYTAGK | 1096.5 | 2y10;2y11;3y6;3y7 | Type A (A+/S+) | 12.17 | 12.17 | 11.38 | 11.38 |
| LTF-2 | YYGYTGAFR | 1826.87 | 2y6;2y7;2y8;3y3;3y4 | Type A (A+/S+) | 5.52 | 3.34 | 9.12 | 5.63 | |
| Serum amyloid A3 | SAA3-1 | ADQFANEWGR | 1360.72 | 2y5;2y6;2y7;2y8;2y9;3y4 | Type A (A+/S+) | 4.71 | 4.06 | 4.03 | 3.82 |
| SAA3-2 | ETIQGITDPLFK | 1192.53 | 2y6;2y7;2y8;2y9;3y7 | Type A (A+/S+) | 7.34 | 5.07 | 8.2 | 8.18 | |
| Tumor necrosis factor alpha |
TNFα-1 | AGGPQGSR | 1359.68 | 2y4;2y5;2y6;2y7;3y3 | Type B (A−/S+) | 4.05 | 4.05 | 5.24 | 5.24 |
| TNFα-2 | DVELAEEVLSEK | 728.36 | 2y10;2y6;2y7;2y8;2y9;3y4 | Type B (A−/S+) | 4.28 | 4.28 | 2.55 | 2.55 | |
| Vanin 1 | VAN1-1 | DSAPNTLSDLTTQALR | 1701.85 | 2y10;2y11;2y8;2y9;3y5;3y6 | Type A (A+/S+) | 4.32 | 4.32 | 6.65 | 6.65 |
| VAN1-2 | NLDLLEGAVTSASK | 1416.75 | 3y5;3y6;3y7;3y8;3y9 | Type A (A+/S+) | 7.26 | 7.26 | 9.2 | 9.2 | |
Selection of quantotypic peptides for QconCAT construction
Proteotypic peptides for the 20 target proteins were selected from the Bovine PeptideAtlas (www.peptideatlas.org) based on their suitability score which takes into account both observations from our previous shot-gun proteomics experiments and predicted observability derived from physiochemical properties.17 Only unique and true tryptic peptides with a length of 7-20 amino acids which mapped to single genome locations were chosen. Detailed criteria for selecting target peptides are given in supplementary file 1.
Five to six peptides (either observed or predicted in PeptideAtlas) from each protein were selected and synthesized using inexpensive Spot-synthesis technology (JPT Peptide Technologies GmbH, Berlin, Germany) and used without purification for optimizing the SRM assay conditions of the naturally occurring peptide analogues.18 A detailed description of SRM assay optimization using synthetic peptides can be found in supplementary file 1. The two most reproducible peptides for the QconCAT protein design and peptide synthesis were chosen based on these SRM assays. According to previous findings19 two independent proteotypic peptides per protein should be sufficient to provide a robust quantification of the specific proteins. For CXCL3, TNF-α, and IL1R only a few proteotypic peptides were suited as target peptides. From these three proteins four of the selected peptides originated from signal sequences, hence, our assays for CXCL3, TNF-α, and IL1R quantification is expected to be more robust in analyses of tissue samples than body fluids.
Isotope labeled heavy QconCAT peptides
The experimental methods for the expression of QconCAT proteins have previously been described.6 A QconCAT protein is concatamer of tryptic standard (quantotypic or Q-) peptides used for quantification. It is the product of a synthetic gene that uses highly abundant codons to maximize expression. In addition, the predicted transcript is scanned for features that might impede translation and if such features are found, the codon usage pattern or sequence of tryptic peptides is adjusted to remove them. Within the QconCAT the quantotypic peptides are present in a 1:1 stoichiometric ratio.6 In addition to the 40 proteotypic peptides, a [Glu1]-FibrinopeptideB (Glufib) peptide and a His-tag was included in the QconCAT. A more detailed description of the QconCAT construction is provided in supplementary file 1. MALDI-TOF mass spectrometry of the purified QconCAT confirmed that the protein was intact and that most of the tryptic peptides were readily discernible (Supplementary Figure 1). The concentration of purified QconCAT protein was determined using the Pierce BCA Protein Kit (Bie and Berntsen, Denmark) with bovine serum albumin (BSA) as standard, according to manufacturer’s manual. Purified QconCAT protein (4 fmol) was added to 1μg of every protein sample before tryptic digestion, hence 1 μg protein digest spiked with four fmol QconCAT peptides was injected on every SRM analysis. This QconCAT plasmid is available free of charge for research applications, and is distributed upon request from DNASU http://dnasu.asu.edu/DNASU/Home.jsp
Mammary gland tissue samples
The experimental procedures for inducing mastitis in dairy cows and obtaining mammary gland tissue have previously been described20. Briefly, a cow was challenged by inoculating a high dose of Streptococcus uberis (S. uberis) in the right frontal quarter of the udder. Simultaneously the left frontal quarter was inoculated with sterile milk (control). The inoculation resulted in a peracute S. uberis mastitis with a marked inflammatory response.20 Eight hours after inoculation, the cow was euthanized and the udder samples were collected. Approximately 5 g of tissue were collected from both infected and control mammary glands (distal site), frozen in liquid nitrogen and stored at −80 °C until use. Tissue samples were homogenized in 5μl/mg TES buffer (10mM Tris-HCl, pH 7.6; 1mM EDTA, 0.25M sucrose) using a TissueLyser (Qiagen, Switzerland). Homogenized samples were centrifuged at 10,000 × g for 30 min at 4 °C. Supernatants were collected for further analysis. Protein concentrations in the supernatants were determined using the Pierce BCA Protein Kit as described above for the concentration determination of the QconCAT protein. All procedures involving animals were approved by the Danish Animal Experiments Inspectorate and complied with the Danish Ministry of Justice Law concerning animal experimentation and care of experimental animals.
Protein digestion
Protein (100 g) from each sample plus 400 fmol (22 ng) QconCAT protein was precipitated using ice-cold acetone and resuspended in 20 μL digestion buffer (0.5M triethylammonium bicarbonate (TEAB), 0.1% (v/v) SDS). Cysteine residues were reduced with 2.5 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) (60 °C for 1 h) and then alkylated with 10 mM methylmethanethiosulfate-labeled (MMTS) (room temperature, 10 min in the dark). Proteins were then digested with trypsin (1:10 w/w) (ABSciex, CA, USA) at 37 °C over night and all samples were passed through a VWR Centrifugal Filter (pore size 0.2 μm) (VWR International, West Chester, PA, USA), for 10 minutes at 10,000 × g.
