Abstract
The extraembryonic ectoderm (ExE) of the mouse conceptus is known to play a role in embryo patterning by signaling to the underlying epiblast and surrounding visceral endoderm. Bmp4 is one of the key ExE signaling molecules and has been recently implicated to participate in regulating development and migration of the anterior visceral endoderm (AVE). However, it remains unclear when exactly BMP4 signaling starts to regulate AVE positioning. To examine this, we have chosen to affect BMP4 function at two different time points, at embryonic day 5.25 (E5.25), thus before AVE migration, and E5.75, just after AVE migration. To this end, an RNAi technique was used, which consisted of the injection of Bmp4 dsRNA into the proamniotic cavity of the egg cylinder followed by its targeted electroporation into the ExE. This resulted in specific knockdown of Bmp4. It was found that Bmp4 RNAi at E5.25, but not at E5.75, led to an abnormal pattern of expression of the AVE marker Cerberus-like. Thus, BMP4 signaling appears to affect the expression of Cer1 at a specific time window. This RNAi approach provides a convenient means to study spatial and temporal function of genes shortly after embryo implantation.
Keywords: anterior visceral endoderm, anterior-posterior axis, Bmp4, Cerberus-like, electroporation, mouse embryo, RNA interference
Introduction
On the fourth day after fertilization (embryonic day 4.5 [E4.5]), the mouse blastocyst implants and undergoes a series of morphological changes that leads to a cup-shaped embryo, the egg cylinder. The epiblast, which will give rise to the embryo proper, is located in the distal region of the egg cylinder, whereas the proximal region is occupied by the extraembryonic ectoderm. Both of these tissues are enveloped by another extraembryonic tissue, the visceral endoderm (VE). At E5.5, the embryo is clearly polarized. Thus, for example, Cer1 and Lefty-1 transcripts are restricted to the cells in the distal tip of the VE (Beddington & Robertson 1998, 1999). These distal cells then move towards the prospective anterior of the embryo to establish the anterior visceral endoderm (AVE) (Thomas & Beddington 1996; Thomas et al. 1998; Srinivas et al. 2004), a signaling center responsible for the formation of the anterior structures of the embryo. The position of the AVE will thus determine the position of the anterior-posterior axis.
The extraembryonic ectoderm (ExE) is known to signal to the epiblast and overlying VE (Rodriguez et al. 2005; Richardson et al. 2006). Bone morphogenetic protein 4 (BMP4), a member of the transforming growth factor-β (TGF-β) superfamily of secreted signaling molecules (Hogan 1996), is expressed throughout the ExE (Coucouvanis & Martin 1999) and acts as one of such ExE signaling molecules (Beddington & Robertson 1999). By E6.0 Bmp4 transcripts become localized to a ring-like structure in the distal-most part of the ExE (Lawson et al. 1999) denoting its role in signaling to the underlying epiblast. Loss of BMP4 in knockout embryos leads to an arrest at gastrulation and lack of mesoderm formation, concomitant with the loss of expression of genes in posterior structures (Winnier et al. 1995; Lawson et al. 1999). Downregulation of Bmp4 elicited by dsRNA electroporation at the blastocyst stage has implicated this molecule in the regulation of gene expression in the AVE (Soares et al. 2005). However, because in this approach BMP4 has been downregulated in the whole embryo, it has remained unclear which Bmp4 (i.e. expressed in the ICM [inner cell mass][Coucouvanis & Martin 1999] or ExE) was responsible for such regulation; nor was it established at which point in time it is required for the correct localization of AVE markers.
To address these questions we wished to establish a method that would allow temporal- and tissue-specific targeting of gene expression at early postimplantation stages by RNAi, an effective tool to study gene function in a mouse embryo (Svoboda et al. 2000; Wianny & Zernicka-Goetz 2000; Calegari et al. 2002; Mellitzer et al. 2002; Plusa et al. 2005; Soares et al. 2005). To this end we have combined microinjection of dsRNA into the newly formed proamniotic cavity with targeted electroporation to deliver the dsRNA to this specific region of the embryo. The directed delivery of Bmp4 dsRNA to the ExE of E5.25-E5.75 mouse embryos resulted in a specific knockdown of Bmp4 mRNA levels. These experiments showed that RNAi-mediated knockdown of Bmp4 at E5.25, but not at E5.75, results in an expansion of the Cer1 expression domain, indicating a mislocalization of the AVE.
