Abstract
Equine infectious anemia virus (EIAV) is a lentivirus that causes persistent infections in horses. We hypothesized that high-avidity CTL specific for nonvariable epitopes might be associated with low viral load and minimal disease in EIAV-infected horses. To test this hypothesis, memory CTL (CTLm) responses were analyzed in two infected horses with high plasma viral loads and recurrent disease (progressors), and in two infected horses with low-to-undetectable viral loads and mild disease (nonprogressors). High-avidity CTLm in one progressor recognized an envelope gp90 epitope, and the data documented for the first time in EIAV that viral variation led to CTL escape. Each of the nonprogressors had high-to-moderate avidity CTLm directed against epitopes within Rev, including the nuclear export and nuclear localization domains. These results suggested that the epitope specificity of high- and moderate-avidity CTLm was an important determinant for disease outcome in the EIAV-infected horses examined.
Keywords: EIAV, Lentivirus, Horse, CTL, CTL escape, Functional avidity, Rev, Viral variation, Viral load, Nonprogressor
Introduction
Horses infected with equine infectious anemia virus (EIAV) usually develop recurrent episodes of plasma viremia and associated acute clinical disease (fever, inappetence, lethargy, thrombocytopenia, and anemia) during the first few months of infection (Cheevers and McGuire, 1985; Montelaro et al., 1993; Sellon et al., 1994). In contrast to other lentiviral infections, including human immunodeficiency virus-1 (HIV-1), most, but not all, horses control EIAV replication within a year and remain persistently infected inapparent carriers (Cheevers and McGuire, 1985; Montelaro et al., 1993; Sellon et al., 1994). Results of EIAV infection in severe combined immunodeficient (SCID) Arabian foals, and recently, immune reconstitution in a SCID foal prior to EIAV challenge, indicate that this control of viral replication is mediated by viral-specific immune responses (Perryman et al., 1988; Crawford et al., 1996; Mealey et al., 2001).
Although the precise immunological mechanisms by which horses control EIAV infection are unknown, evidence exists that CD8+ major histocompatibility complex (MHC) class I restricted viral-specific cytotoxic T Lymphocytes (CTL) are important. Neutralizing antibodies undoubtedly play an antiviral role, but the initial plasma viremia in acute EIAV infection is often terminated prior to the appearance of neutralizing antibody (McGuire et al., 1990), but concurrent with the appearance of CTL (McGuire et al., 1994). Similarly, CTL are associated with control of the primary viremia in HIV-1-infected people (Borrow et al., 1994), while neutralizing antibody is not detected for up to 2 months after the primary viremia subsides (Pantaleo and Fauci, 1995). The adoptive transfer of autologous CTL clones to HIV-1-infected patients results in functional CTL activity in infected tissues and transiently reduces the levels of circulating productively infected CD4+ T lymphocytes (Brodie et al., 1999). Additionally, transfer of CTL clones to HIV-1-infected (hu)-PBL-SCID mice reduces plasma viremia (McKinney et al., 1999). Direct evidence for CD8+ lymphocyte control of simian immunodeficiency virus (SIV) in infected rhesus monkeys is provided by in vivo depletion of CD8+ lymphocytes with monoclonal antibody (Schmitz et al., 1999; Jin et al., 1999). Moreover, vaccines that induce high-frequency SIV-specific CTL responses in rhesus monkeys result in reduced viral loads and prevention of clinical AIDS following pathogenic SIV challenge (Barouch et al., 2000, 2001a,b). Horses recently infected with EIAV have CD8+ effector CTL (CTLe) in peripheral blood mononuclear cells (PBMC), which do not require in vitro stimulation for effector function (McGuire et al., 1994). These CTL recognize epitopes in Env and Gag proteins (McGuire et al., 1994). Later in infection, when the horses have controlled viremic episodes and are inapparent carriers, memory CTL (CTLm) to Env and Gag proteins which require in vitro stimulation are present (Hammond et al., 1997; McGuire et al., 1997). These CTLm have a mean frequency of 293 per million PBMC (McGuire et al., 1997). CTLm epitopes have been identified in EIAV Gag, Pol, Env, and in the protein encoded by the S2 open reading frame (Lonning et al., 1999; McGuire et al., 2000; Zhang et al., 1998).
Following acute infection, recurrent viremic episodes are associated with the emergence of antigenically distinct EIAV variants as defined by neutralizing antibody (Kono et al., 1973; Rwambo et al., 1990). Studies suggest that immune pressure exerted by neutralizing antibody selects for antigenic variation in envelope glycoprotein epitopes, resulting in escape from humoral responses (Leroux et al., 2001; Montelaro et al., 1984; Payne et al., 1987; Ball et al., 1992). Ongoing envelope variation continues during long-term asymptomatic infection (Leroux et al., 2001). However, because variant EIAV isolates can be detected prior to neutralizing antibody, it is unlikely that humoral immune selection is the only factor in their genesis (Carpenter et al., 1987; Hussain et al., 1987; Rwambo et al., 1990), and it is possible that lymphocyte-mediated immune selection is involved in the appearance of these genetically distinct virus isolates. The fact that recurrent episodes of viremia and clinical disease occur in the face of both neutralizing antibody and CTL suggests that EIAV variants arise that must escape both neutralizing antibody and CTL responses. It has been well documented that CTL epitope escape variants occur during HIV-1 infection (Borrow et al., 1997; Harrer et al., 1998) and that variation in HIV-1 CTL epitopes results in loss of cytotoxic activity and is associated with clinical progression to AIDS (Goulder et al., 1997a; Koenig et al., 1995).
Some EIAV-infected horses, designated nonprogressors, control the initial viremia and subsequently maintain low or undetectable plasma viral loads and remain free of episodic clinical disease (Leroux et al., 2001). In HIV-1-infected nonprogressors, high levels of CTL activity have been associated with the lack of disease progression (Harrer et al., 1996; Rinaldo et al., 1995). In addition, it has been shown in HIV-1 and lymphocytic choriomeningitis virus (LCMV) models in mice that the ability of CTL to clear virus in vivo cannot be predicted simply by the ability of CTL to kill infected targets. Rather, in vivo efficacy is dependent on functional avidity: the ability of CTL to kill targets expressing low antigen density (Alexander-Miller et al., 1996; Derby et al., 2001a; Gallimore et al., 1998a). Interestingly, recent evidence suggests that CTL with high functional avidity rapidly select for CTL escape SIV variants(O’Connor et al., 2002).