Mixed-mode cation exchange (MCX) purification of sample peptides
50 μg dried peptides were dissolved in 2% (v/v) ACN, 0.1% (v/v) formic acid and purified with mixed-mode ion-exchange. An Oasis MCX μElution Plate (Waters, Milford, MA, USA) was preconditioned with 800 μl methanol and equilibrated with 3 × 800 μl 0.1% (v/v) trifluoroacetic acid (TFA) in water. The 50 μg peptide samples were loaded on the plate and the wells were washed with 2 × 800 μl 0.1% TFA followed by 80% (v/v) acetonitrile/0.1% (v/v) TFA and Millipore water. The peptides were eluted from the wells with 2 × 800 μl 10 % (v/v) NH4OH/90% (v/v) methanol and the eluate was evaporated to dryness in a speed vacuum centrifuge and stored at −80 °C.
TIQAM transitions
SRM transitions specific for the 40 selected peptides were generated in silico with the Targeted Identification for Quantitative Analysis by MRM (TIQAM) Digestor software21 (from Seattle Proteome Center, http://tools.proteomecenter.org/TIQAM/TIQAM.html) with the following settings: precursor charges 2+ and 3+, 10 fragment ions from y-ion series with precursor Q1 m/z < 1800, Q3 m/z < 1250 and Q3 - Q1 m/z > + 20. Collision energies (CE) were calculated in TIQAM according to the following formulas: CE (2+) = 0.044 × m/z + 5.5 and CE (3+) = 0.051 × m/z + 0.5. The five transitions with highest intensity optimized according to supplementary file 1 were used for the final LC-SRM analysis.
LC-SRM analysis of mammary gland tissue digests
Prior to LC/MS analysis dried tissue digests were reconstituted in 2% (v/v) ACN, 0.1 % (v/v) formic acid to 1 μg/μl and 1 μl (corresponding to 1μg protein equivalent of peptides loaded on column) were injected on the Eksigent TempoTM nano MDLC LC system equipped with a peptide captrap column (Microm Bioresources Inc.). Peptides were loaded in 0.1 % (v/v) formic acid in water for 5 min at a flow rate of 3 μl/min. The samples were separated by a 70-min linear gradient of 2-25% (v/v) acetonitrile with 0.1 % (v/v) formic acid at a flow rate of 300 nl/min on a Acclaim PepMap 100 C18 analytical column (0.75 × 150 mm, 3 μm particles, Dionex, Sunnyvale, CA, USA). An ABSciex 5500 QTRAP fitted with the NanoSpray® III Assembly and controlled by Analyst 1.5 software was used for all LC-SRM analyses of tissue samples. The following parameters were used: 2400 V ion spray voltage, a curtain gas setting of 20 p.s.i., a 150 °C interface heater temperature, a declustering potential of 70 and Q1 and Q3 set to unit resolution. In SRM mode a dwell time of 20 ms was used and in the scheduled SRM mode a 2 s cycle time and a 6 min SRM detection window was applied.
SRM data analysis
All SRM data were processed using MultiQuant 1.2 with the MQL algorithm for peak integration. A 6 min retention time window and a one-point smooth with a peak-splitting factor of 4 were used. For noise percentage and the baseline subtraction window default MultiQuant values of 40 % and 2 min were used. All data were manually inspected to ensure correct peak detection and accurate integration. Transitions not detected or with low resolution were excluded from further data analysis. SRM data from this study is deposited and freely available in the publically accessible SRMAtlas (www.srmatlas.org) and SRM chromatographic data will be freely available in the PASSEL database (www.peptideatlas.org/passel/).
Statistical analysis
The R statistical software (R Development Core Team, 2010) was used for data handling and statistical analysis of variation estimates - CV%, t-tests, protein concentrations and fold changes. The p-value for each protein fold change was obtained from one-sample t-tests with null hypothesis: mean ratio (+/_) equals zero in log2 space. The protein concentration was calculated from all Q3 ion information within each replicate as the back-transformed average ion ratios (i.e., exp(2)) with known denominator = 4 fmol of purified QconCAT protein.
Results and Discussion
The aim of this study was to develop a targeted quantification method applicable for future large scale multiplexed analyses of specific proteins that are involved in the pathogenesis of bovine mastitis. We chose to test this in mammary udder tissue, where we expected that the host response proteins we wanted to quantify were differentially expressed upon pathogen challenge.22 The QconCAT method for manufacturing isotope labeled standard peptides was chosen because this method can provide reference peptides needed for quantitative analyses. Once the QconCAT-gene has been synthesized, repeated production of the QconCAT is quick and inexpensive, and the access to quantifiable standard peptides is effectively unlimited. Despite this advantage, only a few QconCATs have yet been reported and to the best of our knowledge, none have been reported for analyses of bovine proteins. This may reflect the novelty of molecular biology approaches to generation of standards (compared, for example, to commercial peptide synthesis) or the perceived cost. We estimated that at a coverage of two peptides per target protein, a QconCAT becomes cost-effective compared to synthetic stable isotope labeled peptides when quantification is required for more than five different proteins simultaneously. We designed the current study to include 20 proteins, and thus, the QconCAT approach was favored. An overview of the 40 selected peptides that were assembled into the present QconCAT is summarized in Table 1 together with 210 selected transitions. Detailed transition-level information is given in supplementary data Table S1.
The 40 peptides were selected for their structural and chemical properties, as described in the Methods section and in supplementary file 1. In order to investigate their quantotypic properties the QconCAT peptides were expressed as heavy labeled protein concatamers, added and co-digested with crude protein fractions extracted from mammary gland tissues sampled from a healthy and an inflamed mammary gland. A MALDI-TOF MS spectrum of the heavy QconCAT peptides after trypsin digestion of the purified QconCAT protein is presented in Supplementary figure 1.
The gland infected with S. uberis showed clear signs of induced mastitis inflammation20, hence the selected 20 proteins are anticipated to be up-regulated in this infected tissue sample. Observing the quantotypic properties of QconCAT peptides within these two different samples was therefore expected to provide a physiologically relevant set of test samples that could closely reflect the experimental conditions for which these standard peptides typically will be used in future biological experiments. QconCAT peptides act here as internal standards, supporting quantification of the endogenous peptides in the healthy and infected samples.
Manual validation and curation of transition peak detection
For the first pass of data analysis we relied on automated spectral peak detection and integration methods of the MultiQuant software. Further manual verification of peak detection was made to exclude transitions that exceeded the selected retention time window and transitions in the noise level. Transitions with partially overlapping peaks that could be manually distinguished were edited and corrected.