Materials and Methods
Embryo collection
F1 (C57/BL6 × CBA), TgN(Cer1PGFP)328Belo (referred to in the text as Cer1-GFP, Mesnard et al. 2004) and Bmp4+/−mice (Winnier et al. 1995) were bred in a C57/Bl6 background using a 06.00–18.00 12-h light cycle. Postimplantation embryos were recovered from F1 × Cer1-green fluorescent protein (GFP) crosses. For the purposes of staging, E5.25 embryos were dissected at 07.00 hours, E5.5 embryos at 12.30 hours, and E5.75 embryos were collected at 17.00 hours on the fifth day after plugging. For reference, E5.25 embryos were 128.89 μm long (SD = 24.54 μm, n = 5) and E5.75 embryos 194 μm long (SD = 34.39 μm, n = 6) (see also Rivera-Perez et al. 2003; Perea-Gomez et al. 2007). The expression of GFP driven by the Cer1 promoter has been shown to be a faithful marker of Cer1 expression (Mesnard et al. 2004). All experimental procedures with live animals were conducted in accordance with UK Government Home Office Licensing regulations.
Dissection and culture
Embryos were dissected in Dulbecco’s modified eagle medium without phenol red (DMEM, Life Sciences, Paisley, United Kingdom), containing sodium pyruvate and non-essential amino acids, supplemented with 10% fetal calf serum (FCS) at 37°C. Fine forceps were used to extract deciduae from the uterus and remove embryos from the uterine crypt as described (Hogan et al. 1994). The parietal endoderm was removed with fine syringe needles. Embryos were cultured in pre-equilibrated DMEM medium supplemented with 45% human cord serum (HCS) and non-essential amino acids at 37.5°C under a 5% CO2 atmosphere (Jones et al. 2002; Richardson et al. 2006). The culture system used in this work was conducive to normal growth and development (Richardson et al. 2006).
dsRNA synthesis and labeling
RNAs were in vitro transcribed from polymerase chain reaction (PCR)-generated templates using the Ribomax Large-Scale RNA Production System (Promega, Southampton, UK). Chimeric primers containing the T7 promoter sequence and gene-specific sequences were used to amplify Bmp4 (nt 2–601, 599 bp), and GFP (nt 98–638, 540 bp) templates. In vitro transcription reactions and subsequent DNAse treatment and RNA purification were carried out in a 100 μL reaction according to the manufacturer’s instructions but with 75 mM aminoallyl-UTP (Sigma, Dorset, United Kingdom). The purified RNA was denatured at 70°C for 10 min and allowed to anneal at room temperature for several hours. Following annealing, the dsRNA was passed through Bio-Rad P30 MicroBiospin columns (Bio-Rad, Hemel Hempstead, UK) previously buffer-exchanged to 100 mM NaHCO3 (pH 7.5), and mixed at RT (room temperature), dark O/N with 33 μL of 10 mg/mL AlexaFluor 594-succinimidyl ester (Molecular Probes, Eugene, OR, USA) in dimethylsulfoxide (DMSO). After phenol/chloroform extraction and isopropanol precipitation, the dsRNA was further purified through Bio-Rad P30 MicroBiospin columns (10 mM Tris pH 7.4) and an additional phenol/chloroform and isopropanol step. The quality and concentration of dsRNA were determined by gel electrophoresis and spectrophotometry. AlexaFluor-594-labeled dsRNAs were used in all experiments to allow monitoring of the microinjection and subsequent electroporation procedures.
Microinjection and electroporation
Embryos were microinjected in DMEM supplemented with 10% FCS using a Leica micromanipulator and glass needles back-filled with 1.5 μg/μL AF594-Bmp4 or GFP dsRNA, and/or 1.5 μg/μL pCMV-EGFP (Clontech, Palo Alto, CA, USA). The injection needle was introduced into the proamniotic cavity and the dsRNA solution was injected using air pressure with an Eppendorf Transinjector, allowing the cavity to expand slightly. Microinjected embryos were immediately transferred onto ~200 μL of HBS (hepes-buffered saline) solution laid down on a flat electrode chamber (1 mm gap between electrodes) (BTX Inc., San Diego, CA, USA); oriented with the proximal end towards and perpendicular to the cathode for extraembryonic ectoderm targeting, with a slight tilt (~30–45°) to the right side for the first set of pulses and to the left for the second set of pulses. Electric pulses (two sets of three pulses of 1 ms at 30 V) were delivered using a 2001 Electro Cell Manipulator (BTX Inc.). Following electroporation, embryos were cultured as described above and monitored by fluorescence microscopy 11 and 24 h later (for the E5.25 and E5.75 electroporations, respectively).