Given the above observations, we hypothesized that CTLm with high functional avidity directed against nonvariable epitopes would occur in EIAV-infected nonprogressor horses. To test this hypothesis, the CTLm responses in four EIAV-infected Arabian horses were analyzed over a 1.7-year period. Two horses were considered nonprogressors based on minimal clinical disease and low-to-undetectable plasma viral loads as measured by real-time quantitative RT-PCR. In these two horses, CTLm with high and moderate avidities directed against epitopes in Rev were identified. Interestingly, the Rev epitopes included portions of the nuclear export and nuclear localization domains. The other two horses were considered progressors based on high-plasma viral loads and recurrent episodes of disease. Unexpectedly, high-to-moderate avidity CTLm were also identified in both horses. In one progressor, high-avidity CTLm were directed against an epitope in the highly variable putative principle neutralizing domain of envelope gp90, and viral variation within the epitope led to loss of CTLm recognition. These results suggest that in addition to functional avidity, the EIAV proteins targeted by CTLm are critical in determining disease outcome.
Results
EIAV-induced disease progression and plasma viral loads
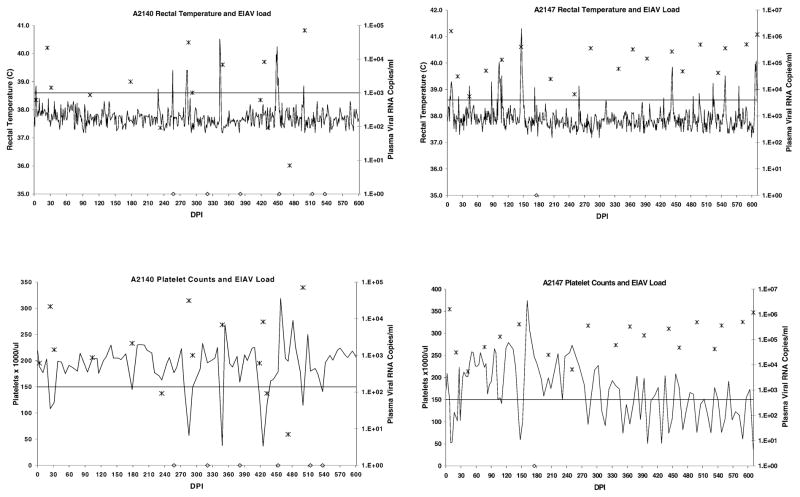
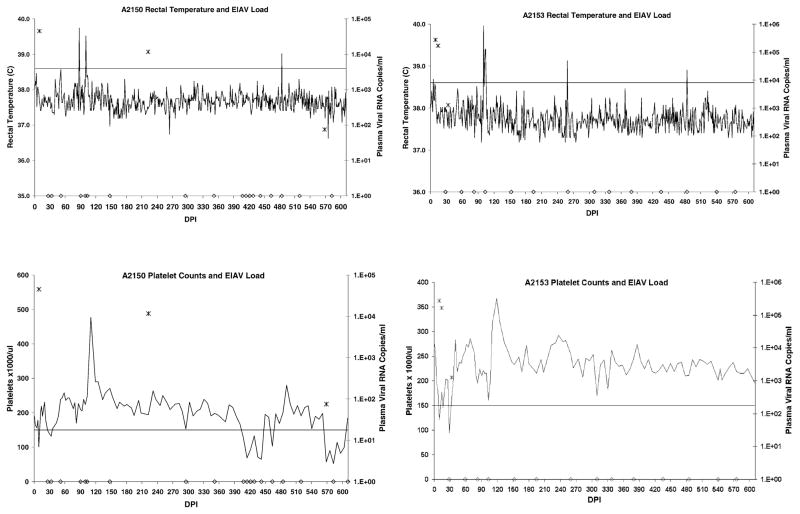
Clinical disease status and plasma viral loads over a 1.7-year period revealed that two horses (A2140 and A2147) experienced recurrent disease (Fig. 1), while the other two horses (A2150 and A2153) experienced minimal disease following resolution of the first viremic episode (Fig. 2).
Figs. 1.
Clinical disease progression and plasma viral load in horses A2140 and A2147. Rectal temperatures above the horizontal line were considered febrile (>38.6°C), while platelet counts below the horizontal line were considered thrombocytopenic (<150,000 platelets/μl whole blood). Clinical disease was defined as thrombocytopenia with or without fever, concurrent with detectable plasma virus. Plasma EIAV RNA copies/ml are indicated with “*”. Samples in which virus was not detected are indicated with “◇” on the x-axis.
Fig. 2.
Clinical disease progression and plasma viral load in horses A2150 and A2153. Rectal temperatures above the horizontal line were considered febrile (>38.6°C), while platelet counts below the horizontal line were considered thrombocytopenic (<150,000 platelets/μl whole blood). Clinical disease was defined as thrombocytopenia with or without fever, concurrent with detectable plasma virus. Plasma EIAV RNA copies/ml are indicated with “*”. Samples in which virus was not detected are indicated with “◇” on the X-axis.
The first of the two horses considered progressors, horse A2140, had six disease episodes (V1–V6) beginning on days postinoculation (DPI) 24, 178, 285, 348, 418, and 500. The other progressor, horse A2147, had 16 disease episodes (V1–V16) beginning on DPI 9, 110, 145, 282, 309, 351, 386, 400, 428, 442, 470, 498, 519, 540, 575, and 610. For both A2140 and A2147, thrombocytopenic disease episodes were almost always associated with fever and other clinical signs of EIAV-induced disease (inappetence and lethargy).
The first of the two horses considered nonprogressors, A2150, had only two disease episodes (V1 and V2) beginning on DPI 9 and 568. An episode of thrombocytopenia occurred on DPI 26 that was not associated with detectable plasma viral RNA (nor fever) and was not considered a distinct disease episode. The thrombocytopenia may have been due to residual bone marrow suppression associated with V1. Another thrombocytopenic episode occurred on DPI 407, which again was not associated with detectable plasma viral RNA (nor fever), and therefore, was not considered a distinct disease episode. Plasma virus was detected on DPI 222, but was not associated with any clinical disease. None of the episodes of thrombocytopenia (including V1 and V2) in horse A2150 were associated with fever or other signs of clinical disease. The other nonprogressor, horse A2153, had only two disease episodes (V1 and V2), early in infection, beginning on DPI 9 and 28. Horse A2153 had only mild fever (38.7°C) associated with V1 only, and never experienced any other clinical signs attributable to EIAV.