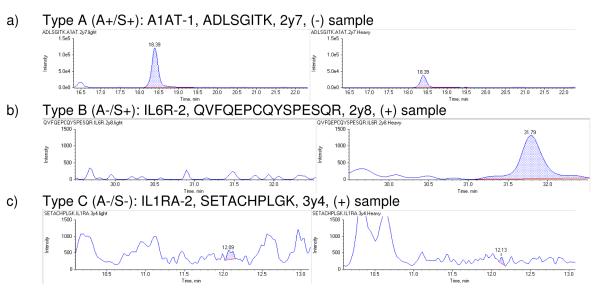
Since the errors in the automated data processing affect both the analyte (‘A’, light) and the standard (‘S’, heavy) transitions, we classified the transitions in three categories. Type A (A+/S+): good analyte detection and standard detection; Type B (A-/S+): weak or no analyte detection but good standard detection, and Type C (A−/S−): weak analyte and standard detection. Figure 1 shows typical transitions from each class in panels a, b and c, respectively.
Figure 1.
Selected reaction monitoring (SRM) chromatograms of three pairs of transitions that exemplify extracted Q3 ions from the analyte peptides (light transitions) in the left panels and the standard peptides (heavy transitions) in the right. When clear, sharp peaks were acquired from both standard and analyte peptides they were classified as a Type A (A+/S+) peptide. Weak detection of the analyte is classified as a Type B (A−/S+) peptide, while poor detection of both standard and analyte is classified as a Type C (A−/S−) peptide. The y-axis ranges (signal intensity) are presented at [0, 1.5e5] in panel a) and [0, 1.5e3] in panels b) and c).
Based on the quality of transition detection we categorized all peptides into quantotypic classes Type A (A+/S+), Type B (A−/S+) and Type C (A−/S−). In all, 25 peptides were of Type A (A+/S+), 10 of Type B (A−/S+) and five of Type C (A−/S−) (Table 1). Supplementary table 1 includes complete peptide and transition data from the three classes. Peptides of Type A have excellent analytical properties and thus are the best candidates for quantification. Type B peptides imply that a good standard signal could be detected, but the weak signal in the present samples prohibited quantification of the analyte. Nevertheless, type B peptides allow estimating the maximum quantity of low abundant proteins in mammary gland, as discussed in more detail in the following text. Peptides of Type C were excluded from further analyses in this work, but it may be expected that they could provide robust quantotypic traits if spiked and analyzed within different tissue extracts, or if spiked in at higher concentration to at least promote these to Type B peptides. After manual curation, 53 out of the 210 initial transitions were excluded from further data analysis (shown in plain text in Table 1; the 157 retained transitions are in bold text)
Reproducibility/stability of QconCAT peptide detection
A primary requirement for using internal standards in SRM based quantification is that the standard peptides can be monitored robustly in repeated analyses and in multiple types of samples. Hence a first step in our investigation was to check the reproducibility of the heavy QconCAT peptides among four repeated injections on the Q-trap instrument. The QconCAT construct was spiked into and co-digested with both healthy and challenged tissue samples and tested for stability of detection by monitoring the variation of signal intensity.
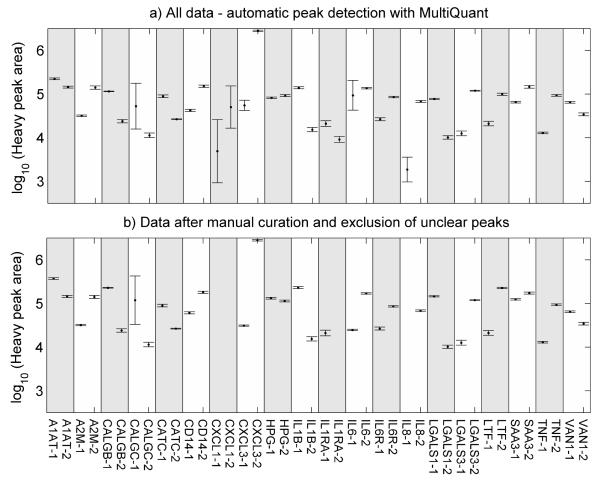
Transition ion chromatograms were obtained and processed for all 40 QconCAT peptides. Peak areas of the 40 heavy QconCAT peptides co-digested and co-analyzed with the healthy mammary gland tissue is presented in Figure 2. Notwithstanding the selection of ‘optimal’ peptides, the peak areas cover at least two orders of magnitude of response, reflecting the variation in ionization efficiency of different peptides. The data are expressed on a logarithmic scale for better visualization. The coefficients of variation (CV %) for all QconCAT peptides, both raw and manually filtered, are given in Table 1. However, correct peak detection of standard QconCAT peptides is compromised when only low intensity ion signals are present. Manual inspection of the data revealed that in this study four QconCAT peptides, namely CXCL1-1, CXCL1-2, Interleukin 1 receptor antagonist (ILRA)-2, and Interleukin 8 (IL8)-1 were all type C peptides and represented by low peak intensities which could not be correctly integrated using MultiQuant, and these peptides were therefore excluded from further analyses in this study. Consequently the analyses of the CXCL1 protein could not be supported by the current SRM method. Figure 2b shows the effect of manual editing of MultiQuant based peak detection. It indicates that the above peptides are not present within the intensity range that allow reproducible detection, and are prone to interfering peaks that make peak integration ambiguous. Manual editing was not necessary for 21 target peptides, mapping to 14 proteins, as determined by the identical values for CV in the control columns “total transition data “(−)” and “filtered transition data ”(−)” in Table 1. Detection of Chemokine (C-X-C motif) ligand 3 (CXCL3)-1 and Interleukin 6 (IL6)-1 was greatly improved but in general, only marginal improvements (e.g., LTF-2) or even deteriorations (e.g.CD14-1) were achieved by critical filtering and exclusion. Except for CALGC-1, all standard peptides have a CV <15 % in the control (−) sample. Similar variation is observed in the challenged (+) sample, with only 3 peptides showing CV > 20 %. As an example, the LGALS3 peptide GNDVAFHFNPR has a CV = 53.83 %. This is caused by small, but relevant retention time (RT) variation from replicate to replicate, due to the performance limitation of the Eksigent-Tempo liquid chromatography (LC) system used in this study. Here, two of the four replicate analyses, the peptide eluted at the threshold of the selected RT window limit, thus the integration of the peak could not be performed as accurately as in the other two replicates. Usually the selected 6 min retention time window would accommodate minor RT variations of the LC system.
Figure 2.
Peak areas of the 40 heavy QconCAT peptides co-digested and co-analyzed with the healthy mammary gland tissue. Each point symbol represents the mean area of a transition group of every separate injection/SRM analysis. All peptides with good standard detection (including both Type A and Type B peptides from Table 1) were used to calculate the mean. The vertical error bars define ± 1 SD about the mean from four repeated measurements of the areas (log10) of all fragment ions that represent the QconCAT standard peptides. Each transition was inspected and manually curated, whereby in a) peptide quantifications are based on automatic peak detection in MultiQuant before manual curation, and includes all data, while b) presents data after manual curation and exclusion of unclear peak data.