Reverse transcription–polymerase chain reaction
Each embryo was separately washed in phosphate-buffered saline (PBS) and lyzed in 200 μL TRIzol (Invitrogen, Paisley, UK). Total RNA was isolated and retro-transcribed by standard procedures. PCR amplification of the cDNA was carried out with the following primers: Bmp4 (5′-TGGACTGTTATTATGCCTT-3′; 5′-GGAGATCACCTCATTT TCTGG-3′) and Glyceraldehyde 3-phosphate dehydrogenase (Gap3dh) (5′-GCATGGACTGTGGTCATGAG-3′; 5′-CCATCACCATCTTCCAGGAG-3′). For each gene, the PCR parameters were optimized to detect differences in mRNA levels: Bmp4 (43 cycles, 60°C annealing); Gap3dh (27 cycles, 60°C annealing). PCR products were resolved by gel electrophoresis and quantified using The Discovery Series Quantity One 1-D Analysis Software Version 4.4.1 (Bio-Rad).
In situ hybridization
E5.75 embryos were collected and processed for in situ hybridization as described (Wilkinson et al. 1990) except that the proteinase K treatment was omitted. The proximal-most part of the embryo was cut and used for genotyping.
Genotyping
The genotyping of the embryos derived from Bmp4 mutant heterozygous crosses was carried out as described (Dunn et al. 1997).
Imaging
All micrograph images were taken using a Nikon inverted microscope, and processed using IPLab software.
Results
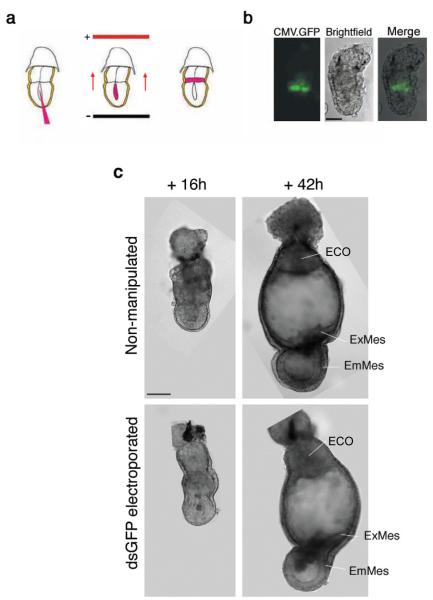
We first wished to examine the delivery efficiency of our method that consists of microinjection of nucleic acids into the proamniotic cavity followed by directed electroporation (Fig. 1a), and its effect on development. To this end we microinjected a CMV-GFP plasmid into the proamniotic cavity of E5.25 embryos, electroporated it towards the ExE, and monitored the GFP-positive cells following culture (n = 20). After 8–10 h of culture, GFP-positive cells were observed in the proximal ExE abutting the epiblast (Fig. 1b). The number of targeted cells in the ExE ranged from 10 to 19, indicating that the targeting of the cells in the extraembryonic tissue had been successful (Fig. 1b). In some embryos (2/20) GFP-positive cells were also seen in the VE of the extraembryonic region (not shown), suggesting that the electroporated DNA had reached not only the layer of cells lining the ExE, but also the adjacent VE cells. However, GFP fluorescence was never observed in the epiblast. The electroporated embryos were allowed to develop in culture for a further 42 h. During this period, they displayed the same typical morphology and developmental rate as those of non-electroporated control embryos cultured under the same conditions (Fig. 1c), indicating that the electroporation does not prevent embryo development at these stages. Thus, we concluded that this combined method of microinjection and directed electroporation is an efficient technique for delivering nucleic acids to specific regions of the postimplantation embryo before gastrulation.
Figure 1.
