Abstract
In female mouse embryos, somatic cells undergo a random form of X chromosome inactivation (XCI), while extraembryonic trophoblast cells in the placenta undergo imprinted XCI, silencing exclusively the paternal X chromosome. Initiation of imprinted XCI requires a functional maternal allele of the X-linked gene Rnf12 which encodes the ubiquitin ligase Rnf12/RLIM. Here we find that knockout (KO) of Rnf12 in female mammary glands inhibits alveolar differentiation and milk production upon pregnancy. Alveolar cells lacking Rnf12/RLIM undergo apoptosis as soon as they differentiate from mammary epithelia. Genetic analyses demonstrate that these functions are mediated primarily by the paternal Rnf12 allele due to non-random XCI in mammary epithelial cells which primarily silence their maternal X chromosomes. These results identify paternal Rnf12/RLIM as a critical survival factor for milk-producing alveolar cells and provide strong evidence for an imprinted XCI pattern in mammary epithelial cells opposite to that found in extraembryonic trophoblast cells.
Introduction
To ensure appropriate dosage compensation, female cells selectively inactivate one of their two X chromosomes in a process called X chromosome inactivation (XCI), a form of epigenetic regulation. During embryogenesis of female mice sex-specific regulation has been discovered in extraembryonic placental trophoblast tissues which exclusively silence the paternal X chromosome (Xp), while somatic female tissues are thought to display a random pattern of XCI (Heard and Disteche, 2006; Payer and Lee, 2008). However, it has recently been reported that specific regions in the adult female brain display a bias to silence the paternal X chromosome (Gregg et al., 2010; Wang et al., 2010), although the physiological importance of this bias is unclear.
Milk-producing alveolar cells in the mammary gland are generated during breast differentiation in pregnant female mice (Visvader, 2009). Prolactin (Prl) signaling via Jak2 and Stat5 is essential for the differentiation and expansion of alveolar cells in pregnant and lactating females (Hennighausen and Robinson, 2008). Upon weaning, mammary alveolar cells undergo apoptosis in a process called involution (Sutherland et al., 2007b). While genes have been identified that promote alveolar differentiation, factors that trigger involution are still obscure.
The X-linked gene Rnf12 encodes the RING finger LIM domain-interacting protein (RLIM), a nuclear ubiquitin ligase that regulates the activity of specific transcription factors in part by controlling the levels of various cofactors (Ostendorff et al., 2002; Kramer et al., 2003; Gungor et al., 2007; Johnsen et al., 2009). Targeting a conditional knockout (KO) of Rnf12 in mouse oocytes, we have shown that the maternal transmission of an Rnf12 KO allele results in early embryonic lethality specifically of female embryos due to defective imprinted XCI, precluding development of embryonic trophoblast tissues. Moreover, the gender ratio of pups born by mothers carrying an Rnf12 mutation is significantly biased towards males. Indeed, males carrying a germline KO of Rnf12 (Δ/Y) appear healthy and are fertile (Shin et al., 2010).
To investigate functions of RLIM/Rnf12 in adult mice, we targeted the KO of Rnf12 to mammary epithelia and find inhibited alveolar differentiation and milk production in pregnant and lactating females. Indeed, alveolar cells lacking Rnf12/RLIM undergo apoptosis as soon as they differentiate indicating crucial survival functions for RLIM. We show that RLIM levels in alveolar cells dramatically decrease within hours upon forced weaning suggesting important roles for triggering involution. Genetic analyses demonstrate that these functions are mediated mainly by the paternal Rnf12 allele. Moreover, we find that mammary epithelia are composed primarily of cells with an active Xp. These results identify sex-specific epigenetic regulation of murine mammary gland biology by RLIM/Rnf12 with implications for development, differentiation, evolution and disease.
Results
Paternal RLIM/Rnf12 regulates alveolar morphogenesis and milk production
RLIM-encoding mRNA is widely expressed in embryonic and adult mouse tissues while RLIM protein expression is more restricted (Bach et al., 1999; Ostendorff et al., 2006). Using immunohistochemistry, we detected robust expression of RLIM in virgin, pregnant and lactating mammary epithelia, in myoepithelial and luminal cell layers, as indicated by its co-localization with cytokeratin 14 and 18 (CK14; CK18), respectively (Fig. S1A, B). We targeted the Rnf12 KO to mammary glands using transgenic mice that express Cre recombinase (Cre) under the control of the mouse mammary tumor virus long terminal repeat (MMTV-LTR) (Wagner et al., 1997). By crossing MMTV-Cre (MC) mice to Rosa26-loxP-stop-loxP-lacZ (R26R) animals (Soriano, 1999), Cre expression in early ducts was confirmed (Fig. S1C). Indeed, RLIM expression was not detectable in mammary epithelia of virgin Rnf12fl/fl × MC females (fl/fl × MC) (Fig. S1D), indicating correct targeting.