There is a clear correlation between expected cellular protein abundance and robustness of detection. The 20 targeted proteins represent both high and low abundance proteins, and although their absolute amounts have not yet been characterized in mammary gland, we can estimate likely ranges of their relative cellular abundances in healthy and challenged glands from studies of their concentrations and fluctuations, which typically have been measured in bovine and human milk and serum. All 20 proteins must be regarded as relatively low abundant proteins compared to e.g., structural proteins and metabolic enzymes.23 The five acute phase proteins: A1AT, A2M, LTF, HP, and SAA3 are typically secreted in the range of μg/ml to mg/ml to body fluids as a primary response to e.g., pathogens and to cytokine signaling.7,10,20 The proteins estimated to be present at μg/ml level in milk include CD14,24 the calgranulins (CALGB and CALGC) secreted by neutrophils and macrophages,25 cathepsin C (CATC) secreted from a variety of granulated immune cells,26 and the immune regulatory galectins (LGALS1 and LGALS3).27 The groups of very low abundant proteins include cytokines (IL1B, IL6, IL8, TNF-α), cytokine receptors (IL1RA, IL6R) and chemokines (CXCL1, CXCL3) which all are highly potent mediators of inflammation. These mediators elicit biological responses at very low concentrations often in the femtomolar to nanomolar range in both bovine7 and human28,29 tissues, hence their abundances may be below our current detection or quantification limit. Our study clearly reflects that there is a correlation between expected protein abundance and the robustness of quantotypic traits, as will be discussed below.
Effect of sample background on QconCAT standard detection
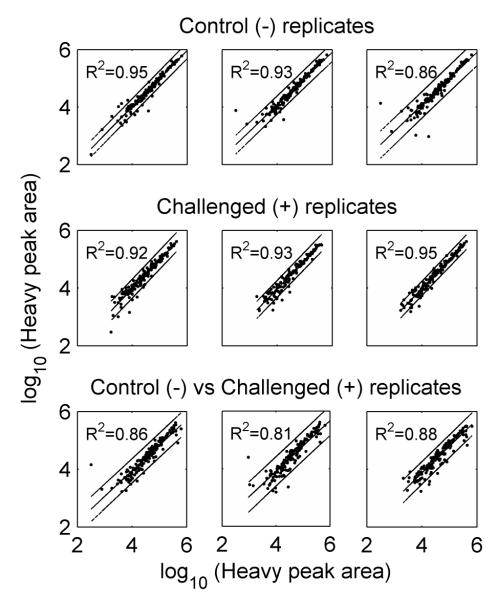
In order to be efficient, the heavy isotopically labeled QconCAT standard peptides must be monitored with similar stability irrespective of the subject sample matrix. Hence, to estimate whether different tissue samples influenced the detection of QconCAT standard peptides, the heavy labeled peptide peak areas of all transitions were measured in both a healthy control (−) mammary gland sample as well as in an infected (+) sample, and the two sets of peak intensities were compared. For this evaluation we used the transitions from Type A and Type B peptides with good standard detection (157 transitions, Table 1). There was no significant difference in the variances of the heavy peak areas between the two sets (P = 0.47, sample estimate ratio of variances = 1.06, CI = [0.9, 1.24]) and there was no significant replicate effect (P > 0.4). Figure 3 shows the strong correlations of the measured light peptide transition intensities, irrespective of the sample type and thus clearly reveals that the differences in the background protein expression levels does not significantly interfere with the variation of the signal intensity of the spiked-in QconCAT peptides. The coefficients of determination (R2) are around 0.9 with slightly better fits among the replicates of the same sample types (control or challenged) than across samples (control versus challenged). Hence we concluded that these peptides are suited for targeted comparative quantification across biological samples that are expected to have different expression levels of proteins.
Figure 3.
Pair-wise correlations of the standard (heavy) peak area of individual transitions within control (−), challenged (+), and between (+/−) replicates. Linear regression and the 95% prediction intervals are shown for each pair. The entire sets of 157 manually curated transitions from both Type A and Type B peptides from Table 1 were used.
Quantotypic properties of QconCAT peptides
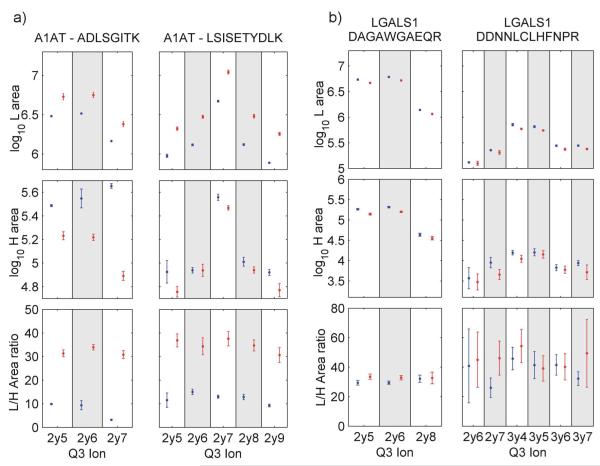
The quantitative properties of the 40 presented QconCAT peptides were evaluated at multiple levels as each component (measured peak areas or calculated ratios) is a source of variation for quantification/quantotypic traits. Figure 4a gives an example of robust and reliable protein quantification based on the transitions of the two peptides from the A1AT protein. All 8 transition ratios reflect approximately a 3-fold up-regulation of A1AT in the challenged sample which is in line with previously reported changes of A1AT in milk during mastitis.14 The LGALS1-1 peptide indicates a small but still significant up-regulation induced by the S. uberis challenge in (Figure 4b) However, the LGALS1-2 peptide while confirming this increase, also shows that the quantification is greatly affected by insufficient heavy QconCAT area detection, which implicitly gives rise to a large variation in the L/H area ratio (Figure 4b). A complete list of peak detection data for all peptides and transitions is presented in the supplementary table 1.
Figure 4.
Quantitative properties of concordant peptides A1AT and LGALS1. Peak area of light (L) and heavy (H) peptides (log10) and H/L area ratio of a) the two A1AT peptides and b) two LGALS1 peptides. All selected transitions from the control sample (blue) and challenged sample (red) are shown. Error bars span ±1 SD around the mean of 4 replicates.