Viral load was determined in plasma samples obtained during clinical disease episodes, and at various other time points between episodes (Figs. 1 and 2). Plasma viral RNA was detected in progressor horses A2140 and A2147 plasma on 70% (14/20) and 95% (19/20) of the days assayed, respectively. Plasma viral load was variable in A2140, ranging from undetectable to 7.1 × 104 RNA copies/ml, whereas in A2147, the viral load appeared to reach a steady state of approximately 3 × 105 RNA copies/ml after V4. In contrast, plasma virus was detected in nonprogressor horses A2150 and A2153 plasma on only 13.6% (3/22) and 17.6% (3/17) of the days assayed, respectively. For A2150, viral load values were 2.7 × 105 and 7.5 × 101 RNA copies/ml for V1 and V2, respectively. The only other positive sample was obtained on DPI 222, which contained 1.1 × 104 RNA copies/ml. For A2153, viral load values ranged from 1.2 × 103 to 2.7 × 105 RNA copies/ml during V1 and V2, but viral RNA could not be detected thereafter.
CTLm analysis in nonprogressors A2140 and A2147
CTLm epitope mapping in A2140
To determine the EIAV proteins recognized by CTLm, equine kidney (EK) cells were transduced with retroviral vectors expressing either whole EIAV proteins or polypeptides (McGuire et al., 2000) and used as CTL targets. CTLm in PBMC obtained after V1 from A2140 recognized targets expressing the N-terminal third of the envelope protein which includes a part of the gp90 surface unit (SU), and targets expressing the Gag p15 protein (McGuire et al., 2000).
To further analyze the envelope epitope, CTL assays were performed using EK cell targets transduced with retroviral vectors expressing envelope polypeptide truncations (McGuire et al., 2000). The epitope was contained in SU, and to identify the minimal epitope, overlapping peptides were synthesized and used to sensitize EK target cells. The 12-mer CTLm epitope (RVEDVT-NTAEYW) spanned amino acids (aa) positions 195 to 206 in the EIAVWSU5 envelope protein and was designated Env195-206RW12 (Table 1). The arginine in position 1 was considered a probable anchor residue, since its omission resulted in a 4 log10 reduction in target cell recognition efficiency. CTL assays using peptide-pulsed ELA-A1 and ELA-w11 half-matched EK target cells indicated that presentation of Env-RW12 was associated with the ELA-A1 haplotype.
Table 1.
CTLm epitopes (and synthesized epitope variant peptides) identified in EIAVWSU5-infected horses A2140, A2147, A2150, and A2153
| CTLm epitope designation (abbreviated) | No. of aa | aa Sequence | ELA-A restriction | EIAV protein (aa positions) | Overall avidity (nM) | Synthesized variant peptide designation (abbreviated) | Synthesized variant peptide aa sequence | Horse | Progressor/nonprogressor |
|---|---|---|---|---|---|---|---|---|---|
| Env195–206RW12 Env-RW12 |
12 | RVEDVTNTAEYW | A1 | Env 195–206 | 0.21 | Env195–206HW12v Env-HW12v |
HVKDETNTTEYW | A2140 | Progressor |
| Gag21–32GW12 Gag-GW12 |
12 | GSQKLTTGNCNW | A1 | Gag 21–32 | 155 | None assigned | GSQKLSTGNCNW GSQKLNTGNCNW |
A2140 | Progressor |
| Gag288–297FK10 Gag-FK10 |
10 | FVDRLLSQIK | A4 | Gag 288–297 | 18.7 | None | None | A2147 | Progressor |
| Rev57–67QW11 Rev-QW11 |
11 | QAEVLQERLEW | A1 | Rev 57–67 | 0.29 | None | None | A2150 | Nonprogressor |
| Rev120–131PR12 Rev-PR12 |
12 | PRVLRPGDSKRR | A1 | Rev 120–131 | 5658 | None synthesized | None synthesized | A2150 | Nonprogressor |
| Rev24–33R10 Rev-IR10 |
10 | IPSQTCITRR | Not determined | Rev 24–33 | 47.8 | None synthesized | None synthesized | A2153 | Nonprogressor |
| Rev104–112GR9 Rev-GR9 |
9 | GGYQRAQER | Not determined | Rev 104–112 | 25.1 | None synthesized | None synthesized | A2153 | Nonprogressor |
A similar strategy was used to identify the A2140 CTLm epitope within Gag p15. The epitope (GSQKLTTGNCNW) spanned aa positions 21 to 32 in the EIAVWSU5 Gag protein and was designated Gag21-32 GW12 (Table 1). CTL assays using peptide-pulsed ELA-A1 and ELA-w11 half-matched EK target cells indicated that Gag-GW12 was restricted by the ELA-A1 haplotype.
EIAV CTLm epitope escape variants in A2140
Because the Env-RW12 epitope was located within the putative principle neutralizing domain (PND) of EIAV gp90, a region known to be highly variable (Ball et al., 1992), it was possible that viral variation within the epitope could have resulted in loss of CTLm recognition. In addition, natural variation in EIAV Gag regions that contain CTL epitopes is known to occur (Zhang et al., 1999), raising the possibility that variation within the Gag-GW12 epitope could also have resulted in loss of CTLm recognition.
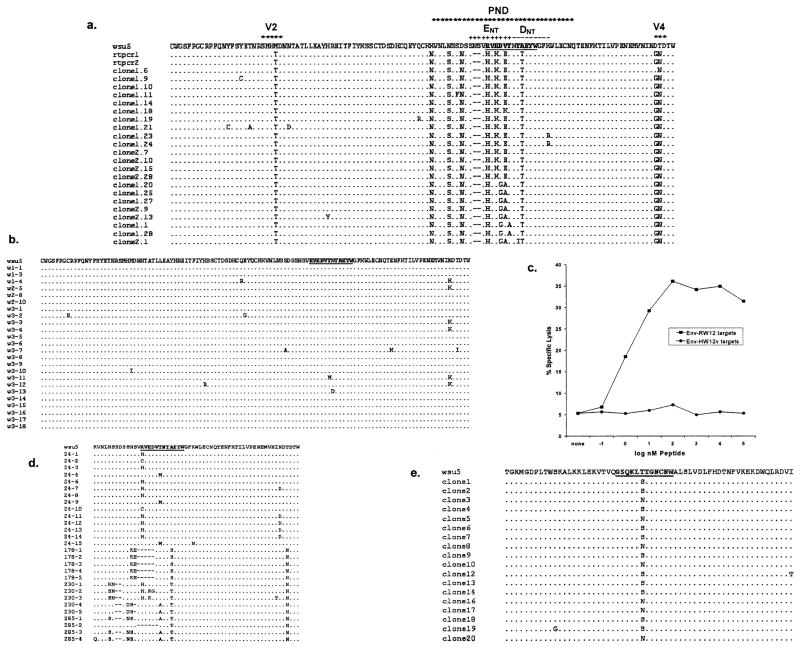
Analysis of deduced aa sequences of EIAV clones obtained from A2140 V5 plasma indicated that viral variation occurred in the Env-RW12 epitope (Fig. 3a). None of the observed aa changes were present in the virus stock used for inoculation (Fig. 3b). The majority of the Env-RW12 variants (59%; 13/22 clones) had the sequence HVKDETTTEYW, which was designated Env195-206 HW12v (Table 1). The aa substitution at position 1 (R → H) involved a suspected anchor residue. The other aa changes within the epitope involved residues likely important for T cell receptor (TCR) binding and included both conservative and nonconservative substitutions. This variant peptide was synthesized and used to sensitize EK target cells for CTL assays. A2140 CTLm obtained after V5 failed to lyse targets pulsed with Env-HW12v (Fig. 3c). The result was the same when CTLm obtained between V1 and V2 (at 47 DPI) were used (data not shown). In addition, Env-HW12v was incapable of stimulating CTLm to lyse either Env-RW12- or Env-HW12v-sensitized target cells.