Method for tissue- and time-specific delivery of dsRNA by directed electroporation. (a) Diagram depicting the method used for the combined microinjection and electroporation. Embryos were recovered and injected into the proamniotic cavity with AlexaFluor594-labeled dsRNA. Immediately after, they were transferred to the electroporation chamber with the extraembryonic region towards the cathode with a tilt of minus 45 degrees (first set of pulses) and plus 45 degrees (second set of pulses) to achieve directed electroporation to the extraembryonic ectoderm. Following electroporation, the embryos were cultured in vitro. (b) Effective delivery of nucleic acids to the targeted region by directed electroporation. Green fluorescent protein (GFP) expression in the distal extraembryonic ectoderm of a wild type embryo subjected to microinjection and directed electroporation of a CMV-EGFP plasmid, 8–10 h after electroporation. Bar, 50 μm. (c) Injection and electroporation of dsRNA into early postimplantation embryos have no detrimental effect on embryo development. Representative brightfield micrographs of non-manipulated and injected/electroporated embryos with GFP dsRNA, imaged 16 h and 42 h after electroporation. The embryos subjected to experimental manipulation displayed the same typical morphology and developmental rate as those of non-electroporated control embryos. ECO, ectoplacental cone; EmMes, embryonic mesoderm; ExMes, extraembryonic mesoderm. Bar, 50 μm. The type of embryos shown in the upper panel pictures represent the best culture results using the current available whole embryo culture techniques.
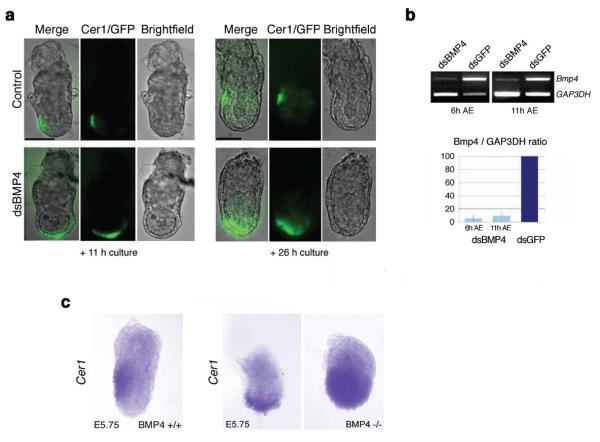
We then wished to establish whether the same technique would result in efficient delivery of dsRNA and consequent downregulation of gene expression in this region. To examine whether BMP4 expressed in the ExE regulates the positioning of the AVE cells, we directed dsRNA for Bmp4 into the ExE at different developmental times. To monitor the effects on the development of AVE, the experiments were carried out in a transgenic line expressing GFP under the control of the Cer1 promoter (Mesnard et al. 2004). Cer1 expression is a reliable indicator of the integrity of the DVE (distal visceral endoderm)/AVE (Soares et al. 2005; Richardson et al. 2006). GFP expression in this line has been shown to faithfully reflect the expression of Cer1, which is found in the same domain as other AVE markers such as Lefty-1 (Torres-Padilla et al. 2007). AlexaFluor594-labeled Bmp4 dsRNA was microinjected into the proamniotic cavity at E5.25, followed by directed electroporation to target specifically the distal region of the ExE of Cer1-GFP transgenic embryos. As a control, we used AF594-labeled GFP dsRNA to electroporate the embryos using the same approach. In both cases, dsRNA fluorescence could be seen in the ExE, but it was mostly overshadowed by the very bright fluorescence emanating from the dsRNA within the proamniotic cavity, where the solution had been previously injected (not shown). After 11 h of culture, a time equivalent to E5.75, the embryos microinjected and electroporated with the control GFP dsRNA at E5.25 showed normal AVE localization, as judged by the pattern of Cer1-GFP expression (n = 4, Fig. 2a). In contrast, the domain of Cer1-GFP-expressing cells in embryos electroporated with Bmp4 dsRNA extended towards both the future ‘anterior’ and ‘posterior’ poles of the embryo adopting a circumferential distribution from its original distal position at the same time point (n = 5, Fig. 2a). After 26 h of culture, the control embryos showed Cer1-GFP positive cells laterally, and confined to the AVE, as expected. In contrast, Cer1-GFP expression remained distal and mislocalized in embryos electroporated with Bmp4 dsRNA (Fig. 2a).
Figure 2.