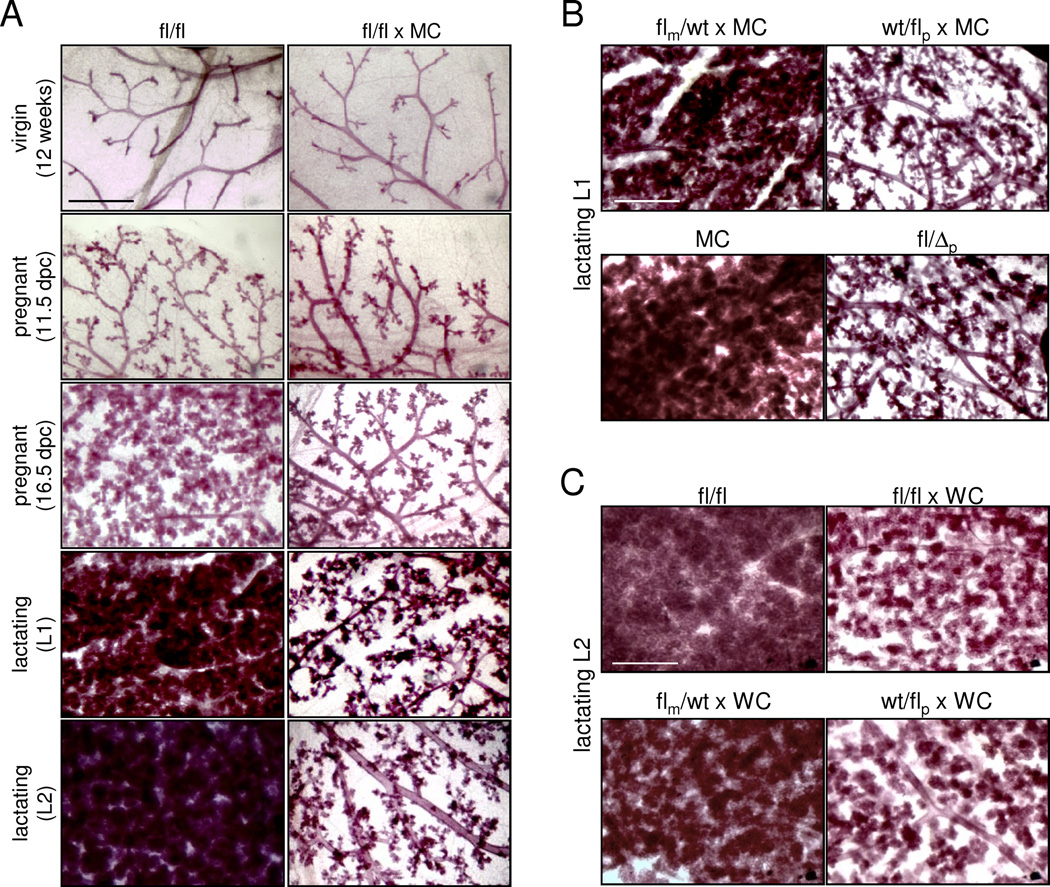
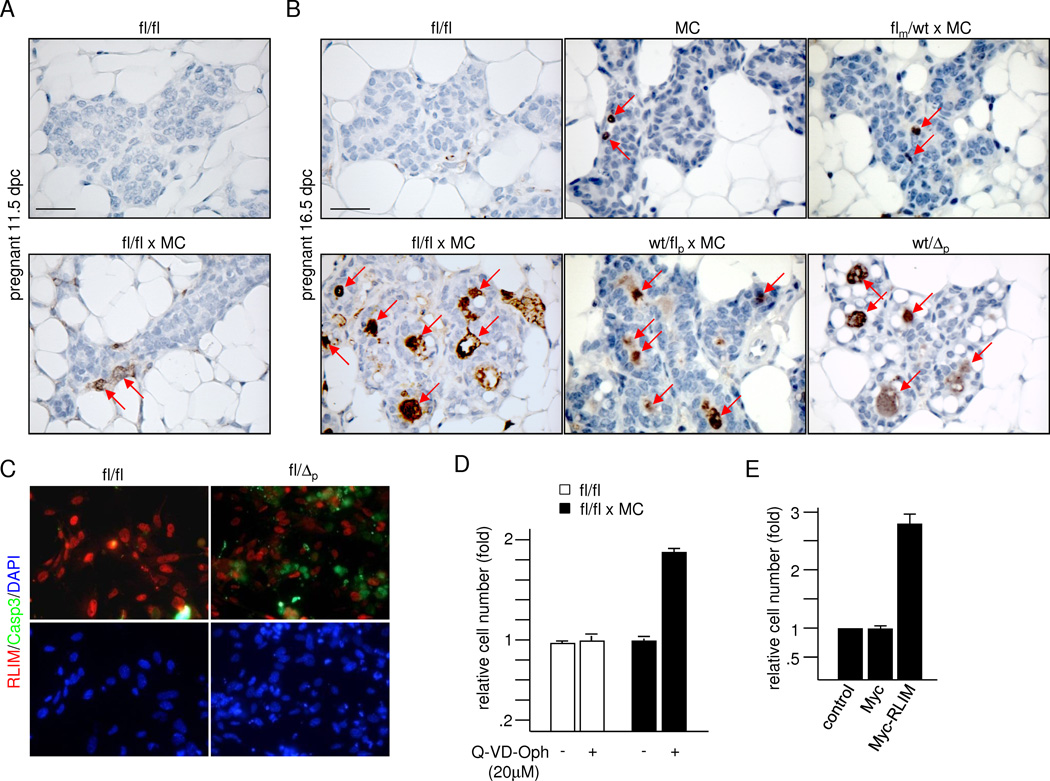
Examining mammary glands that lack RLIM (fl/fl × MC) revealed a dramatic inhibition in the differentiation of alveolar cells, while ductal development and branching appeared normal in virgin females (Figs. 1A; S2A). Examining various mammary stages in first-time pregnant and lactating females showed that the phenotype became apparent around 16.5 days post-coitum (dpc) and was highly pronounced at lactating stages (Fig. 1A). Because the maternal Rnf12 allele drives development of extraembryonic placental trophoblast cells (Shin et al., 2010), we compared mammary phenotypes of heterozygous females with either a maternal or a paternal deletion of the Rnf12 gene (flm/wt × MC and wt/flp × MC, respectively). Surprisingly, while we detected minimal mammary defects in flm/wt × MC lactating females, wt/flp × MC females displayed a phenotype comparable to that observed in fl/fl × MC females (Fig. 1B). This was not due to indirect effects of Cre expression because (1) mammary phenotypes generated with a maternally or paternally transmitted MC transgene were similar (data not shown), ruling out parental effects of the MC driver, (2) induced Cre levels were similar in all animals tested (Fig. S1E), (3) alveolar differentiation and milk production appeared comparable in control females with and without Cre expression (Fig. 1B) and (4) Cre-negative females carrying a paternal germline deletion of Rnf12 (fl/Δp) displayed a similar phenotype than wt/flp × MC or fl/fl × MC females (Fig. 1B). However, some fl/fl × MC, wt/flp × MC and fl/Δp females escaped a clear mammary phenotype at a low rate (less than 1 out of 8 of more than 30 animals tested per genotype), suggesting that additional factors may compensate for RLIM loss in a minority of animals. We also detected inhibited alveolar differentiation in mammary glands of lactating wt/flp females using K14-Cre (Dassule et al., 2000) or WAP-Cre (WC) (Wagner et al., 1997) as drivers (Figs. S2B; 1C) that induce early and late mammary expression of Cre, respectively.
Fig. 1.
Paternal Rnf12 is required for alveolar morphogenesis. A) Whole mount mammary tissues of virgin, pregnant and lactating fl/fl × MC and fl/fl females are shown at various stages. B) The paternal Rnf12 allele mainly contributes to alveolar differentiation. Comparison of mammary phenotypes at L1 in flm/wt × MC, wt/flp × MC and fl/Δp females. MMTV-Cre (MC) serves as control. C) Comparison of mammary phenotypes induced by targeting Rnf12 via WAP-Cre (WC) in fl/fl × WC, flm/wt × WC and wt/flp × WC females at L2. Scale bars a–c: 0.5 mm. See also Fig. S1.
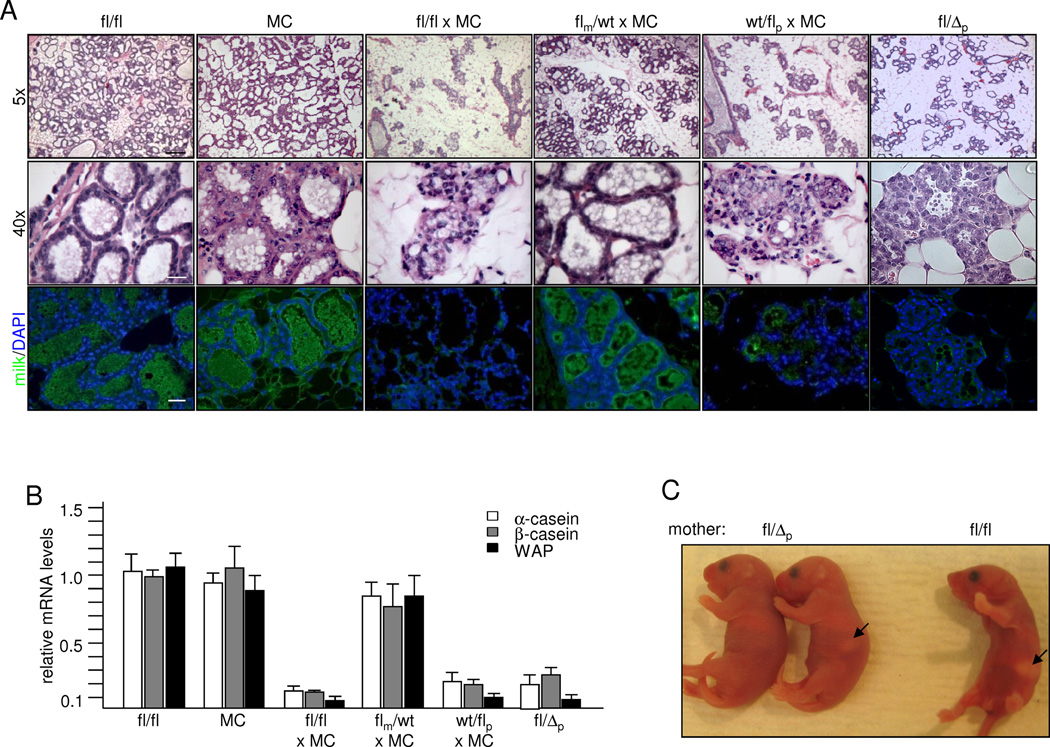
Analyses of H/E stained mammary sections of first-time pregnant females after parturition (L1) showed a reduced alveolar density smaller alveoli that appeared disorganized in fl/fl × MC, wt/flp × MC and wt/Δp glands when compared to flm/wt × MC, fl/fl and MC females (Fig. 2A). Examining milk production using antibodies directed against general milk proteins (Asselin-Labat et al., 2007) revealed reduced staining intensity in females carrying a paternal Rnf12 mutation (Fig. 2A) and RT-qPCR performed on RNA isolated from L1 glands confirmed significantly lower levels of mRNAs encoding milk proteins Whey Acidic Protein (WAP), α-casein and β-casein (Fig. 2B). Indeed, these females were often unable to support their litters resulting in a significantly lower 3-day survival rate of pups (Figs. 2C, S3C). These results demonstrate a critical role of the paternal Rnf12 allele for differentiating alveolar cells. As mice were bred in a congenic C57BL/6 background to eliminate strain-specific influences, these results indicate a sex-specific parent-of-origin effect.
Fig. 2.
Regulation of milk production by paternal Rnf12. A) Comparison of mammary gland sections at L1 from fl/fl and MC control and fl/fl × MC, fl/fl × MC and heterozygous flm/wt × MC, wt/flp × MC and fl/Δp females. Upper and middle panels: H&E stainings; lower panel: Immuno-stainings with milk antibodies. Scale bars: 120 µm upper panels; 20 µm middle and lower panels. B) RT-qPCR analysis of mRNA levels of milk proteins in L1 mammary glands. C) Less milk in pups born by fl/Δp mothers when compared to pups born by control mothers. Arrows point at milk as seen in stomachs of representative newborn pups. See also Fig. S2.