Robustness of QconCAT based protein quantification
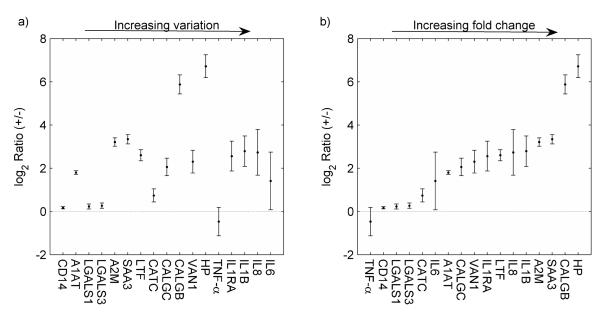
The ultimate aim of developing the present QconCAT SRM assay was to provide an efficient, accurate and affordable method that can support protein quantifications which are needed for testing hypotheses on bovine host response to mastitis pathogens, exemplified here with S. uberis. CXCL3 and Interleukin 6 receptor (IL6R) which both were represented by two type B peptides, could not be quantified because the endogenous peptides were not detectable in our extraction of mammary gland tissues. The relative abundances of the 17 mastitis-related proteins that were successfully quantified are shown in Figure 5. Protein fold changes are plotted by increasing CV in panel a) and by increasing average ratios in panel b). Ratios were calculated from the mean of both concordant peptides from each protein except for the quantification of CALGC, IL6, IL8 and IL1RA where ratios could only be based on data from a single peptide.
Figure 5.
The log2 challenged/control (+/−) protein ratios displayed in increasing order of the CV % in panel a), and in increasing order of average ratios in panel b). Relative quantification included only Type A peptides (Table1) that provide both good standard and analyte detection. Error bars span the 95% confidence interval for the mean of each protein from four repeated measurements. No correlation between the CV and the average ratios can be observed (r=0.1).
Table 2 presents estimated abundance of these 17 proteins in udder tissues, as well as summary statistics (p-values and CV %) and fold changes. Concentration estimates are based on the equivalent of 4 fmol QconCAT protein added to the 1μg of total protein sample that was loaded for each injection on the Q-TRAP instrument. The 1μg protein represents extraction from 0.05 mg wet weight tissue. The first 12 proteins have CV < 25 % indicating robust reproducibility of quantification with QconCAT standard peptides. The largest ranges of variation i.e., CV > 25 % is observed for the low intensity fragment ions of the low abundance analytes (e.g., IL6, IL8, and tumor necrosis factor alpha (TNF-α)). As expected, the observed fold changes show that 16 of the 17 proteins i.e. all proteins except TNF-α were significantly up regulated (p < 0.05) in mammary gland after challenge with S. uberis. Also CD14, LGALS1 and LGALS2, measured with relatively small fold changes, were significantly increased after pathogen challenge. To our knowledge, only one previous study has provided quantitative analyses of the inflammatory response in mammary gland tissues upon S. uberis challenge30. This study presented transcriptome profiles using a combination of microarray and quantitative real-time reverse transcription PCR (qRT-PCR), reported fold changes that were dependent on the RNA quantification technology used and did not correlate well with each other. However, they demonstrated considerable increases in the transcripts for SAA3: 6.91/55.0, HP: 2.37, LTF: 3.39, CALGB: 1.61, CALGC: 7.26/20.0, LGALS3: 1.91, CD14: 1.75, IL8: 7.9, IL1B: 15.9, IL6: 6.0, and TNF-α: 3.4 (bold numbers refer to qRT-PCR data while numbers in plain font refer to microarray data) when measured in mammary gland tissue 60 hours after S. uberis challenge.30 Although fold changes of mRNA and proteins are not guaranteed to be directly comparable,31 the consistent overall trend indicates the ranges of relative protein expression that might be expected in the mammary gland tissue. The fold changes we observed for SAA3 (10.13), LTF (6.07), CALGC (4.16), LGALS3 (1.20), CD14 (1.12), IL8 (6.65), IL1B (6.91), and IL6 (2.65) are similar to the transcriptome fold change values whereas proteins HP (105.36) and CALGB (58.73) are significantly higher than the transcriptome fold change. TNF-α (0.72) is slightly down regulated at the protein level when compared to the published transcriptome fold change.30 The difference in fold change may reflect the quite large span in sampling time in these two experiments, as our samples were collected 8 h after infection, rather than the 60 h in the transcriptomics study. Two other studies measuring TNF-α using ELISA after S. uberis challenge were not able to detect increased concentrations of TNF-α in milk until ≥66 h after infection. Therefore TNF-α have probably not been present in a high enough concentration to be detected. Furthermore TNF-α is a cytokine that is expressed in low copy numbers, and the robustness by which it can be quantified is likely limited by its low abundance in mammary gland tissues. Taken together, the general close agreement between the measured protein fold changes by our SRM quantification and previous transcriptome data30 supports the robustness of our QconCAT-SRM based quantification method, allowing detection and relative quantification of these 17 mastitis related bovine proteins. To date, there is very limited information about expression patterns and cellular abundances of these important host-defense proteins in bovine samples. The QconCAT-SRM method presented here provides for the first time a truly rapid method for the quantification of these proteins in a wide range of bovine tissues.
Table 2.
Summary of relative (+/−) and absolute (fmol/μg) quantification in challenged/control (+/−) samples shown in figure 6. The number of supporting peptides and transitions, absolute concentrations in fmol/μg protein load (which reflects extraction from 50 μg tissues) in the control and challenged samples, the mean fold changes, p-values and CVs are provided. The proteins are listed according to increasing CV.