Bone morphogenetic protein 4 (Bmp4) RNAi at embryonic day 5.25 (E5.25) results in the mislocalization of the anterior visceral endoderm (AVE) as determined by Cer1-green fluorescent protein (GFP) expression. (a) Bmp4 RNAi in the extraembryonic ectoderm at E5.25 results in an abnormal Cer1-GFP expression in the visceral endoderm. E5.25 embryos from F1xCer1-GFP crosses were electroporated with Bmp4 dsRNA and GFP dsRNA (control) as illustrated in Fig. 1 (a). Brightfield and fluorescence micrographs are representative of four independent experiments showing the same embryos imaged 11 h and 26 h after electroporation. Bar, 50 μm. (b) Specific Bmp4 mRNA knockdown by RNAi at E5.25. Reverse transcription–polymerase chain reaction (RT–PCR) of single embryos cultured in vitro for 6 h and 11 h following microinjection and electroporation (AE) at E5.25. Glyceraldehyde 3-phosphate dehydrogenase (Gap3dh) was used as an internal control for complementary DNA (cDNA) loading. Quantification of the average Bmp4 mRNA levels (Bmp4/Gap3dh ratio) of embryos electroporated with Bmp4 dsRNA is shown as a percentage of those of dsGFP-treated controls. Pale blue, 2 dsBmp4-electroporated embryos cultured for 6 h AE (control embryos: 3, dark blue); and 3 dsBmp4-electroporated embryos cultured for 11 h AE (control embryos: 3). Standard deviation bars are indicated. (c) Whole-mount in situ hybridization with Cer1 antisense probe in wild type and Bmp4−/– embryos. Cer1 expression in the visceral endoderm of wild type embryos is localized anteriorly, reaching the embryonic/extraembryonic border, as expected at this stage. In contrast, the expression domain of Cer1 is mislocalized in Bmp4−/– embryos: in some instances, Cer1 is expressed distally in a semi-circumferential distribution; in others it shows a widespread expression covering most of the visceral endoderm overlying the epiblast. Bar, 50 μm.
To assess the efficiency of the RNAi procedure, we carried out semiquantitative reverse transcription–polymerase chain reaction (RT–PCR) in whole embryos after electroporation. This showed that Bmp4 dsRNA electroporation had produced a knockdown of Bmp4 of over 95%, which was evident as early as 6 h after electroporation (the earliest time point analyzed) and persisted throughout time in culture (~90% knockdown, 11 h after electroporation) (Fig. 2b). Levels of Gap3dh (Fig. 2b) and Stat3 mRNA (not shown) were unaffected, suggesting that the Bmp4 dsRNA electroporation resulted in sequence-specific knockdown of Bmp4 expression.
To further validate these findings, we compared the pattern of Cer1 expression of our dsRNA electroporated embryos to that of Bmp4 null mice. E5.75 embryos derived from Bmp4 heterozygous crosses were collected and processed for in situ hybridization for Cer1, and had the proximal part removed for genotyping. The expression of Cer1 in wild type embryos was found to be restricted to the AVE, which had reached the embryonic/extraembryonic boundary at this time (Fig. 2c). In contrast, in Bmp4−/– embryos, the expression of Cer1 appeared mislocalized and presented rather a circumferential distribution along the embryonic region of the embryo (Fig. 2c). The distribution of Cer1 transcripts in the mutants varied from a slight expansion of the domain of expression of Cer1 to a widespread expression covering most of the VE overlying the epiblast (compare middle and right panels in Fig. 2c). This variation appears consistent with the phenotypic variability and incomplete penetrance in Bmp4−/– mutant mice (Winnier et al. 1995; Dunn et al. 1997). These results indicate that the mislocalization of Cer1 observed in BMP4 RNAi embryos is indeed due to the downregulation of the ExE-expressed Bmp4.
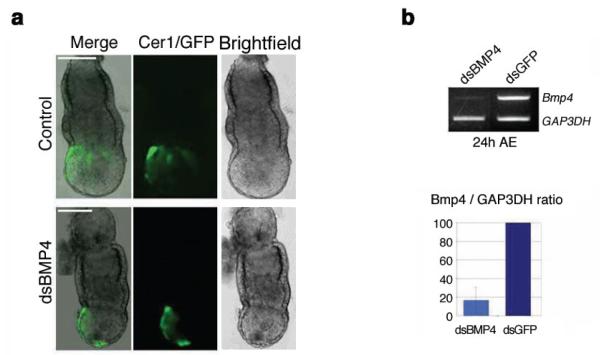
It has been found that the position of the AVE is influenced by signals emanating from the ExE at the onset of the AVE migration, whereas at E5.75 – after migration has commenced – such an influence has no longer been observed (Richardson et al. 2006). To explore whether BMP4 could also affect the AVE positioning after the onset of its migration, we carried out the same Bmp4 RNAi experiments at E5.75. Following microinjection and electroporation at this stage both the control (n = 10) and the Bmp4 dsRNA-treated embryos (n = 12) showed the expected distribution of GFP-positive cells in the AVE after 24 h of culture (Fig. 3a). It is noteworthy, however, that some remnants of GFP expression were still observed in the DVE of some Bmp4 dsRNA-treated embryos, despite the apparently normal positioning of the AVE. The BMP4 mRNA knockdown level was again determined by semiquantitative RT–PCR. This showed that 24 h after electroporation Bmp4 expression was decreased by 82% (Fig. 3b). Thus, RNAi-mediated knockdown of Bmp4 at E5.25, but not at E5.75, results in an abnormal expansion of the Cer1 expression domain.