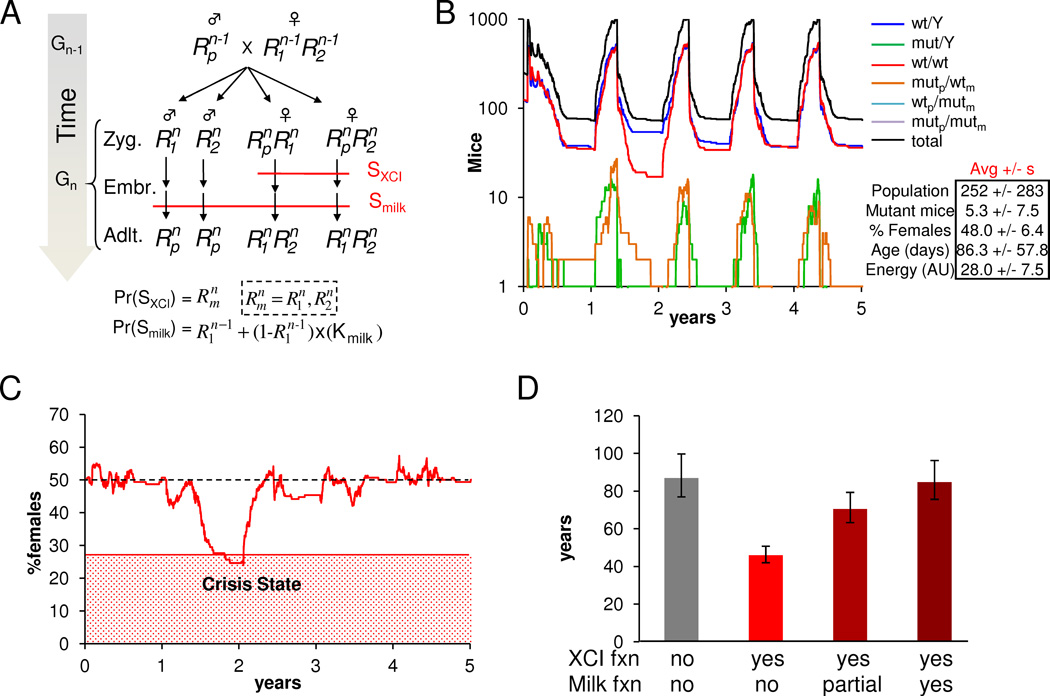
We next investigated the significance of the paternal function of Rnf12 for milk production in relation to its maternal function during imprinted XCI. Given that (1) failure of the XCI process leads to lethality specifically in females, (2) maternal but not paternal Rnf12 is required for imprinted XCI, and (3) Rnf12 KO males are viable and fertile (Shin et al., 2010), only the maternal Rnf12 allele should be subject to selection pressure in females. This suggests that loss-of-function mutations in the Rnf12 gene could lead to a gender bias towards males, which represents an unfavorable strategy for reproduction in mice (Hamilton, 1967). To explore the long-term genetic interactions of maternal and paternal Rnf12 functions in affecting gender bias for populations of feral mice, we pursued a stochastic agent-based modeling approach (Grimm et al., 2005). We constructed the simplest possible model that incorporated important factors such as age-dependent checkpoints for mating and death, seasonal variation in food growth, and density-dependent predation. Furthermore, we incorporated known constants for mouse maturation age, litter size, metabolism, and lifespan, and manually adjusted the free parameters to ensure that simulation behavior matched expected values for average colony size and average mouse age (Table S1). Each simulation was initialized with equal numbers of males and females wt for Rnf12 and allowed to iteratively progress through time (Figs. 3A, B; S3A, B), keeping track of each mouse’s age, Rnf12 genotype, energy level (a function of food consumption and constant metabolic rate), gestation time, and seasons (Tables S1). Since individual simulations show a high degree of variability in gender bias as a function of time, we calculated the average probabilities of encountering a shortage of females that may be considered a crisis event in different scenarios (≤ 20%, 27.5% and 35% females) (Fig. 3C; S3C; Table S2). Results show that given random mutation in each generation of either allele, the maternal XCI function led to a gender bias at all population sizes and significantly reduced the average time until a given colony encountered a gender crisis (Fig. 3D). Introducing the paternal Rnf12 functions for milk production in mammary glands in the model mitigated the gender bias, even at the experimentally observed 30% penetrance. Gender bias introduced by the maternally imprinted XCI function was almost completely mitigated by the paternally imprinted milk function in colony sizes of 250 animals, and partially with sizes of 100 and 500 animals (Figs. 3D; S3D). Importantly, the effect of the maternal XCI function as well as the mitigation of gender biasing by the paternally imprinted milk function was qualitatively independent of all simulation parameters including: seasonal food fluctuations, variations in food consumption, the magnitude of predation, the length of the breeding season, the propensity for mating, colony size, and choice of crisis threshold (not shown). However, both effects were quantitatively most dramatic for colonies of moderate size, high average birth frequency, and high average survival time. Examining hypothetical dominant or recessive regimes showed that only the imprinted regime resulted in rescue (Fig. S3B, D). Thus, while these simulations represent only one of many natural selection conditions found in nature, our qualitative predictions suggest that the paternal functions of Rnf12 in milk production tends to balance out biases caused by maternal Rnf12 functions by providing selection pressure on the paternal allele and that this may have evolutionary relevance when considering crisis situations in fluctuating populations.
Fig. 3.
Functions of paternal Rnf12 compensate for effects on gender ratio in mouse populations introduced by maternal functions. A) Stochastic computational model propagation scheme and the logical rules associated with imprinted XCI and mammary gland-dependent survival of mice under the imprinting regime. For each new generation, Gn, zygotes are generated according to the Mendelian laws of inheritance from randomly-selected parents sampled from Gn-1. Female zygotes must undergo XCI in order to survive to the embryo stage. This survival is dependent on Rnf12 (survival checkpoint SXCI). RLIM requirement for milk production and survival of born pups was implemented as a separate survival checkpoint (Smilk) during the embryo-adult transition. B) Example of a 10-year simulation run with a high mutation rate of 1% under the imprinting regime and Kmilk=100% for a colony of 250 mice (first 5 years shown). Rnf12 genotype counts are indicated as a function of time. Oscillatory count patterns arise due characteristic times for mice to mature, gestate, and die, and to seasonal oscillation in available food stores and reproduction cycles. Average statistics for the fraction of females, total mice counts, total non-wild type mice counts, average mouse ages, and average mouse energy levels are summarized +/− sample standard deviation. C) Sample Trajectory of the fraction of females in a population of 250 mice on average with the red region indicating a gender crisis state of <27.5% females. D) Average simulation T1/2 values for populations of 250 mice. Gray bar represents Rnf12-independent simulations. Red bars represent simulations with a 1% mutation rate and Kmilk = 100% (no), 30% (partial), and 0% (yes). Error bars were derived by recalculating T1/2 for crisis probability rates +/− probability SEM. See also Fig. S3 and Tables S1 and S2.