Fig. 3.
(a) Deduced EIAV envelope gp90 amino acid sequences (aa 122–238) obtained from A2140 V5 plasma (GenBank Accession Nox. AY288526-AY288549) as compared to the EIAVWSU5 stock used for inoculation. Only those aa different from those in EIAVWSU5 are shown. rtpcr1 and rtpcr2 were two different directly sequenced RT-PCR products. Clones designated with a “1.” were derived from the rtpcr1 product, while clones designated with a “2.” were derived from the rtpcr2 product. The Env-RW12 CTLm epitope is in bold and underlined. The hypervariable regions V2 and V4, the putative principle neutralizing domain (PND), and the two murine monoclonal neutralizing antibody epitopes ENT and DNT (Ball et al., 1992) are indicated. (b) Deduced EIAV envelope gp90 amino acid cloned sequences (aa 122–238) obtained from the EIAVWSU5 stock used for inoculation (GenBank Accession Nos. AY288550-AY288559). Only those aa different from the EIAVWSU5 consensus are shown. The Env-RW12 CTLm epitope is in bold and underlined. (c) A2140 CTLm were added to homologous EK targets pulsed with either peptide Env-RW12 or Env-HW12v at the concentrations indicated. CTLm were from PBMC obtained after V5 and stimulated for 7 days with EIAVWSU5. Effector:target cell ratio was 20:1. (d) Deduced EIAV envelope gp90 amino acid sequences (aa 182–238) obtained from A2140 V1 (DPI 24), V2 (DPI 178), between V2 and V3 (DPI 230), and V3 (DPI 285) plasmas (GenBank Accession Nos. AY288560-AY288587) as compared to the EIAVWSU5 stock used for inoculation. Only those aa different from those in EIAVWSU5 are shown. Clones are designated by the DPI from which the RT-PCR product was obtained, followed by the clone number. The Env-RW12 CTLm epitope is in bold and underlined. (e) Deduced EIAV Gag p15 amino acid sequences (aa −3–57) obtained from A2140 V5 plasma (GenBank Accession Nos. AY288588-AY288605) as compared to the EIAVWSU5 stock used for inoculation. Only those aa different from those in EIAVWSU5 are shown. The Gag-GW12 CTLm epitope is in bold and underlined.
Because two possible neutralizing antibody binding sites (ENT and DNT) (Ball et al., 1992) span the Env-RW12 epitope, it was of interest to attempt to determine whether Env-HW12v was the result of selection pressure exerted by neutralizing antibody or CTL. Neutralizing antibody activity directed against the EIAVWSU5 inoculum was present in DPI 285 (V3) and DPI 45 (between V1 and V2) plasma (100% and 95% virus reduction, respectively), but was absent in DPI 24 (V1) plasma (<4% virus reduction). Pre-incubation of DPI 45 plasma with peptides containing both putative neutralizing antibody epitopes was done using serial 10-fold dilutions of both peptides in plasma starting with a concentration of 1 mg/ml. These peptides failed to block neutralizing antibody activity in DPI 45 plasma, suggesting either that the Env-RW12 region was not a target for neutralizing antibody or that additional neutralizing antibody epitopes were present outside this region. Analysis of cloned EIAV sequences from plasma obtained during V1 (DPI 24), V2 (DPI 178), between V2 and V3 (DPI 230) and V3 (DPI 285) indicated that aa changes within the Env-RW12 epitope, which included a change in the suspected N-terminal anchor residue, occurred as early as DPI 24, before neutralizing antibody was detected (Fig. 3d). CTLm from before DPI 47 were not available for study. The aa changes within the epitope became more pronounced at the later time points, and it is possible that both CTL and neutralizing antibody were involved in their genesis. Regardless of the cause, significant aa variation occurred within Env-RW12 that abrogated the CTLm response and likely contributed to the recurrent disease experienced by A2140.
Deduced aa sequences of EIAV clones obtained from A2140 V5 plasma were also analyzed to determine whether viral variation occurred within the Gag-GW12 CTLm epitope (Fig. 3e). Of these clones, 61% (11/18) had the sequence GSQKLSTGNCNW, while 39% (7/18) had the sequence GSQKLNTGNCNW. The aa substitutions at position 6 were conservative in both variants. These two CTLm epitope variant peptides were synthesized (Table 1) and used to sensitize EK target cells. CTLm from between V1 and V2 (DPI 47) recognized Gag-GW12 as well as both variants. CTLm obtained after V5 and stimulated with peptide Gag-GW12 actually recognized both variants more efficiently than Gag-GW12, and each of the variants were capable of stimulating CTLm, with the GSQKLSTGNCNW variant being most efficient (data not shown). It was concluded that even though viral variation through the Gag-GW12 epitope occurred, CTLm responses were not diminished and were eventually enhanced.
CTLm epitope mapping in A2147
The same panel of retroviral vectors was used to transduce target cells for initial identification of EIAV proteins with epitopes recognized by A2147 CTLm. Overlapping Gag p26 peptides (Zhang et al., 1998) were then used to sensitize target cells, and the optimal CTLm epitope (FVDRLLSQIK) spanned aa positions 288 to 297 in the EIAVWSU5 Gag protein and was designated Gag288-297 FK10 (Table 1). This epitope was restricted by the ELA-A4 haplotype.