Figure 3.
Bone morphogenetic protein 4 (Bmp4) RNAi at embryonic day 5.75 (E5.75) has no effect on the anterior visceral endoderm (AVE) positioning as determined by Cer1-GFP expression. (a) Bmp4 RNAi in the extraembryonic ectoderm at E5.75 does not alter the Cer1 pattern of expression in the visceral endoderm. E5.75 embryos from F1xCer1-GFP crosses were electroporated with Bmp4 dsRNA and green fluorescent protein (GFP) dsRNA (control). Brightfield and fluorescence micrographs are representative of four independent experiments taken 24 h after electroporation. Bar, 50 μm. (b) Specific Bmp4 mRNA knockdown by RNAi at E5.75. Reverse transcription–polymerase chain reaction (RT–PCR) of single embryos cultured in vitro for 24 h following microinjection and electroporation (AE) at E5.75. Glyceraldehyde 3-phosphate dehydrogenase (Gap3dh) was used as an internal control for complementary DNA (cDNA) loading. Quantification of the average Bmp4 mRNA levels (Bmp4/Gap3dh ratio) of embryos electroporated with dsBmp4 RNA is shown as a percentage of those of dsGFP-treated controls. Light blue, 6 dsBmp4-electroporated embryos cultured for 24 h AE; dark blue, 5 control embryos. Standard deviation bars are indicated.
Discussion
We have found that targeting Bmp4 in the ExE by RNAi leads to the mislocalization of AVE-specific gene expression as determined by the misexpression of Cer1. This phenotype was observed when RNAi was applied at E5.25 but not at E5.75, suggesting that BMP4 signaling regulates Cer1 expression before the onset of the AVE migration. These results are in accordance with Cer1 expression changes observed upon ablation of the prospective posterior ExE at E5.25 by Richardson et al. (2006). Although the AVE appeared to be properly established upon Bmp4 RNAi at E5.75, it is conceivable that BMP4 downregulation at this stage is not always entirely innocuous for development, as residual expression of Cer1 was observed in the DVE in some instances. Thus, the RNAi technique described here can discern between relatively short periods of time of gene function.
Using a different RNAi approach through electroporation before implantation, Soares et al. (2005) had previously implicated BMP4 in regulating gene expression in the AVE. However, unlike the method used in this paper, Bmp4 downregulation had been elicited at the blastocyst stage and the expression of this gene has been affected in all lineages of the embryo. In contrast, here we report that the downregulation of Bmp4 restricted to the ExE alone disrupts the correct expression of the AVE marker Cer1. This indicates that expression of BMP4 in the ExE at approximately E5.25 is influencing gene expression important for the AVE positioning. Hence, the function of signaling molecules confined to specific regions can be studied with increased spatial precision and so can the effects of their downregulation be attributed to spatial and temporal patterns of gene expression.
The early postimplantation stages of mouse development entail a complex cascade of signaling events that pattern the egg cylinder and establish the animal body plan. As shown here, the methodology of combining microinjection and electroporation to target gene expression by RNAi in a temporal- and tissue-specific manner could be a valuable tool to dissect key molecular interactions in early postimplantation development.
Acknowledgements
We would like to thank Brigid Hogan for the Bmp4 mutant mice, Stephen Frankenberg for the Cer1 probe and Susana Chuva de Sousa Lopes for invaluable technical advice. This work was supported by grants from the Welcome Trust and BBSRC to M.Z.G., and from Cancer Research UK. M.Z-G. is a Wellcome Trust Senior Research Fellow.