RLIM/Rnf12 is a survival factor specifically for milk-producing alveolar cells
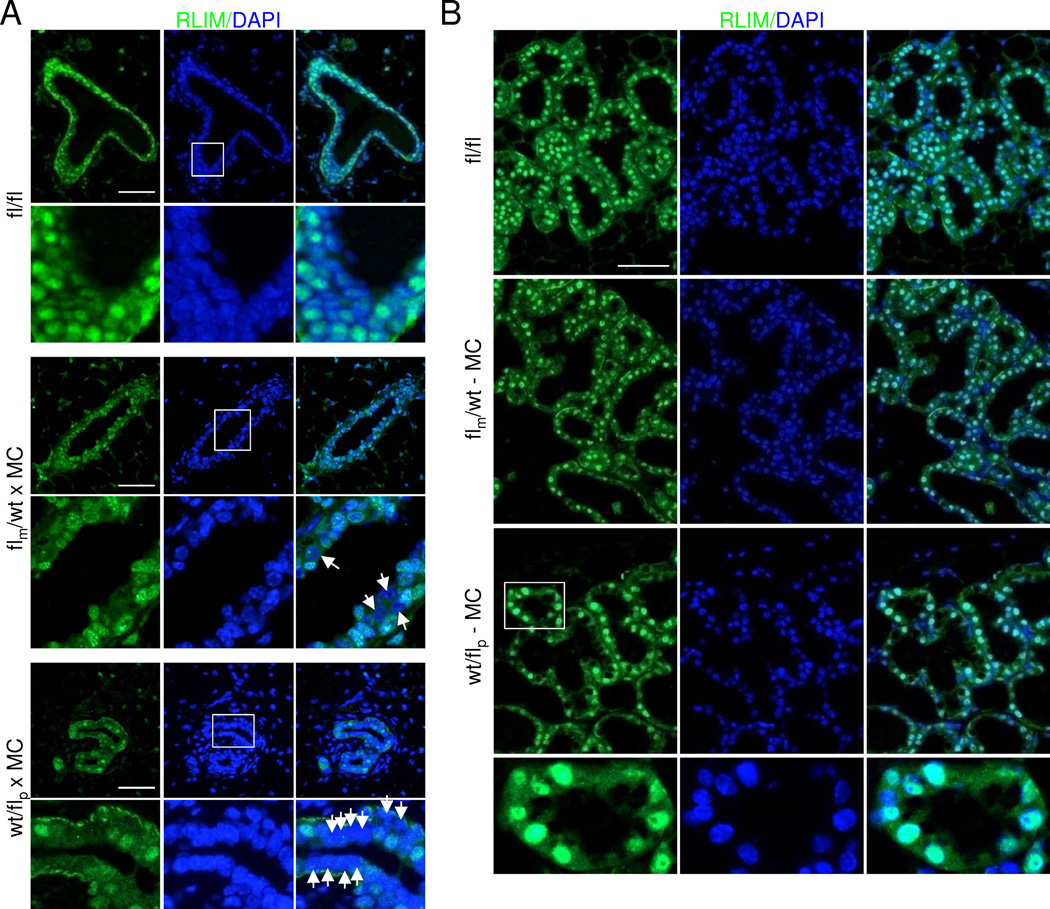
Next, we examined the contributions of the Rnf12 alleles to RLIM protein levels by comparing RLIM expression in virgin mammary tissues of heterozygous wt/flp × MC and flm/wt × MC females. Whereas RLIM was expressed in more than 95% and around 80% of mammary epithelial cells in fl/fl and flm/wt × MC females, respectively, only around 20% of cells stained RLIM-positive in wt/flp × MC mammary glands (Figs. 4A, Fig. S4A). Co-stainings with RLIM and CK14 or CK18 antibodies of differentiating primary mammary epithelial cells isolated from virgin females showed that the majority of CK14 or CK18-positive cells isolated from flm/wt × MC animals also expressed RLIM. However, most wt/flp × MC epithelial cells positive for CK14 or CK18 stained negative for RLIM (Fig. S4B). These data show that RLIM expression in myoepithelial and luminal mammary cells originates mainly from transcription of the paternal Rnf12 allele. Importantly, while we detected many living RLIM-negative epithelial cells in mammary glands of virgin wt/flp × MC females that stained positive for DAPI (Fig. 4A), in lactating glands only RLIM-positive cells participated in alveolar epithelia (Fig. 4B). This indicates that cells lacking RLIM are able to participate in the formation of ductal but not alveolar structures, suggesting crucial functions of RLIM specifically for differentiating alveolar cells.
Fig. 4.
RLIM/Rnf12 is expressed primarily from the paternal Rnf12 allele in mammary epithelia. Boxed regions are shown below in higher magnification. A) Expression of RLIM in virgin fl/fl, flm/wt × MC and wt/flp × MC glands. Arrows point at RLIM-negative cells. B) Mammary expression of RLIM in lactating flm/wt × MC and wt/flp × MC glands at L2. Note that in wt/flp × MC females mainly RLIM-positive alveolar cells are present while many RLIM-negative cells exist in ducts of virgin animals (A). Scale bar = 50 µm. See also Fig. S4.
To investigate the effects underlying the observed mammary phenotype we examined proliferation rates via immunostainings using antibodies directed against phospho-S10 histone H3 (P-H3), specifically recognizing cells in mitosis (Cui et al., 2004). We detected no significant difference in the percentage of mitotic cells in mammary tissues of pregnant fl/fl × MC and fl/fl controls at 16.5 dpc (Fig. S5A), consistent with similar levels of estrogen, progesterone and prolactin (Prl) in blood samples at L1 (not shown). However, stainings with antibodies against cleaved Caspase 3, a marker for apoptotic cells (Gown and Willingham, 2002), showed massive cell death occurring in mammary glands of pregnant females lacking RLIM (Fig. 5). Apoptotic cells were initially detected at 11.5 dpc, in significantly increased numbers at 16.5 dpc (Fig. 5A, B) and were still detectable at lactating stages (Fig. S5B). Indeed, while we counted only around 1% and 2% of caspase 3-positive cells in alveoli of MC and flm/wt × MC females at 16.5 dpc, respectively, more than 10% of cells stained positive in alveoli of females with a paternal Rnf12 deletion (Figs. 5B; S5C). In addition, cultured mammary epithelial cells isolated from fl/Δp virgin females showed mostly mutually exclusive staining when co-stained with cleaved caspase 3 and RLIM antibodies, indicating that apoptosis occurred primarily in cells lacking RLIM (Fig. 5C). This result was corroborated by the fact that only RLIM-positive cells from fl/fl × MC females were present after 10 days in culture, which were mainly composed of Cre-negative cells of non-epithelial origin (e.g. fibroblasts; not shown). Co-stainings of differentiating primary mammary cells revealed that both CK14 and CK18-positive cells were affected by cell death (Fig. S5D). Addition of pan-caspase inhibitor Q-VD-OPh led to significantly increased cell numbers in differentiating primary mammary epithelial cells from fl/fl × MC but not wt/wt animals when compared to untreated cells (Fig. 5D), demonstrating that cell death occurred via apoptosis. Moreover, while numbers of differentiating RLIM-negative mammary cells isolated from fl/fl × MC animals did not change upon lentiviral expression of a Myc-tag alone when compared to uninfected cells, physiological levels of exogenous Myc-RLIM (Fig. S6A) resulted in significantly increased cell numbers (Figs. 5E; S6B). This rescue reflected a decrease in apoptosis because Myc-RLIM-infected and control-infected cells displayed similar rates of proliferation (Figs. S6C). These data show that RLIM acts as a survival factor in milk-producing alveolar cells and suggest that the main cause of the mammary phenotype in females with a mutated Rnf12 gene is due to apoptosis.
Fig. 5.
RLIM/Rnf12 is a critical survival factor for alveolar cells. A) Staining of mammary sections from 11.5 dpc pregnant fl/fl and Rnf12fl/fl × MC females using antibodies directed against cleaved Caspase 3. B) Staining of sections of 16.5 dpc fl/fl, MC, fl/fl × MC and heterozygous flm/wt × MC, wt/flp × MC and wt/Δp females using antibodies against cleaved Caspase 3. C) Mammary cells isolated from virgin fl/fl and fl/Δp females were differentiated for 60h before co-staining with RLIM (red) and cleaved Caspase 3 (green) antibodies. D) Increased numbers of differentiating mammary cells isolated from fl/fl × MC females upon treatment with pan caspase inhibitor Q-VD-Oph. Equal numbers of cells were grown for 24h in culture in the presence or absence of 20 µM Q-VD-Oph. E) Rescue from apoptosis by exogenous RLIM. Equal numbers of differentiating mammary epithelial cells isolated from virgin fl/fl × MC females were infected with lentivirus expressing Myc-RLIM-FL or Myc alone. Uninfected cells served as control. 60h after infection cell numbers were counted. Scale bars A, B = 50 µm. See also Figs. S5 and S6.