| Protein | Number of peptides |
Number of transitions |
Conc. (−) fmol/μg |
Conc. (+) fmol/μg |
Mean ratio (+/−) |
log2 mean ratio |
p-value | CV(%) |
|---|---|---|---|---|---|---|---|---|
| CD14 | 2 | 9 | 0.88 | 0.98 | 1.12 | 0.17 | 1.86E-03 | 2.20 |
| A1AT | 2 | 8 | 19.13 | 66.59 | 3.48 | 1.80 | 6.30E-06 | 3.59 |
| LGALS1 | 2 | 9 | 67.31 | 78.83 | 1.17 | 0.23 | 8.51E-03 | 5.21 |
| LGALS3 | 2 | 11 | 7.30 | 8.78 | 1.20 | 0.26 | 6.56E-03 | 5.55 |
| A2M | 2 | 11 | 17.49 | 162.00 | 9.26 | 3.21 | 1.49E-05 | 8.38 |
| SAA3 | 2 | 7 | 0.72 | 7.30 | 10.13 | 3.34 | 1.80E-05 | 9.24 |
| LTF | 2 | 6 | 31.43 | 190.81 | 6.07 | 2.60 | 6.65E-05 | 10.91 |
| CATC | 2 | 10 | 0.63 | 1.05 | 1.67 | 0.74 | 4.53E-03 | 13.95 |
| CALGC | 1 | 7 | 6.80 | 28.28 | 4.16 | 2.06 | 5.31E-04 | 16.40 |
| CALGB | 2 | 7 | 1.91 | 111.90 | 58.73 | 5.88 | 2.85E-05 | 18.84 |
| VAN1 | 2 | 11 | 0.45 | 2.23 | 4.94 | 2.30 | 8.02E-04 | 21.88 |
| HP | 2 | 7 | 0.20 | 20.95 | 105.36 | 6.72 | 3.31E-05 | 25.00 |
| TNF-α | 2 | 11 | 0.20 | 0.15 | 0.72 | −0.47 | 1.06E-01 | 25.60 |
| IL1RA | 1 | 6 | 0.29 | 1.69 | 5.89 | 2.56 | 1.31E-03 | 27.01 |
| IL1Β | 2 | 9 | 0.06 | 0.41 | 6.91 | 2.79 | 1.07E-03 | 33.96 |
| IL8 | 1 | 2 | 0.04 | 0.25 | 6.65 | 2.73 | 3.73E-03 | 47.41 |
| IL6 | 1 | 4 | 0.13 | 0.35 | 2.65 | 1.41 | 4.37E-02 | 59.73 |
We conclude that the expression levels of the five proteins TNF-α, IL1RA, IL1B, IL8 and IL6 that were quantified with uncertainty (CV > 25 %) and the two proteins CXCL3 and IL6R that could not be quantified are below our current limit of quantification in mammary gland tissues. Nevertheless, the robust standard peptide signals (the A and B type peptides in table 1) allow us to estimate the maximum amounts (upper limits of expression) of these low abundant proteins in mammary gland tissues. Assuming that 5% of standard peptide signal intensity represents the lower limit of quantification, 0.2 fmol of these low abundant proteins would allow stable detection, and therefore robust quantification in the 1μg protein extracts. Hence, we estimate that there is less than 0.2 fmol of TNF-α, IL1RA, IL1B, IL8, IL6, CXCL3, and Interleukin 6 receptor (IL6R) peptides in the analyzed protein load (1μg), and consequently less than 4 fmol of these proteins in 1 mg udder tissue.
The seven proteins TNF-α, IL1RA, IL1B, IL8, IL6, CXCL3, and IL6R are all a part of the cytokine/chemokine family. Cytokines and chemokines take part in the innate immune response signaling, and are well known to be expressed in only low abundance in mammary gland tissue as well as milk.7,32 Therefore, further studies are needed to evaluate the quantotypic properties of their target peptides. A limitation in depth of quantification is the quantity of protein that can be applied to the reversed phase chromatography column as column overloading leads to peak broadening, RT shifts and results in impaired quantification. Prior fractionation with increased sample concentration (e.g., OFFGEL electrophoresis33,34) can enhance the signal of low abundance peptides without overloading the column. Optimally, future validation in other tissue samples should be done by analyzing protein extracts from cells that are major sources of cytokines during infection. These could include lymphocytes, macrophages and neutrophils that can be isolated from milk and blood.7 SRM based protein quantification may become particularly advantageous for species where detailed antibody availability is very low, which particularly includes most farm animals. For investigation of the immune response in cows, only a limited number of bovine-specific antibodies are commercially available and likewise, very few commercial and non-commercial ELISA methods have been developed so far including assays for SAA3, BSA, LTF, C5a, IFN-γ, IL1B, IL8, IL10, LBP, sCD14, and TNF-α.24,35-38, (Bethyl Laboratories Inc. Montgomery, TX).
The advantages of mass spectrometry based protein quantification are becoming increasingly obvious, as these methods provide specific, fast and affordable protein quantification at an ever increasing speed.2 In particular, the rapidly growing field of SRM-based targeted protein quantification provides interesting alternatives to current analytical and diagnostic methods in farm animals. For improving analyses of pig and cattle proteins/proteomes, the accumulation of LC-MS/MS proteotypic peptide data by the development of pig and cattle peptide data in the PeptideAtlas repository (www.peptideatlas.org) have greatly improved the resources for design and development of SRM mass spectrometry based protein assays for use in farm animal sciences.
Conclusion
The aim of this study was to develop a robust, inexpensive, highly sensitive and specific quantitative assay system based on QconCAT coupled SRM methodology that can support future studies and particularly allow verification of specific and absolute protein abundances, including proteins that are of major interest for understanding molecular mechanisms in bovine host response to pathogens. We present a fast and cost efficient method that will allow frequent sampling and multiple measurements to be made, and will support in depth temporal characterization of tissues involved in host response, and particularly support studies where including many biological replicates is of key interest. The method presented here will support studies of biological variation and fluctuation of these inflammatory proteins, and may prove useful for diagnostics and surveillance in dairy herds. Additional studies using the presented QconCAT construct to characterize the level of these 20 proteins in other tissues, body fluids and cells may provide important additional information about the quantotypic traits of the target peptides presented in this work.
Our results clearly demonstrated that the QconCAT construct provides an excellent and robust targeted relative quantification method for 12 of the selected proteins in the specific mammary gland tissues studied. The QconCAT based spike peptides showed robust quantotypic performance for 17 of the targeted proteins, and only five peptides were classified Type C (A−/S− ;meaning weak analyte and standard signal) peptides which cannot yet support routine quantification using this current approach. These might be successfully substituted by other proteotypic peptide candidates in new QconCAT constructs or by adding additional fractionation or anti-peptide immunopurification. The 10 Type B (A−/S+) peptides all belong to the group of low abundant proteins and although their stable detection makes them suited for routine quantification; their quantotypic performance remains to be verified in tissue extracts and body fluids where the abundance of these targeted proteins is higher than in the bovine udder tissues which were chosen for this study. The relative quantification methods we provide for 17 of the targeted proteins will be particularly useful for characterizing tissue samples where in vivo labeling methods such as stable isotope labeling by amino acids in cell culture (SILAC) are not feasible.
The QconCAT-SRM approach described here has revealed, at the protein level, similarities in protein fold changes when compared to transcriptome studies and significant differential fold changes for three proteins (HP, CALGB and TNF-α) using easily established quantitative direct measurement assays. These targeted QconCAT-SRM based assays can circumvent the considerable cost and effort that is associated with developing species-specific ELISA assays which have prohibited their development and use within farm animal studies. Hence, we expect that SRM based mass spectrometry will be a valuable approach for both research and diagnostics in farm animals, and will in the future allow protein quantification at much larger scales than what has so far been feasible. In this study we present a first subset of SRM based protein assays targeting the bovine inflammatory response using a QconCAT method to provide stable isotopically heavy labeled peptides for the targeted quantification of 17 inflammatory proteins.