References
- Beddington RSP, Robertson EJ. Anterior patterning in mouse. Trends Gen. 1998;7:277–284. doi: 10.1016/s0168-9525(98)01499-1. [DOI] [PubMed] [Google Scholar]
- Beddington RSP, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- Calegari F, Haubensak W, Yang D, Huttner WB, Buchholz F. Tissue-specific RNA interference in postimplantation mouse embryos with endoribonuclease-prepared short interfering RNA. Proc. Natl Acad. Sci. USA. 2002;99:14 236–14 240. doi: 10.1073/pnas.192559699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucouvanis E, Martin GR. BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development. 1999;126:535–546. doi: 10.1242/dev.126.3.535. [DOI] [PubMed] [Google Scholar]
- Dunn NR, Winnier GE, Hargett LK, Schrick JJ, Fogo AB, Hogan BL. Haploinsufficient phenotypes in Bmp4 heterozygous null mice and modification by mutations in Gli3 and Alx4. Dev. Biol. 1997;188:235–247. doi: 10.1006/dbio.1997.8664. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Hogan BL, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. 2nd edn Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York, USA: 1994. p. 475. [Google Scholar]
- Jones EA, Crotty D, Kulesa PM, et al. Dynamic in vivo imaging of postimplantation mammalian embryos using whole embryo culture. Genesis. 2002;34:228–235. doi: 10.1002/gene.10162. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellitzer G, Hallonet M, Chen L, Ang SL. Spatial and temporal ‘knock down’ of gene expression by electroporation of double-stranded RNA and morpholinos into early postimplantation mouse embryos. Mech. Dev. 2002;118:57–63. doi: 10.1016/s0925-4773(02)00191-0. [DOI] [PubMed] [Google Scholar]
- Mesnard D, Filipe M, Belo JA, Zernicka-Goetz M. The anterior-posterior axis emerges respecting the morphology of the mouse embryo that changes and aligns with the uterus before gastrulation. Curr. Biol. 2004;14:184–196. doi: 10.1016/j.cub.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Perea-Gomez A, Meilhac SM, Piotrowska-Nitsche K, Gray D, Collignon J, Zernicka-Goetz M. Regionalization of the mouse visceral endoderm as the blastocyst transforms into the egg cylinder. BMC Dev. Biol. 2007;7:96. doi: 10.1186/1471-213X-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusa B, Frankenberg S, Chalmers A, et al. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J. Cell Sci. 2005;118:505–515. doi: 10.1242/jcs.01666. [DOI] [PubMed] [Google Scholar]
- Richardson L, Torres-Padilla ME, Zernicka-Goetz M. Regionalised signalling within the extraembryonic ectoderm regulates anterior visceral endoderm positioning in the mouse embryo. Mech Dev. 2006;123:288–296. doi: 10.1016/j.mod.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Rivera-Pérez JA, Mager J, Magnuson T. Dynamic morphogenetic events characterize the mouse visceral endoderm. Dev. Biol. 2003;261:470–487. doi: 10.1016/s0012-1606(03)00302-6. [DOI] [PubMed] [Google Scholar]
- Rodriguez TA, Srinivas S, Clements MP, Smith JC, Beddington RS. Induction and migration of the anterior visceral endoderm is regulated by the extra-embryonic ectoderm. Development. 2005;132:2513–2520. doi: 10.1242/dev.01847. [DOI] [PubMed] [Google Scholar]
- Soares ML, Haraguchi S, Torres-Padilla ME, et al. Functional studies of signaling pathways in peri-implantation development of the mouse embryo by RNAi. BMC Dev. Biol. 2005;5:28. doi: 10.1186/1471-213X-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Rodriguez T, Clements M, Smith JC, Beddington RS. Active cell migration drives the unilateral movements of the anterior visceral endoderm. Development. 2004;131:1157–1164. doi: 10.1242/dev.01005. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNAi. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- PubMed,CAS,Web of Science® Times Cited: 229Camsfx
- Thomas P, Beddington R. Anterior primitive endoderm may be responsible for patterning the anterior neural plate in the mouse embryo. Curr. Biol. 1996;6:1487–1496. doi: 10.1016/s0960-9822(96)00753-1. [DOI] [PubMed] [Google Scholar]
- Thomas PQ, Brown A, Beddington RS. Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development. 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Richardson L, Kolasinska P, Meilhac SM, Luetke-Eversloh MV, Zernicka-Goetz M. The anterior visceral endoderm of the mouse embryo is established from both preimplantation precursor cells and by de novo gene expression after implantation. Dev. Biol. 2007;309:97–112. doi: 10.1016/j.ydbio.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]