RLIM/Rnf12 is involved in triggering involution
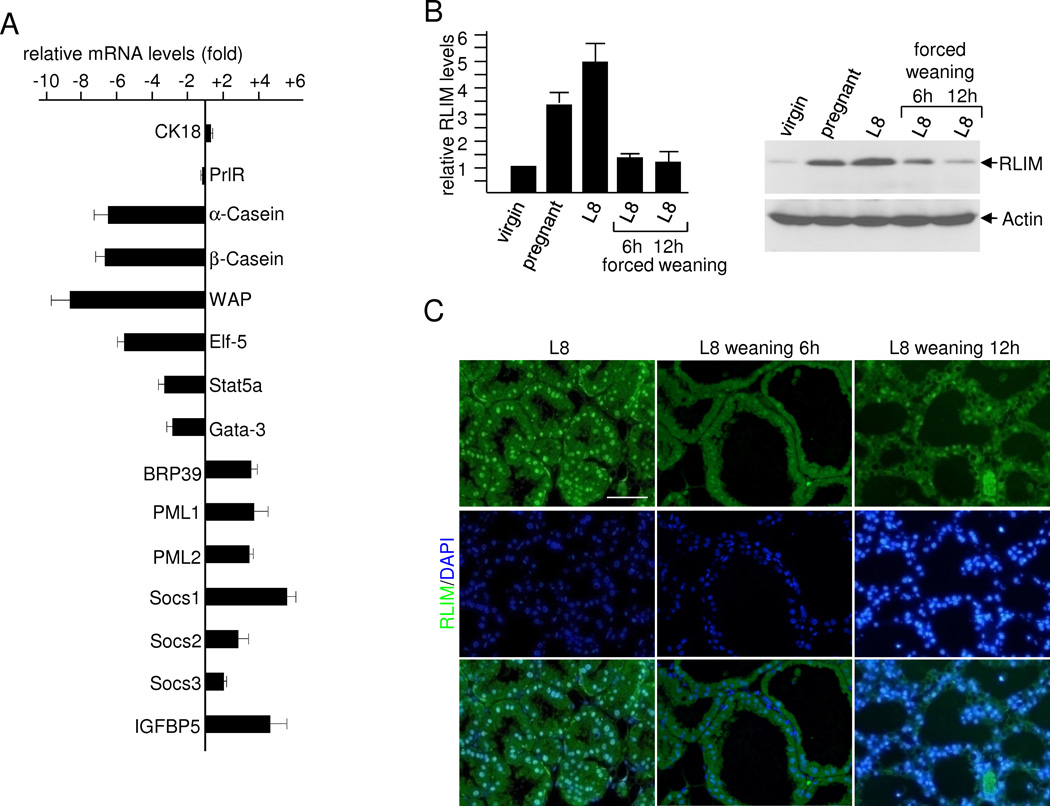
To investigate the expression of genes affected by Rnf12, we performed quantitative RT-PCR examining known regulators of alveolar differentiation including genes that either promote or inhibit Prl signaling (Brisken and Rajaram, 2006; Hennighausen and Robinson, 2008; Visvader, 2009). We examined mammary glands of first-time pregnant females at 16.5 dpc when the effects of the conditional Rnf12 KO become apparent (Fig. 1A). While levels of mRNAs encoding CK18 and prolactin receptor (PrlR) did not change significantly, those encoding milk proteins were significantly decreased in fl/fl × MC animals when compared to fl/fl animals, validating this assay (Fig. 6A). Expression of known effectors of alveolar differentiation such as Stat5a, Elf-5 and Gata-3 were strongly decreased in mammary glands of fl/fl × MC animals when compared to control animals. In contrast, mRNAs encoding proteins that are normally up-regulated during involution such as BRP39 (Mohanty et al., 2009), PML1, and PML2 (Li et al., 2009), Socs1 and Socs2 (Sutherland et al., 2007a) and IGFBP-5 (Flint et al., 2008), were also up-regulated in mammary glands lacking RLIM of pregnant females already at 16.5 dpc (Fig. 6A). Because Socs 1, Socs 2 and IGFBP-5 represent major inhibitors of Prl and IGF alveolar survival pathways (Sutherland et al., 2007b), respectively, their upregulation is likely to contribute to the premature apoptosis of alveolar cells lacking RLIM.
Fig. 6.
RLIM/Rnf12 is a key regulator of mammary involution. A) Down-regulation of factors promoting alveolar differentiation and up-regulation of factors that promote involution in RLIM-lacking mammary epithelia of pregnant females at 16.5 dpc. RT-qPCR analysis of whole mammary tissue isolated from fl/fl × MC females compared with fl/fl. Values were determined from three independent experiments. Mammary RNA of 5 different females was pooled for each experiment. B) RLIM expression profile in mammary glands. Left panel: Summary of RLIM levels as measured in Western blots of cell extracts prepared from mammary glands from virgin (8 weeks), pregnant (16.5 dpc) and lactating (L8) females and L8 females 6h and 12h after forced weaning. Graph represents data from 4 independent experiments using Image J software. Right panel: Representative Western blot hybridized with RLIM (upper) and Actin (lower) antibodies. C) Mammary sections of L8 females before and 6h and 12 hours after forced weaning were stained with RLIM antibodies. Note the dramatic decrease in RLIM staining in alveolar epithelia upon forced weaning. Scale bar = 50 µm.
To examine if RLIM could be part of a trigger that induces involution, we compared its overall expression in virgin, pregnant and lactating mammary glands, including females in which weaning was forced via removal of pups. In cell extracts prepared from whole mammary glands we detected increased levels of RLIM in pregnant and lactating females likely due to the expansion of alveolar cells (Fig. 6B). Importantly, overall mammary RLIM levels sharply decreased within 6h upon forced weaning and immunohistochemical analysis showed that this occurred in mammary epithelia (Fig. 6C). These results are consistent with a crucial role of RLIM in preventing a premature involution of alveolar cells and suggest that its downregulation is a key event in triggering involution.
Epigenetic regulation of mouse mammary gland biology
Our results show that the paternal Rnf12 allele is expressed in around 80% of mammary epithelial cells (Figs. 4; S4A). This was surprising because Rnf12 is X-linked and somatic tissues in females undergo random XCI. To investigate the basis for the skewed paternal expression of RLIM/Rnf12 we examined the possibilities that either Rnf12 selectively escapes silencing from the inactivated Xp, or that mammary epithelia display an imprinted XCI pattern and are mainly composed of cells with a paternal active X chromosome (Xa). We first used RNA FISH on primary mammary epithelial cells and co-hybridized with established probes for Xist, which is exclusively expressed from and paints the inactive X chromosome (Xi), and Rnf12. We observed mostly a single Rnf12 transcription focus per cell that was non-overlapping with the Xist, indicating that Rnf12 was expressed from the Xa but silenced on the Xi (Fig. S7A). Moreover, co-hybridization of an Rnf12 probe with probes for X-linked genes Pgk1, Atrx, G6pd, Pdha1 and Utx spanning much of the X chromosome revealed single transcription foci that were generally side by side in nuclei (Fig. S7B–F), indicating expression from the same active X chromosome (Xa).
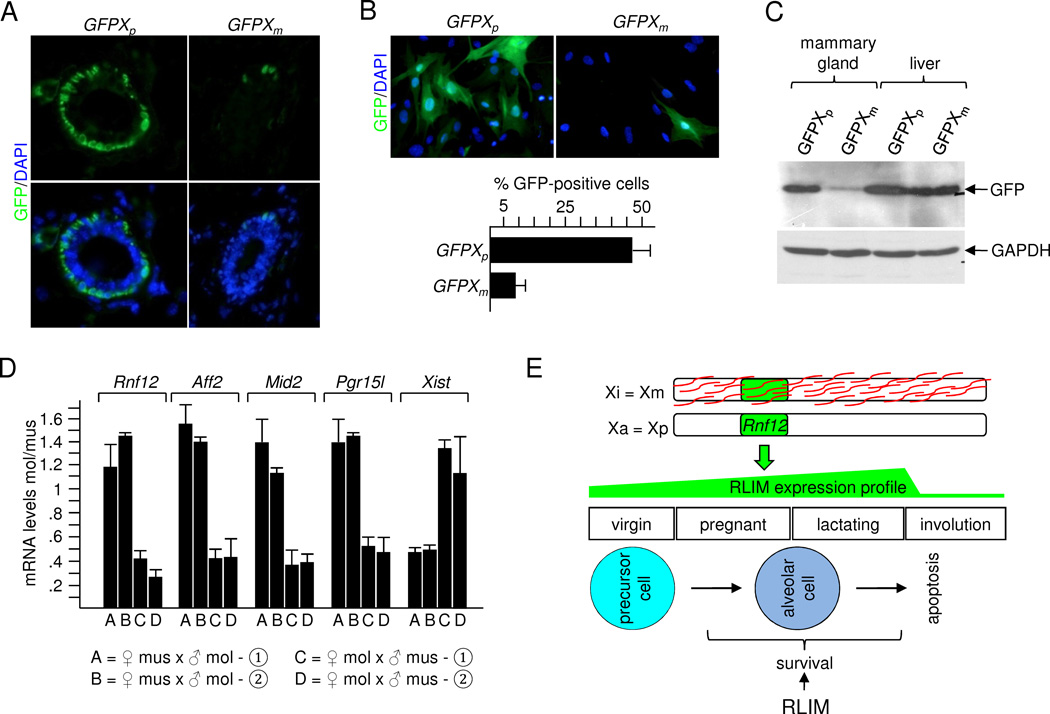
To further address this issue we used transgenic mice that carry an X-linked EGFP transgene (GFPX) driven by chicken β-actin promoter and CMV intermediate early enhancer (Hadjantonakis et al., 2001). Immunostainings comparing virgin mammary glands of females carrying a paternally or maternally transmitted transgene (GFPXp and GFPXm, respectively) revealed a much higher percentage of GFP-positive cells in GFPXp glands (8/10 mice) when compared to GFPXm glands (6/7 mice; Fig. 7A). Co-stainings with GFP and CK14 or CK18 antibodies revealed GFP expression mainly in myoepithelial cells (Fig. S7H; data not shown), consistent with high actin expression in this cell layer. Quantification of primary cells in culture co-stained with CK14 and GFP antibodies showed that 4–5x fold more CK14-positive cells stained positive for GFP in cells harboring a GFPXp, compared to those with a GFPXm (Fig. 7B). Western blotting confirmed much higher GFP expression from the paternal allele in extracts prepared from L8 mammary glands, whereas in liver extracts of the same mice similar GFP levels were detected (Fig. 7C).