Supplementary Material
Synopsis.
There is great need to develop methods for monitoring bovine host response to pathogens that cause mastitis. In this paper we present a targeted proteome approach based on QconCAT heavy standard peptides to provide a fast and cost efficient quantification of 20 proteins which are key players in bovine host response. We present quantotypic properties of 40 proteotypic peptides, and discuss their application to research in host pathogen interactions.
Acknowledgements
We are grateful for excellent technical assistance from Dorte Thomassen and Laura Jeppesen.
This work was carried out in a BIOSENS Consortium project and was financed by the Danish Ministry of Food Agriculture and Fisheries, Lattec I/S, The Milk Levy Fund, the Faculty of Science and Technology and the Graduate School of Agriculture, Food and Environment at Aarhus University.
UK was supported by a fellowship within the PostDoc-Program of the German Academic Exchange Service, the PM50 GM076547/Center for Systems Biology, NHGRI ARRA HG005805 and the Luxembourg Centre for Systems Biomedicine and the University of Luxembourg (to RM). The involvement of RJB and VH was supported by the Biotechnology and Biological Science research Council, UK (BB/G009112/1).
Footnotes
Supporting Information Supplementary table 1, Supplementary file 1: Detailed methods and Supplementary figure 1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Gstaiger M, Aebersold R. Applying Mass Spectrometry-Based Proteomics to Genetics, Genomics and Network Biology. Nat. Rev. Genet. 2009;10(9):617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- (2).Huttenhain R, Malmstrom J, Picotti P, Aebersold R. Perspectives of Targeted Mass Spectrometry for Protein Biomarker Verification. Curr. Opin. Chem. Biol. 2009;13(5-6):518–525. doi: 10.1016/j.cbpa.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Schiess R, Wollscheid B, Aebersold R. Targeted Proteomic Strategy for Clinical Biomarker Discovery. Mol. Oncol. 2009;3(1):33–44. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Viguier C, Arora S, Gilmartin N, Welbeck K, O’Kennedy R. Mastitis Detection: Current Trends and Future Perspectives. Trends Biotechnol. 2009;27(8):486–493. doi: 10.1016/j.tibtech.2009.05.004. [DOI] [PubMed] [Google Scholar]
- (5).Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ. Multiplexed Absolute Quantification in Proteomics Using Artificial QCAT Proteins of Concatenated Signature Peptides. Nat. Methods. 2005;2(8):587–589. doi: 10.1038/nmeth774. [DOI] [PubMed] [Google Scholar]
- (6).Pratt JM, Simpson DM, Doherty MK, Rivers J, Gaskell SJ, Beynon RJ. Multiplexed Absolute Quantification for Proteomics Using Concatenated Signature Peptides Encoded by QconCAT Genes. Nat. Protoc. 2006;1(2):1029–1043. doi: 10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- (7).Bannerman DD. Pathogen-Dependent Induction of Cytokines and Other Soluble Inflammatory Mediators During Intramammary Infection of Dairy Cows. J. Anim Sci. 2009;87(13 Suppl):10–25. doi: 10.2527/jas.2008-1187. [DOI] [PubMed] [Google Scholar]
- (8).Seegers H, Fourichon C, Beaudeau F. Production Effects Related to Mastitis and Mastitis Economics in Dairy Cattle Herds. Vet. Res. 2003;34(5):475–491. doi: 10.1051/vetres:2003027. [DOI] [PubMed] [Google Scholar]
- (9).Pyorala S. Indicators of Inflammation in the Diagnosis of Mastitis. Vet. Res. 2003;34(5):565–578. doi: 10.1051/vetres:2003026. [DOI] [PubMed] [Google Scholar]
- (10).Rainard P, Riollet C. Innate Immunity of the Bovine Mammary Gland. Vet. Res. 2006;37(3):369–400. doi: 10.1051/vetres:2006007. [DOI] [PubMed] [Google Scholar]
- (11).Lund MS, Guldbrandtsen B, Buitenhuis AJ, Thomsen B, Bendixen C. Detection of Quantitative Trait Loci in Danish Holstein Cattle Affecting Clinical Mastitis, Somatic Cell Score, Udder Conformation Traits, and Assessment of Associated Effects on Milk Yield. J. Dairy Sci. 2008;91(10):4028–4036. doi: 10.3168/jds.2007-0290. [DOI] [PubMed] [Google Scholar]
- (12).Sorensen LP, Guldbrandtsen B, Thomasen JR, Lund MS. Pathogen-Specific Effects of Quantitative Trait Loci Affecting Clinical Mastitis and Somatic Cell Count in Danish Holstein Cattle. J. Dairy Sci. 2008;91(6):2493–2500. doi: 10.3168/jds.2007-0583. [DOI] [PubMed] [Google Scholar]
- (13).Boehmer JL, Bannerman DD, Shefcheck K, Ward JL. Proteomic Analysis of Differentially Expressed Proteins in Bovine Milk During Experimentally Induced Escherichia Coli Mastitis. J. Dairy Sci. 2008;91(11):4206–4218. doi: 10.3168/jds.2008-1297. [DOI] [PubMed] [Google Scholar]
- (14).Danielsen M, Codrea MC, Ingvartsen KL, Friggens NC, Bendixen E, Rontved CM. Quantitative Milk Proteomics--Host Responses to Lipopolysaccharide-Mediated Inflammation of Bovine Mammary Gland. Proteomics. 2010;10(12):2240–2249. doi: 10.1002/pmic.200900771. [DOI] [PubMed] [Google Scholar]
- (15).Jenner RG, Young RA. Insights into Host Responses Against Pathogens From Transcriptional Profiling. Nat. Rev. Microbiol. 2005;3(4):281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- (16).Lutzow YC, Donaldson L, Gray CP, Vuocolo T, Pearson RD, Reverter A, Byrne KA, Sheehy PA, Windon R, Tellam RL. Identification of Immune Genes and Proteins Involved in the Response of Bovine Mammary Tissue to Staphylococcus Aureus Infection. BMC. Vet. Res. 2008;4:18. doi: 10.1186/1746-6148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Computational Prediction of Proteotypic Peptides for Quantitative Proteomics. Nat. Biotechnol. 2007;25(1):125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- (18).Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. High-Throughput Generation of Selected Reaction-Monitoring Assays for Proteins and Proteomes. Nat. Methods. 2010;7(1):43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- (19).Lange V, Picotti P, Domon B, Aebersold R. Selected Reaction Monitoring for Quantitative Proteomics: a Tutorial. Mol. Syst. Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Pedersen LH, Aalbaek B, Rontved CM, Ingvartsen KL, Sorensen NS, Heegaard PM, Jensen HE. Early Pathogenesis and Inflammatory Response in Experimental Bovine Mastitis Due to Streptococcus Uberis. J. Comp Pathol. 2003;128(2-3):156–164. doi: 10.1053/jcpa.2002.0620. [DOI] [PubMed] [Google Scholar]