Gag protein recognition by CTLm from A2147 was first characterized after V5 (before V6), but the exact time point at which the Gag-FK10 CTLm first appeared is not known. Given the EIAV variation observed previously in Gag CTLm epitopes, plasma EIAV RT-PCR products from several disease episodes (V1, V2, V3, V10, and V14) were sequenced. No aa variation was observed in the Gag-FK10 epitope recognized by CTLm from A2147 (data not shown).
CTLm analysis in nonprogressors A2150 and A2153
CTLm epitope mapping in A2150 and A2153
The same panel of retroviral vectors was used to transduce target cells for initial identification of EIAV proteins with epitopes recognized by A2150 and A2153 CTLm. For both horses, only EK targets expressing Rev were lysed by CTLm obtained shortly after V1. To identify the epitopes within Rev, overlapping peptides were synthesized and used to sensitize EK target cells.
CTLm from A2150 recognized two distinct epitopes within Rev. The first (QAEVLQERLEW) spanned aa positions 57–67 of EIAVWSU5 Rev and was designated Rev57-67QW11 (Table 1). The second (PRVLRPGDSKRR) spanned aa positions 120–131 of EIAVWSU5 Rev and was designated Rev120-131PR12 (Table 1). Both epitopes were restricted by the ELA-A1 haplotype. CTLm from A2153 also recognized two distinct Rev epitopes, and the minimal epitopes were designated Rev24-33IR10 (IPSQTCITRR), spanning aa positions 24–33 of EIAVWSU5 Rev, and Rev104-112GR9 (GGYQRAQER), spanning aa positions 104–112 of EIA-VWSU5 Rev (Table 1). The restricting MHC class I haplotypes were not determined for either of the two Rev-specific CTLm epitopes identified in A2153.
Variation in Rev epitopes recognized by A2150 and A2153
Because genetic and biological variation is known to occur in EIAV Rev during infection (Belshan et al., 2001), it was of interest to determine the plasma viral Rev sequences in A2150 and A2153. Since both horses had low or undetectable plasma viral load, nested RT-PCR was used to amplify EIAVWSU5 Rev sequences from plasma viral RNA in A2150 (DPI 568 plasma) and A2153 (DPI 575 plasma).
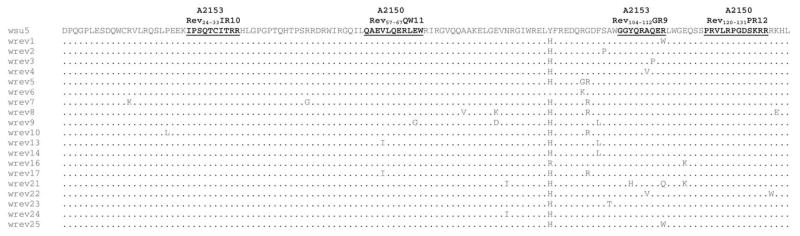
In A2150 DPI 568 plasma, 25 of 25 and 13 of 25 cloned Rev sequences were identical to the inoculum consensus sequence (Fig. 4) through the Rev-QW11 and Rev-PR12 CTLm epitopes, respectively (Fig. 5a). In A2153 DPI 575 plasma, 17 of 23 and 16 of 23 cloned Rev sequences were identical to the inoculum consensus sequence through the Rev-IR10 and Rev-GR9 epitopes, respectively (Fig. 5b).
Fig. 4.
Deduced amino acid sequences for Rev clones (aa 1–135) obtained from the EIAVWSU5 stock used for inoculation (GenBank Accession Nos. AY288606-AY288624). Only those aa different from EIAVWSU5 consensus sequence are shown. The epitopes recognized by CTLm from A2150 and A2153 are indicated. The Y → H substitution at position 91 was present in 17/19 clones and was therefore used as the inoculum consensus for A2150 and A2153 plasma Rev alignments.
Fig. 5.
(a) Deduced amino acid sequences for cloned EIAV Rev variants (aa 1–135) obtained from A2150 DPI 568 plasma (GenBank Accession Nos. AY288625-AY288649) and (b) A2153 DPI 575 plasma (GenBank Accession Nos. AY288650-AY288672) as compared to the EIAVWSU5 stock used for inoculation. Only those aa different from those in EIAVWSU5 are shown. The nuclear export signal (NES) and putative nuclear localization signal (NLS) (Fridell et al., 1993; Harris et al., 1998) are indicated, as are the CTLm epitopes. Sequences designated “b” were from clones derived from a second independent nested RT-PCR reaction.
Functional avidity for Env-RW12-, Gag-GW12-, and Gag-FK10-specific CTLm in progressors A2140 and A2147
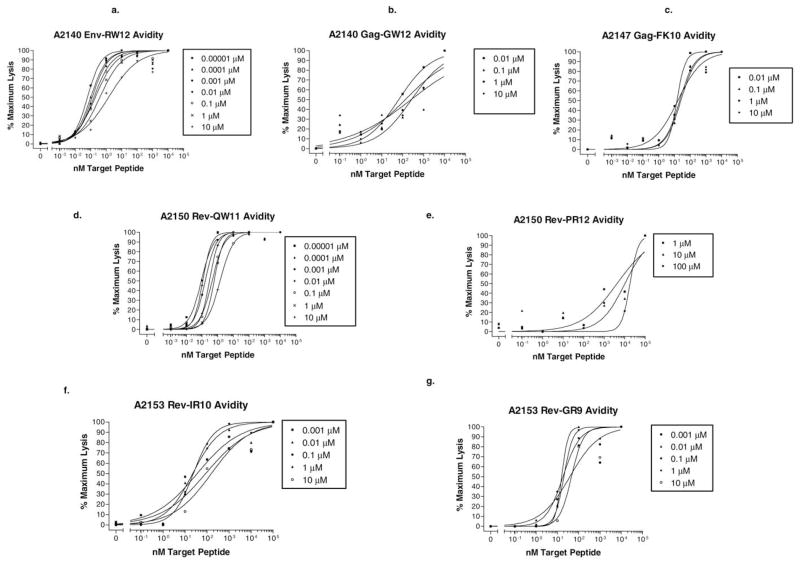
For all CTLm epitopes, the functional avidity values varied with the peptide concentration used for CTLm stimulation. The Env-RW12-specific CTLm exhibited high functional avidity values ranging from 0.06 to 1.5 nM (Fig. 6a), with a geometric mean of 0.21 nM representing the overall functional avidity. For Gag-GW12, CTLm functional avidity was moderate and ranged from 64.7 to 253 nM (Fig. 6b), with a geometric mean of 155 nM. The Gag-FK10-specific CTLm were also of moderate functional avidity, with values ranging from 14.1 to 25.3 nM (Fig. 6c) and a geometric mean of 18.7 nM.