Fig. 7.
Mouse mammary epithelial cells display an imprinted XCI pattern. A) Immunostaining of virgin mammary glands of hybrid females carrying a paternal or maternal GFPX transgene. B) Upper panel: Representative mammary cells in culture prepared from virgin GFPXp or GFPXm animals were stained with a GFP antibody. Lower panel: Summary of GFP-positive cells counting more than 100 cells each out of 3 independent experiments. C) Western blot on protein extracts prepared from L8 GFPXp and GFPXm animals were stained with antibodies against GFP and, as control, GAPDH. Extracts of mammary gland and liver were prepared from the same animal. D) Allele-specific RT-PCR analysis of X-linked genes Rnf12, Aff2, Mid2, Pgr15l and Xist in mammary epithelial cells of F1 hybrid females originating from reciprocal crosses between Mus musculus (mus) and Mus molossinus (mol). Two independent females (➀, ➁) from each cross were examined. E) Schematic representation illustrating the imprinted XCI pattern in mammary epithelia and the survival function of RLIM for alveolar cells. Mainly the maternal X (Xm) is silenced (Xi) and coated by Xist RNAs (red) and Rnf12 is expressed from the paternal X (Xp) which is active (Xa). The profile of RLIM expression and stages of mammary gland development/differentiation is indicated. See also Fig. S7.
To obtain definitive evidence for an imprinted XCI pattern, we performed allele-specific RT-qPCR on mRNA isolated from mammary epithelial cells of F1 hybrid females originating from reciprocal crossings between Mus musculus and Mus molossinus (strains C57BL/6 and JF1, respectively). We used TaqMan probes of X-linked genes Rnf12, Aff2, Mid2, Pgr15l and Xist to distinguish between the two X chromosomes by virtue of single nucleotide polymorphisms (SNPs). Specificity of probes was verified on pure C57BL/6 and JF1 strains. Again, we observed a clear bias towards transcription of the paternally inherited alleles of all genes analyzed except Xist which, consistent with its role during XCI, was mainly expressed from the maternal allele (Fig. 7D). Because the SNP in Aff2 is also present among various Mus musculus strains, we focused on this gene and compared its expression in mammary gland and liver of the same animals. Expression of Aff2 in mammary glands of both mus/mol (BL6/JF1) and mus/mus (BL6/DBA) F1 hybrid females originating from reciprocal crossings displayed a strong bias towards expression of the paternal allele. In contrast, no parental bias was observed in liver (Fig. S7H, I). Furthermore, there was similar contribution in expression from the BL6 chromosome when compared to DBA in mus/mus F1 females in liver, in agreement with a similar propensity of X chromosomes from these strains to become inactivated (Chadwick et al., 2006). We detected a mildly stronger contribution of the BL6 allele in mus/mol F1 hybrids in liver (Fig. S7H), suggesting that random XCI is slightly skewed towards silencing the X chromosome originating from molossinus. Finally, in F1 hybrid females of Mus musculus strains B6C3F1/J, B6D2F1/J, B6SJLF1/J and B6129SF1/J we also detected significantly higher expression of the paternal Aff2 alleles in mammary glands when compared to liver (data not shown). Combined, these results reveal that mouse mammary epithelial tissues are composed mainly of cells with a paternal Xa (Fig. 7E).
Discussion
Mechanisms of RLIM action in the mammary gland
Our results provide strong evidence that RLIM/Rnf12 is required for the survival of differentiated milk-producing alveolar cells in pregnant and lactating female mice. Other reported functions of Rnf12 in females comprise a crucial involvement for the initiation of imprinted XCI in 4-cell-staged embryos, while male mice carrying a Rnf12 germline KO are viable, fertile and appear normal (Shin et al., 2010). Because brain development appeared normal in males and females with a targeted KO in various embryonic brain regions (Michael Bossenz and I.B., unpublished), our results suggest that Rnf12 exerts its main functions during embryonic development and in pregnant/lactating adults. Thus, the major biological functions of RLIM/Rnf12 appear sex-specific although we cannot rule out more subtle functions of Rnf12 under challenged conditions.
RLIM/Rnf12 can inhibit or promote activity of specific transcription factors, depending on the protein context (Bach et al., 1999; Ostendorff et al., 2002; Gungor et al., 2007; Johnsen et al., 2009). Consistent with this, our results indicate that RLIM is involved in the activation of specific sets of genes that promote alveolar differentiation as well as repression of genes that promote involution including Socs 1, Socs 2 and IGFBP5 (Fig. 6A). Because Socs 1, Socs 2 and IGFBP5 inhibit the Prl and IGF-1 alveolar survival pathways, respectively, (Sutherland et al., 2007b), their repression provides a molecular mechanism that likely contributes to RLIMs’ survival function. However, it is unclear whether the repressor and activator functions of RLIM are direct or indirect.
RLIM is a key factor for triggering involution
The fact that RLIM serves as a survival factor specifically for alveolar cells makes it a prime candidate for triggering involution. Indeed, within only hours upon forced weaning, RLIM levels in mammary epithelia dramatically decrease (Fig. 6B; C), suggesting that this depletion may trigger normal involution. In further agreement with this is a short half-life of cellular RLIM which is RING finger-dependent (Ostendorff et al., 2002). Such a scenario is reminiscent to results reported for the inhibitor of apoptosis (IAP) family of RING finger ubiquitin ligases. IAPs promote the survival of thymocytes and cell death by apoptotic stimuli is triggered via IAP downregulation (Vucic et al., 2011). Likewise, it is tempting to speculate that weaning-induced downregulation of RLIM triggers mammary involution. However, the mechanisms by which RLIM and IAP proteins promote cell survival are likely different because IAPs interact with and inhibit a variety of cytoplasmic proteins including caspases and proteins in TNF receptor complexes. In contrast, RLIM is predominantly nuclear (Bach et al., 1999; Ostendorff et al., 2006) and sequence conservation between RLIM and IAPs is restricted to the RING finger, suggesting association with different protein networks. Ubiquitin ligases such as IAP and Mdm2 are involved in the development of cancer (Vucic et al., 2011), illustrating the crucial role that cell death plays during tumorigenesis (Hanahan and Weinberg, 2011). Thus, it will be interesting to elucidate the mechanisms underlying RLIM’s selective survival function for alveolar cells in the future in both normal mammary epithelia and breast cancer.
Mechanisms controlling epigenetic regulation of mammary epithelia
Our findings reveal crucial functions of Rnf12 specifically in female nurturing tissues. Indeed, while the maternal Rnf12 allele regulates development of the extraembryonic placental trophoblast during early embryogenesis (Shin et al., 2010), our results show that the paternal allele is required for the mammary survival function in alveolar cells in pregnant and lactating adult females (Fig. 5). The sex-specific epigenetic effects in mammary epithelia are explained by the observation that RLIM is expressed mainly from the paternal Rnf12 allele (Fig. 4). Results obtained via mathematical modeling indicate that the paternal functions of Rnf12 in the mammary gland can provide a selection pressure that can prevent a gender bias that would otherwise be imposed by the maternal functions during imprinted XCI. Thus, compensatory functions may represent some of the underlying driving forces for the evolution of this unique pattern of epigenetic regulation.
Our results indicate an imprinted XCI pattern in mouse mammary epithelia. This is supported by genetic data on mammary phenotypes induced in females heterozygous for Rnf12, RLIM expression data in heterozygous Rnf12 KO animals, results obtained via RNA FISH, allele-specific RT-PCR on expression of X-linked genes and our data on hybrid females that carry either a maternal or paternal GFPX transgene. The fact that expression of GFPX in mammary tissues is driven to myoepithelial cells by its promoter combined with our finding that RLIM is expressed mainly from the paternal allele in myoepithelial and luminal cells (Figs. 4, S4A) strongly argues that both these cell types display an imprinted XCI pattern. Thus, the mammary phenotypes caused by the deletion of the paternal Rnf12 allele and expression of paternal RLIM are a consequence of the imprinted XCI pattern. Indeed, in glutamatergic neurons in the cortex, a bias to express the maternal X chromosome in female mice has recently been detected (Gregg et al., 2010; Wang et al., 2010). Because the X chromosomes of different mouse strains display different propensities to stay active during random XCI (Chadwick et al., 2006), this is likely to influence bias intensity of the imprinted XCI pattern in mammary glands.