- (21).Lange V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong CH, Deutsch EW, Brusniak MY, Buhlmann P, Bjorck L, Domon B, Aebersold R. Targeted Quantitative Analysis of Streptococcus Pyogenes Virulence Factors by Multiple Reaction Monitoring. Mol. Cell Proteomics. 2008;7(8):1489–1500. doi: 10.1074/mcp.M800032-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Schukken YH, Gunther J, Fitzpatrick J, Fontaine MC, Goetze L, Holst O, Leigh J, Petzl W, Schuberth HJ, Sipka A, Smith DG, Quesnell R, Watts J, Yancey R, Zerbe H, Gurjar A, Zadoks RN, Seyfert HM. Host-Response Patterns of Intramammary Infections in Dairy Cows. Vet. Immunol. Immunopathol. 2011;144(3-4):270–289. doi: 10.1016/j.vetimm.2011.08.022. [DOI] [PubMed] [Google Scholar]
- (23).Boehmer JL, DeGrasse JA, McFarland MA, Tall EA, Shefcheck KJ, Ward JL, Bannerman DD. The Proteomic Advantage: Label-Free Quantification of Proteins Expressed in Bovine Milk During Experimentally Induced Coliform Mastitis. Vet. Immunol. Immunopathol. 2010;138(4):252–266. doi: 10.1016/j.vetimm.2010.10.004. [DOI] [PubMed] [Google Scholar]
- (24).Bannerman DD, Paape MJ, Goff JP, Kimura K, Lippolis JD, Hope JC. Innate Immune Response to Intramammary Infection With Serratia Marcescens and Streptococcus Uberis. Vet. Res. 2004;35(6):681–700. doi: 10.1051/vetres:2004040. [DOI] [PubMed] [Google Scholar]
- (25).Goyette J, Geczy CL. Inflammation-Associated S100 Proteins: New Mechanisms That Regulate Function. Amino. Acids. 2011;41(4):821–842. doi: 10.1007/s00726-010-0528-0. [DOI] [PubMed] [Google Scholar]
- (26).Trivedi NN, Caughey GH. Mast Cell Peptidases: Chameleons of Innate Immunity and Host Defense. Am. J. Respir. Cell Mol. Biol. 2010;42(3):257–267. doi: 10.1165/rcmb.2009-0324RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hokama A, Mizoguchi E, Mizoguchi A. Roles of Galectins in Inflammatory Bowel Disease. World J. Gastroenterol. 2008;14(33):5133–5137. doi: 10.3748/wjg.14.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Bienvenu JA, Monneret G, Gutowski MC, Fabien N. Cytokine Assays in Human Sera and Tissues. Toxicology. 1998;129(1):55–61. doi: 10.1016/s0300-483x(98)00063-8. [DOI] [PubMed] [Google Scholar]
- (29).Zhou X, Fragala MS, McElhaney JE, Kuchel GA. Conceptual and Methodological Issues Relevant to Cytokine and Inflammatory Marker Measurements in Clinical Research. Curr. Opin. Clin. Nutr. Metab Care. 2010;13(5):541–547. doi: 10.1097/MCO.0b013e32833cf3bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Swanson KM, Stelwagen K, Dobson J, Henderson HV, Davis SR, Farr VC, Singh K. Transcriptome Profiling of Streptococcus Uberis-Induced Mastitis Reveals Fundamental Differences Between Immune Gene Expression in the Mammary Gland and in a Primary Cell Culture Model. J. Dairy Sci. 2009;92(1):117–129. doi: 10.3168/jds.2008-1382. [DOI] [PubMed] [Google Scholar]
- (31).Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full Dynamic Range Proteome Analysis of S. Cerevisiae by Targeted Proteomics. Cell. 2009;138(4):795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Boehmer JL, Ward JL, Peters RR, Shefcheck KJ, McFarland MA, Bannerman DD. Proteomic Analysis of the Temporal Expression of Bovine Milk Proteins During Coliform Mastitis and Label-Free Relative Quantification. J. Dairy Sci. 2010;93(2):593–603. doi: 10.3168/jds.2009-2526. [DOI] [PubMed] [Google Scholar]
- (33).Horth P, Miller CA, Preckel T, Wenz C. Efficient Fractionation and Improved Protein Identification by Peptide OFFGEL Electrophoresis. Mol. Cell Proteomics. 2006;5(10):1968–1974. doi: 10.1074/mcp.T600037-MCP200. [DOI] [PubMed] [Google Scholar]
- (34).Keidel EM, Dosch D, Brunner A, Kellermann J, Lottspeich F. Evaluation of Protein Loading Techniques and Improved Separation in OFFGEL Isoelectric Focusing. Electrophoresis. 2011;32(13):1659–1666. doi: 10.1002/elps.201000544. [DOI] [PubMed] [Google Scholar]
- (35).Bannerman DD, Paape MJ, Hare WR, Sohn EJ. Increased Levels of LPS-Binding Protein in Bovine Blood and Milk Following Bacterial Lipopolysaccharide Challenge. J. Dairy Sci. 2003;86(10):3128–3137. doi: 10.3168/jds.S0022-0302(03)73914-9. [DOI] [PubMed] [Google Scholar]
- (36).Bannerman DD, Paape MJ, Lee JW, Zhao X, Hope JC, Rainard P. Escherichia Coli and Staphylococcus Aureus Elicit Differential Innate Immune Responses Following Intramammary Infection. Clin. Diagn. Lab Immunol. 2004;11(3):463–472. doi: 10.1128/CDLI.11.3.463-472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Entrican G, Lunney JK, Rutten VP, Baldwin CL. A Current Perspective on Availability of Tools, Resources and Networks for Veterinary Immunology. Vet. Immunol. Immunopathol. 2009;128(1-3):24–29. doi: 10.1016/j.vetimm.2008.10.291. [DOI] [PubMed] [Google Scholar]
- (38).Rontved CM, Andersen JB, Dernfalk J, Ingvartsen KL. Effects of Diet Energy Density and Milking Frequency in Early Lactation on Tumor Necrosis Factor-Alpha Responsiveness in Dairy Cows. Vet. Immunol. Immunopathol. 2005;104(3-4):171–181. doi: 10.1016/j.vetimm.2004.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.