Fig. 6.
(a–g) CTLm functional avidity. CTLm from each horse were stimulated for 7 days with the indicated peptides at 10 pM (0.00001 μM) to 100 μM and then added at an effector:target cell ratio of 20:1 to homologous EK targets pulsed with the same peptides at 10−1-105 nM. Functional avidity is the peptide concentration that results in 50% maximal target cell lysis and was not determined for CTLm stimulations resulting in maximum specific lysis less than 10% above the % specific lysis of homologous target cells not pulsed with peptide. Peptide concentrations used to stimulate each CTLm subline are shown in boxes.
Functional avidity for Rev-QW11-, Rev-PR12-, Rev-IR10-, and Rev-GR9-specific CTLm in nonprogressors A2150 and A2153
The Rev-QW11-specific CTLm exhibited high functional avidity values ranging from 0.09 to 1.4 nM (Fig. 6d), with a geometric mean of 0.29 nM. For Rev-PR12, CTLm functional avidity was low and ranged from 3177 to 8345 nM (Fig. 6e), with a geometric mean of 5658 nM. The Rev-IR10-specific CTLm were of moderate functional avidity, with values ranging from 35.4 to 156 nM (Fig. 6f) and a geometric mean of 47.8 nM. For Rev-GR9, CTLm functional avidity was also moderate and ranged from 38.2 to 45.5 nM (Fig. 6g), with a geometric mean of 25.1 nM.
Overall CTLm functional avidity comparisons
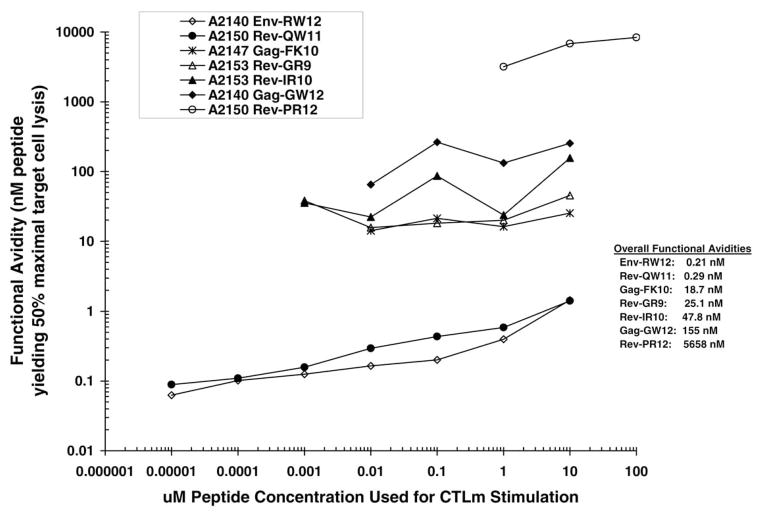
Analysis of functional avidity vs stimulation peptide concentration curves indicated that each peptide-specific CTLm line could be differentiated from the others at most peptide concentrations used for stimulation (Fig. 7). CTLm with high overall functional avidity (geometric mean) occurred in one of the nonprogressors (Rev-QW11 in A2150) and in one of the progressors (Env-RW12 in A2140), while CTLm with moderate-to-low overall functional avidity occurred in both groups (Table 1).
Fig. 7.
Overall functional avidities for each of the CTLm epitopes identified in A2140, A2147, A2150, and A2153. Overall functional avidity was defined as the geometric mean of the range of functional avidity values determined for each peptide-specific CTLm line stimulated for 7 days with peptide concentrations ranging from 10 pM to 100 μM. Functional avidity was not determined for CTLm stimulations resulting in maximum specific lysis less than 10% above the % specific lysis of homologous target cells not pulsed with peptide.
Discussion
Horses infected with EIAV provide a unique outbred model for the study of immunological control mechanisms in a naturally occurring lentiviral disease. Although there is strong evidence that CTL are a particularly important control mechanism, lentiviruses persist in the face of CTL. In EIAV, CTL occur in association with the control of the initial episode of plasma viremia and clinical disease, yet clinical disease recurs during the first year of infection. To further define the role of CTL in EIAV control, a detailed analysis of EIAV-specific CTLm responses was done with respect to epitopes recognized, quality (as measured by functional avidity), and viral evasion, in horses with progressive and nonprogressive disease. Our results indicated that CTLm functional avidity alone did not determine disease status, since high-avidity CTLm were found in the progressor horses as well as in the nonprogressor horses. However, CTLm from both nonprogressors targeted multiple epitopes in Rev, an EIAV protein essential for viral replication. In contrast, high-avidity CTLm in one of the progressors were directed against an epitope within the highly variable envelope gp90 protein, and our data provided the initial demonstration that EIAV variation of a CTL epitope resulted in loss of CTL recognition.
The avidity of the interaction between CTL and their target cells determines the efficiency with which CTL recognize and kill their specific targets (Gallimore et al., 1998a). In comparison to low-avidity CTL, high-avidity CTL of the same specificity recognize target cells expressing lower antigen density and initiate lysis of targets more rapidly at any given antigen density (Derby et al., 2001a). Adoptive transfer experiments in mice infected with LCMV as well as in mice infected with vaccinia virus constructs expressing HIV-1 proteins indicate that the in vivo protective effects of CTL correlate with their avidity (Alexander-Miller et al., 1996; Derby et al., 2001a; Gallimore et al., 1998a). Further studies in mice show that high-avidity CTL are preferentially expanded during the active phase of viral infection, but that low-avidity CTL are always more abundant (Alexander-Miller, 2000). Surprisingly, a selective loss of high-avidity CTL occurs in the CTL memory pool, and thus, the ratio of low:high avidity CTLm may not reflect that of CTLe found during the acute response (Alexander-Miller, 2000). However, the CTL functional avidity established at the peak of the effector response can be maintained throughout the memory phase (Slifka and Whitton, 2001). In the present study, functional avidity was determined using the CTL memory pool, and it is possible that the avidity of CTLe during active infection may have actually been higher. Irregardless, it is likely that the relative pattern of observed avidities of each of the CTLm lines would be unchanged.