It is intriguing that female extraembryonic trophoblast cells in the placenta and milk-producing alveolar cells in the adult mammary gland both require RLIM, but which is supplied via transcription of opposite Rnf12 alleles and display opposing imprinted XCI patterns. How can an imprinted XCI pattern be established in specific adult cell types? Female somatic cells undergo random XCI early during embryogenesis and through mitosis into descendant cells the replicated copies of the Xi and Xa are generally maintained. Thus, to explain the imprinted XCI pattern in mammary epithelial cells, it is likely that in a pool of mammary precursor cells, those with an active Xp have a growth advantage over cells with an active Xm at some point during embryogenesis. The finding that the paternal bias is already established in adult virgin epithelia (Fig. 4A) before the occurrence of a KO mammary phenotype (Fig. 1) indicates that either progenitor cells that will form ducts already display this bias in the mammary Anlage, or that this bias is established during ductal development. Future work to elucidate factors and mechanisms leading to such a selective growth advantage should prove to be exciting. In this context it is interesting to note that an increasing number of imprinted genes have been identified on the X chromosome including certain Pem/Rhox (Maclean et al., 2011) and Xlr genes (Raefski and O'Neill, 2005).
In summary, we propose that RLIM/Rnf12 expressed from the paternal allele acts as a survival factor for milk-producing mammary alveolar cells in pregnant and lactating female mice.
Material and Methods
Mice
Floxed Rnf12 mice and genotyping have been described (Shin et al., 2010). MMTV-Cre (Line F) mice and WAP-Cre mice (Wagner et al., 1997) were obtained from the MMHCC repository (NCI, Frederick), K14-Cre mice (Dassule et al., 2000) were a kind gift of S. Sinha (State University of New York, Buffalo). R26R (Soriano, 1999) and GFPX mice (Hadjantonakis et al., 2001) were purchased from The Jackson Laboratories. Mice were generally kept in a C57BL/6 congenic background except GFPX mice which were in a mixed background. Mus musculus/Mus molossinus and Mus musculus/Mus musculus F1 hybrid females were generated by reciprocal crossings between C57BL/6 and JF1 or C57BL/6 and DBA mice, respectively. F1 hybrid females B6C3F1/J, B6D2F1/J, B6SJLF1/J and B6129SF1/J were purchased from The Jackson Labs. Mice were bred in the animal facility of UMMS according to NIH guidelines, established by the Institute of Animal Care and Usage Committee.
Antibodies and Immunodetections
Antibodies were rabbit RLIM (Ostendorff et al., 2002), guinea pig RLIM (Ostendorff et al., 2006), Cre (Covance), CK 14 (Abcam), CK18 (Abcam), cleaved caspase 3 (Cell Signaling), milk (Nordic Immunological Laboratories), GAPDH (Millipore), GFP (Rockland), Phospho-histone H3 (Ser10) P-H3 (Millipore) and Actin (Sigma). Immunocytochemistry and Western blots were performed as previously reported (Tursun et al., 2005). Signals obtained in Western blots were quantified using Image J software.
Analyses of mammary glands from female mice
For whole-mount analyses, fourth inguinal mammary glands were excised and spread on microscope slides. Tissues were fixed in 25% glacial acid in ethanol, stained o/n in carmine alum stain, dehydrated in ethanol, cleared in Xylene and mounted with Permount (Fisher Scientific). For LacZ stainings, tissues were fixed in 1.5% formaldehyde for 15 min before detection of β-galactosidase activity (Pereira, 2001). Mammary tissues were fixed in 10% formalin and paraffin-embedded sections were stained with Hematoxylin/Eosin. Immuno-staining was performed as reported (Shin et al., 2010). Mammary tissues from pregnant and lactating females were derived from first-time pregnant females. L1 mammary glands were harvested 2–4h after parturition. To analyze L8 glands, adult female mice were maintained with at least six pups.
Primary mammary epithelial cells
Mouse mammary cells were dissociated as described (Shackleton et al., 2006; Stingl et al., 2006). Briefly, fourth inguinal mammary glands from 2 month-old virgin mice were digested using collagenase/hyaluronidase (2 to 6h) before treatment with Trypsin/Dispase/DNaseI followed by filtration through a 40µm cell strainer. Cells were cultured in differentiation medium (DMEM/F12 containing 10% FBS and 800ng/ml of Prl). Fibroblasts were removed as described (Langdon, 2004). To test for apoptosis, primary mammary cells were differentiated for 1 day in the presence of 20 µM caspase inhibitor Q-VD-OPh (R&D Systems) in FBS. Cell numbers were measured using a CellTiter 95R Aqueous One Solution Cell Proliferation Assay Kit (Promega). Results were pooled from 8 individual wells per measuring point in three independent experiments. Lentiviral infections of primary or MCF7 cells were performed using lentiviral vectors (pLenti CMV/TO Puro) as described (Campeau et al., 2009) expressing 6xMyc-tag (Myc) or Myc-RLIM. Two days after infections cells were aliquoted and seeded on 96 well plates for counting. RNA FISH experiments using probes for Rnf12, Pgk1, Atrx, G6pd, Pdha1 and Utx were performed as reported (Shin et al., 2010; Kalantry et al., 2009).
RT-PCR analyses
RT–qPCR was performed as described (Shin et al., 2010). Primer sequences for α-casein, β-casein, Wap, Elf5, K18, Stat5a, Socs2 (Choi et al., 2009) and PML1, PML2 and Gata-3 (Li et al., 2009) have been described. Other primers were β-actin: mAct-F (5’-TGCTGTCCCTGTATGCCTCT-3’), mAct-R (5’- ATGTCACGCACGATTTCC-3’); IGFBP5: IGFBP5-F (5’-TGTGGCGTCTACACGGAGCG-3’), IGFBP5-R (5’-CTTGGTTTGCTCGCCGTA-3’); BRP39: BRP39-F (5’-CAACATCAGCAGCGACAA-3’), BRP39-R (5’-CATAAGAACGCAGGAACG-3’); Socs1: Socs1-F (5’-CTCCGTGACTACCTGAGTTCCT-3’), Socs1-R (5’-ATCTCACCCTCCACAACCACT-3’); Socs3: Socs3-F (5’-CACTCCCACTGCCCAACCT-3’), Socs3-R (5’-GCTACTGCCTCTTCACAACG-3’); PRLR: PRLR-F (5’-GAGGACGAGCGGCTAATG-3’), PRLR-R (5’-TCTCAGGGATGTGGAAGG-3’). Allele-specific RT-PCR on mRNA isolated from L8 mammary cells of F1 hybrid females was performed using TaqMan probes (Applied Biosystems). Total RNA was treated with DNAse I and reverse-transcribed (Transcriptor high fidelity cDNA synthesis kit, Roche). Real-time PCR was performed with gene-specific primers and allele-specific probes based on TaqMan SNP genotyping assay (Applied Biosystems) using ABI Prism 7300 SDS. SNP sites were rs13483812 (Aff2), rs13483897 (Pgr15l), rs29254008 (Mid2), rs29081562 (Rnf12) and rs29080019 (Xist). Relative quantities of mRNA were determined using the ΔCt method (Breen et al., 2000).
Modeling Mouse Colonies Using Stochastic Agents
To investigate long-term effects of Rnf12 function on gender biasing in feral mouse colonies, we constructed stochastic agent-based (Grimm et al., 2005) models using the Java 1.6 programming language. Parameterization was constrained by a three-month average mouse age (Lidicker, 1966), a specified colony size and overall stability of model behavior. While we tested multiple models that included both constant and seasonal food production, reproduction and survival based mechanisms for population control, as well as simple deterministic models, we found that results remained qualitatively identical: the maternally imprinted XCI function resulted in gender biasing, while the added paternally imprinted milk function mitigated the biasing effect. For realistic modeling of feral mice, we selected a stochastic model which incorporated both seasonal fluctuation in energy availability and density-dependent probabilities for mating and predation and because it allowed us to quantify possible Rnf12 effects under variable naturally-occurring extreme conditions (e.g. when periods of relative food abundance and colony growth are punctuated by periods of predation and/or scarcity, during which gender bias may have dramatic effects on survival of the entire colony). For a full list of model parameters and justifications see Table S1. Java 1.6 source code is available upon request. Simulations were initialized with equal numbers of wt males and females and allowed to iteratively progress through time (Δt = 1 day), keeping track of each mouse’s age, Rnf12 genotype, energy level (a function of food consumption and constant metabolic rate) and gestation time. Shared food stores are replenished each day based on time of year, peaking during the summer (Lidicker, 1966; Korslund and Steen, 2006). Each day, mature non-pregnant females select one mature male for reproduction, subject to density-dependent probability of encounter, and time of year (Lidicker, 1966; Pulliam and Danielson, 1991). Successful mating results in a gestation-period delay of 19–21 days after which still-living mothers give birth to 6–8 offspring using Mendelian inheritance, a constant Rnf12 mutation rate, and logical rules for offspring survival to adulthood, determined by Rnf12-dependent X chromosome inactivation (Shin et al., 2010), and milk production in female mice. In addition to death caused by starvation, mice-agents were subject to stochastic death from old age and density-dependent predation. Simulations were repeated 100,000 times for each condition (Table S2) and average probabilities of passing below the bias threshold (gender crisis) were calculated. These probabilities were used to calculate the T1/2 values, the expected time until it is more likely to have already encountered a crisis state given a constant daily probability of being in a crisis state. Error bars show recalculated T1/2 values using average probability of gender crisis +/− SEM. See Fig. 3 for simulations of 250 mice using the proposed imprinting regime. In addition to the Rnf12 imprinting regime, the biasing model was extended in order to compare the effects under other genetic regimes (Fig. S3; Table S2). Furthermore, we tested the dependence of the Rnf12 function on the average mouse colony size for colonies of 50, 100, 250, and 500 mice (Fig. S3; not shown). Simulation workflow and sample mouse count trajectories are shown in Fig. S3.