Although functional avidities of CTL specific for different peptide epitopes have been compared (O’Connor et al., 2002), antigen densities on antigen-presenting cells (APC) or target cells are relative and vary for different peptides depending on MHC binding affinity (Alexander-Miller et al., 1996). In the present study, overall functional avidity was determined for each individual peptide epitope-specific CTLm line, over a range of stimulation peptide concentrations. A range of peptide concentrations was used to stimulate each CTLm line because a single peptide concentration may not have been optimal for every epitope, and alternatively, may have been supra-optimal for others, leading to apoptosis of high-avidity CTLm (Derby et al., 2001b).
Antiviral selection pressure imposed by CTL resulting in CTL escape mutations is well described for HIV-1 and SIV (Allen et al., 2000; O’Connor et al., 2002; Price et al., 1997; Borrow et al., 1997; Chen et al., 2000; Goulder et al., 1997a,b, 2001; Harrer et al., 1998; Kelleher et al., 2001). In HIV-1, a narrow CTL response characterized by an immunodominant clone is more likely to result in antigenic escape and loss of viral control than a broad CTL response directed against multiple epitopes (Wodarz and Nowak, 2000). Moreover, high-avidity CTL are more likely than low-avidity CTL to rapidly select for escape variants following acute SIV infection in rhesus monkeys (O’Connor et al., 2002). Based on the high level of killing and high avidity displayed by the Env-RW12 CTL in A2140, the Env-RW12 CTL response was probably immunodominant. Given the highly variable nature of the gp90 region targeted by these CTL, it is not surprising that the Env-HW12v CTL escape variant arose.
In general, variation in EIAV gp90 has been attributed to neutralizing antibody selection pressure (Ball et al., 1992; Hussain et al., 1987, 1988; Leroux et al., 2001). Because the Env-RW12 CTL epitope occurred within the putative PND, it is possible that Env-HW12v was, in part, a neutralizing antibody escape variant. Two contiguous linear murine monoclonal neutralizing antibody epitopes have been mapped to the EIAV PND (Ball et al., 1992), and they bisect the Env-RW12 CTL epitope. Although serum antibodies from EIAV-infected horses bind these same epitopes (Ball et al., 1992; Hussain et al., 1988), there is no evidence that these antibodies neutralize EIAV infectivity, and the peptide epitopes recognized by EIAV-neutralizing antibodies from EIAV-infected horses are unknown. Moreover, neutralizing antibodies may not be critical in the selection of EIAV antigenic variants (Rwambo et al., 1990). Similarly, neutralizing antibodies are not likely involved in the genesis of gp90 variants in the early stages of EIAV infection, and regions other than the PND probably determine envelope neutralizing properties in the later stages (Leroux et al., 1997). In the present study, peptides containing the Env-RW12 CTL epitope as well as the two known epitopes recognized by murine monoclonal neutralizing antibodies failed to block neutralizing antibody activity in A2140 plasma. Moreover, a probable MHC class I anchor residue changed in the Env-RW12 CTL epitope before the appearance of detectable neutralizing antibody. These observations suggest that the CTL epitope variant Env-HW12v was the result of CTL selection pressure.
The variation observed in the Gag-GW12 CTLm epitope was interesting in that CTLm recognition of one of the variants was more efficient than the original epitope. Natural variation occurs in EIAV Gag proteins containing CTL epitopes, including p15 and p26 (Zhang et al., 1999), and it is possible that these variants are the result of CTL selection pressure. The Gag-GW12 variants observed in the present study could have originated secondary to CTL selection pressure, but variant-specific CTL were subsequently induced. Despite CTL recognition of these variants, disease episodes were not controlled in A2140.
Horse A2147 had progressive disease despite moderate avidity CTLm directed against a nonvariable epitope in Gag p26. The Gag-FK10 CTLm were the only CTL identified in A2147 and it is possible that effective viral control requires higher numbers of CTL, or a CTL response directed against epitopes in multiple viral proteins (Harrer et al., 1996). Lymphocyte proliferation decreases during EIAV viremia (Newman et al., 1991), and a reduced helper T lymphocyte response might have contributed to the inability of the Gag-FK10 CTL to control viral replication. Additionally, CTL responses are inhibited by downregulation of MHC class I expression on APC during HIV-1 infection (Andrieu et al., 2001), and there is preliminary information that this occurs during EIAV infection in vitro (W. Maury, unpublished observation). In A2140, the Gag-GW12 peptide appeared to be subdominant to the Env-RW12 peptide, based on the relative strengths of the corresponding CTL responses (as measured by % specific lysis). Subdominant epitopes can be characterized by inefficient MHC class I/peptide processing, unstable MHC class I/peptide binding, and a CTL response that is not protective (Gallimore et al., 1998b). It is therefore possible that the CTL response in A2147 was ineffective because a subdominant Gag epitope was exclusively targeted by CTL with moderate avidity.
Both nonprogressors had high- and low-avidity CTLm (A2150) or moderate-avidity CTLm (A2153) directed against multiple epitopes in Rev. Within infected cells, Rev mediates the export of incompletely spliced viral RNA from the nucleus and is absolutely required for expression of viral structural genes and production of new virus (Belshan et al., 2001). Discrete functional domains within Rev important for nuclear localization and nuclear export include the nuclear localization signal (NLS) (Harris et al., 1998) and the nuclear export signal (NES) (Fridell et al., 1993). Kinetic studies using HIV-1 Rev-specific CTL clones have shown that cytolysis occurs well before peak virus production and that selection pressure exerted by these CTL in vitro can result in a virus population with mutations in the Rev epitope, abrogating the CTL killing (Van Baalen et al., 1998). It has been suggested that a mutation in the HIV-1 Rev essential activation domain (analogous to the NES of EIAV Rev) may have contributed to the lack of disease progression in a long-term asymptomatic patient (Iversen et al., 1995). Moreover, Rev-specific CTL have been inversely correlated with rapid progression to AIDS (Van Baalen et al., 1997). Rev variants arise during EIAV infection (Alexandersen and Carpenter, 1991; Belshan et al., 1998, 2001; Leroux et al., 1997), and in one EIAV-infected pony a statistical correlation exists between Rev activity and different stages of clinical disease (Belshan et al., 2001).