Research Highlights.
RLIM/Rnf12 is a crucial regulator of mammary alveolar morphogenesis
RLIM is an alveolar survival factor and its downregulation triggers involution
Mammary functions of Rnf12 are mediated mainly by the paternal allele
Opposite imprinted XCI pattern in mammary gland and extraembryonic trophoblast cells
Supplementary Material
Acknowledgments
We thank B. Andersen, E. Baehrecke, T. Fazzio, D. Haig, O. Rando, W. Robinson, L. Shaw and S. Sinha for advice, comments and/or reagents and T. Fazzio for critically reading the manuscript. I.B. is a member of the UMass DERC (DK32520). This work was supported by NIH grants R01CA131158 to I.B. and R01CA89209 to A.M.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der WJ, Lindeman GJ, Visvader JE. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Bach I, Rodriguez-Esteban C, Carriere C, Bhushan A, Krones A, Rose DW, Glass CK, Andersen B, Izpisua Belmonte JC, Rosenfeld MG. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat. Genet. 1999;22:394–399. doi: 10.1038/11970. [DOI] [PubMed] [Google Scholar]
- Breen G, Harold D, Ralston S, Shaw D, St Clair D. Determining SNP allele frequencies in DNA pools. Biotechniques. 2000;28:464–466. doi: 10.2144/00283st03. 468, 470. [DOI] [PubMed] [Google Scholar]
- Brisken C, Rajaram RD. Alveolar and lactogenic differentiation. J. Mammary. Gland. Biol. Neoplasia. 2006;11:239–248. doi: 10.1007/s10911-006-9026-0. [DOI] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS. One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick LH, Pertz LM, Broman KW, Bartolomei MS, Willard HF. Genetic control of X chromosome inactivation in mice: definition of the Xce candidate interval. Genetics. 2006;173:2103–2110. doi: 10.1534/genetics.105.054882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Chakrabarti R, Escamilla-Hernandez R, Sinha S. Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev. Biol. 2009;329:227–241. doi: 10.1016/j.ydbio.2009.02.032. [DOI] [PubMed] [Google Scholar]
- Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- Flint DJ, Tonner E, Beattie J, Allan GJ. Role of insulin-like growth factor binding proteins in mammary gland development. J. MammaryGland. Biol. Neoplasia. 2008;13:443–453. doi: 10.1007/s10911-008-9095-3. [DOI] [PubMed] [Google Scholar]
- Gown AM, Willingham MC. Improved detection of apoptotic cells in archival paraffin sections: immunohistochemistry using antibodies to cleaved caspase 3. J. Histochem. Cytochem. 2002;50:449–454. doi: 10.1177/002215540205000401. [DOI] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm V, Revilla E, Berger U, Jeltsch F, Mooij WM, Railsback SF, Thulke HH, Weiner J, Wiegand T, DeAngelis DL. Pattern-oriented modeling of agent-based complex systems: lessons from ecology. Science. 2005;310:987–991. doi: 10.1126/science.1116681. [DOI] [PubMed] [Google Scholar]
- Gungor C, Taniguchi-Ishigaki N, Ma H, Drung A, Tursun B, Ostendorff HP, Bossenz M, Becker CG, Becker T, Bach I. Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15000–15005. doi: 10.1073/pnas.0703738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Cox LL, Tam PP, Nagy A. An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. Genesis. 2001;29:133–140. doi: 10.1002/gene.1016. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. Extraordinary sex ratios. A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science. 1967;156:477–488. doi: 10.1126/science.156.3774.477. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen SA, Gungor C, Prenzel T, Riethdorf S, Riethdorf L, Taniguchi-Ishigaki N, Rau T, Tursun B, Furlow JD, Sauter G, Scheffner M, Pantel K, Gannon F, Bach I. Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 2009;69:128–136. doi: 10.1158/0008-5472.CAN-08-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korslund L, Steen H. Small rodent winter survival: snow conditions limit access to food resources. J. Anim Ecol. 2006;75:156–166. doi: 10.1111/j.1365-2656.2005.01031.x. [DOI] [PubMed] [Google Scholar]
- Kramer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, Brill B, Groner B, Bach I, Heinzel T, Gottlicher M. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon SP. Isolation and culture of ovarian cancer cell lines. Methods Mol. Med. 2004;88:133–139. doi: 10.1385/1-59259-406-9:133. [DOI] [PubMed] [Google Scholar]
- Li W, Ferguson BJ, Khaled WT, Tevendale M, Stingl J, Poli V, Rich T, Salomoni P, Watson CJ. PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4725–4730. doi: 10.1073/pnas.0807640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidicker WZ., Jr Ecological Observations on a Feral House Mouse Population Declining to Extinction. Ecological Monographs. 1966;36:25–50. [Google Scholar]
- Maclean JA, Bettegowda A, Kim BJ, Lou CH, Yang SM, Bhardwaj A, Shanker S, Hu Z, Fan Y, Eckardt S, McLaughlin KJ, Skoultchi AI, Wilkinson MF. The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone h1 and DNA methylation. Mol. Cell Biol. 2011;31:1275–1287. doi: 10.1128/MCB.00734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty AK, Fisher AJ, Yu Z, Pradeep MA, Janjanam J, Kaushik JK. Cloning, expression, characterization and crystallization of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis. Protein Expr. Purif. 2009;64:213–218. doi: 10.1016/j.pep.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Ostendorff HP, Peirano RI, Peters MA, Schlüter A, Bossenz M, Scheffner M, Bach I. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002;416:99–103. doi: 10.1038/416099a. [DOI] [PubMed] [Google Scholar]
- Ostendorff HP, Tursun B, Cornils K, Schluter A, Drung A, Gungor C, Bach I. Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev. Dyn. 2006;235:786–791. doi: 10.1002/dvdy.20669. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Pereira FA. Whole-mount histochemical detection of beta-galactosidase activity. Curr. Protoc. Mol. Biol. 2001;Chapter 14(Unit) doi: 10.1002/0471142727.mb1414s50. [DOI] [PubMed] [Google Scholar]
- Pulliam HR, Danielson BJ. Sources, Sinks, and Habitat Selection: A Landscape Perspective on Population Dynamics. The American Naturalist. 1991;137:S50–S66. [Google Scholar]
- Raefski AS, O'Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat. Genet. 2005;37:620–624. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shin J, Bossenz M, Chung Y, Ma H, Byron M, Taniguchi-Ishigaki N, Zhu X, Jiao B, Hall LL, Green MR, Jones SN, Hermans-Borgmeyer I, Lawrence JB, Bach I. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature. 2010;467:977–981. doi: 10.1038/nature09457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Sutherland KD, Lindeman GJ, Visvader JE. Knocking off SOCS genes in the mammary gland. Cell Cycle. 2007a;6:799–803. doi: 10.4161/cc.6.7.4037. [DOI] [PubMed] [Google Scholar]
- Sutherland KD, Lindeman GJ, Visvader JE. The molecular culprits underlying precocious mammary gland involution. J. Mammary. Gland. Biol. Neoplasia. 2007b;12:15–23. doi: 10.1007/s10911-007-9034-8. [DOI] [PubMed] [Google Scholar]
- Tursun B, Schluter A, Peters MA, Viehweger B, Ostendorff HP, Soosairajah J, Drung A, Bossenz M, Johnsen SA, Schweizer M, Bernard O, Bach I. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 2005;19:2307–2319. doi: 10.1101/gad.1340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat. Rev. Mol. Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- Wagner KU, Wall RJ, St Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Soloway PD, Clark AG. Paternally biased X inactivation in mouse neonatal brain. Genome Biol. 2010;11:R79. doi: 10.1186/gb-2010-11-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.