Analysis of deduced aa sequences from DPI 568 plasma Rev clones indicated that no variation occurred through the Rev-QW11 epitope recognized by CTLm from A2150, while 12 of 25 clones contained two aa substitutions in the carboxyl-terminus of epitope Rev-PR12 (also recognized by A2150 CTLm). It is probable that the high-avidity CTLm directed against the nonvariable Rev-QW11 epitope played a role in limiting viral load and clinical disease in A2150. In addition, the two aa substitutions within Rev-PR12 involve the amino-terminal portion of the EIAV Rev NLS, and it is possible that these variants had reduced replication efficiency. These variants were not present among the clones analyzed from the virus stock used for inoculation, and it is possible that the Rev-PR12-specific CTL were involved in their selection.
A low percentage of plasma Rev clones from A2153 DPI 575 plasma had aa substitutions within the Rev-IR10 (6 of 23 clones; 26%) and Rev-GR9 (7 of 23 clones; 30%) CTL epitopes. The lack of significant CTL epitope variation, coupled with the fact that the EIAV Rev NES was partially contained within the Rev-IR10 epitope, may have contributed to the low viral load and lack of progressive disease in A2153.
In summary, of four EIAVWSU5-infected horses, two developed progressive disease while two maintained low viral loads and had nonprogressive disease. In one progressor horse, CTLm with high functional avidity directed against a variable EIAV epitope likely led to CTL escape, while moderate-avidity CTLm directed against a nonvariable epitope were not protective in the other progressor horse. In the nonprogressor horses, CTLm with various functional avidities were directed against several different epitopes within EIAV Rev, a protein essential for viral replication. Importantly, high-avidity CTLm targeted a non-variable Rev epitope in one of the nonprogressor horses, while moderate-avidity CTLm were directed against two Rev epitopes in the other nonprogressor horse. A moderate-to high-avidity CTL response directed against multiple non-variable viral peptides which are expressed early in infection and are required for viral replication is likely to be critical for lentiviral control. This study provided evidence that in some EIAV-infected horses, the viral proteins targeted by CTLm are as critical as functional avidity in the control of viral load and clinical disease.
Materials and methods
Horses, EIAV infection, clinical evaluation, and sample collection
Four yearling Arabian horses (A2140, A2147, A2150, and A2153) were inoculated intravenously with 107 50% tissue culture infective doses (TCID50) of the WSU5 strain of EIAV (O’Rourke et al., 1988). The equine leukocyte alloantigen (ELA)-A types of the four horses (Table 2) were determined serologically (Bailey, 1980). The ELA-A locus is the best-defined polymorphic MHC class I locus in the horse (Antczak et al., 1986), but other polymorphic loci likely exist (Ellis et al., 1995). Rectal temperature and clinical status were recorded daily, and for the first month postinfection, platelet counts were made on whole blood collected every 2 days and then weekly thereafter. For determination of viral load, plasmas were collected and stored at −80°C every 2 days for the first month postinfection and then weekly thereafter. Clinical EIAV disease episodes were defined when thrombocytopenia (platelets < 150,000/μl blood) occurred with or without fever (rectal temperature ≥38.6°C), concurrent with detectable plasma viral RNA.
Table 2.
ELA-A class I serotypes of horses A2140, A2147, A2150, and A2153
The second allele could not be determined serologically.
PBMC stimulations
PBMC were isolated (Zhang et al., 1998) from each horse at various time points throughout the experiment and stimulated in vitro for 7 days using autologous EIAVWSU5-infected or peptide-pulsed monocytes (McGuire et al., 2000; Zhang et al., 1998). Briefly, EIAVWSU5 in 17% fetal bovine serum was added to the isolated PBMC in amounts equal to a multiplicity of monocyte infection of 1, assuming that PBMC were 5% monocytes. When PBMC were pulsed with peptides, a peptide concentration of 40 μM was used. Virus (or peptide) and PBMC were incubated for 2 h at 37°C with occasional mixing before centrifugation at 250 g for 10 min. PBMC were then resuspended at 5 × 106/ml RPMI 1640 medium with 10% fetal bovine serum (FBS), 20 mM HEPES, 10 μg/ml gentamicin, and 10 μM 2-mercaptoethanol. Tissue culture flasks (175 cm2) were seeded with 40 ml/flask and incubated at 37° C with 5% CO2 for 7 days at 37°C. PBMC were also isolated, suspended in FBS with 10% DMSO, and frozen in liquid nitrogen for use in later assays. When frozen PBMC were used, stimulations were performed as above except that 10 U/ml recombinant human IL-2 and irradiated autologous PBMC stimulators were added (Zhang et al., 1998).
CTLm epitope mapping
To determine the EIAV proteins with CTL epitopes, CTL activity in the stimulated PBMC was first determined in 51Cr release CTL assays using equine kidney target cells transduced with retroviral vectors containing inserts that span the entire EIAVWSU5 genome (Lonning et al., 1999; McGuire et al., 2000; Zhang et al., 1998). Transduced EK cells were selected with G418 sulfate before use as target cells and then seeded into 96-well plates. Target cells were labeled with 50 μl/well DMEM with 5% calf serum and 25 μCi 51Cr/ml for 2 h at 37°C. After washing the plates, the stimulated PBMC effectors were added at effector:target cell ratios of 20:1 and incubated for 17 h, and 100 μl supernatant was removed to determine 51Cr release. Percent specific lysis was calculated using the following formula: percent specific lysis = [(E − S)/(M − S)] × 100, where E = the mean of three or six test wells, S = the mean spontaneous release from three or six target cell wells without effector cells, and M = the mean maximal release from three or six target cell wells with 2% Triton X-100 (McGuire et al., 1994). The formula used to calculate standard error of the percent specific lysis accounts for the variability of E, S, and M. After regions of the viral genome containing CTL epitopes were known, peptides 20–25 aa in length and overlapping by 8–11 aa, corresponding to the epitope-containing regions, were synthesized by the Laboratory for Biotechnology and Bioanalysis (Washington State University, Pullman, WA). Target cells were sensitized by pulsing normal EK cells with the synthetic peptides at 105 nM for 2 h at 37°C in 96-well plates containing 50 μl/well DMEM with 5% calf serum and 25 μCi 51Cr/ml. Minimal epitopes were defined by sensitizing target cells with truncated overlapping peptides based on the patterns of recognition of the longer peptides.
CTLm functional avidity
PBMC were stimulated as above, except that PBMC were seeded into 24-well plates at 2 × 106/ml. Tenfold dilutions of peptides (ranging from 10 μto 10 pM) corresponding to the minimal CTL epitopes were used to stimulate CTLm. CTL assays were then performed using target cells pulsed with 10-fold dilutions of peptides ranging from 100 μM to 100 pM. CTLm functional avidity was defined as the peptide concentration that resulted in 50% maximal target cell specific lysis (Alexander-Miller et al., 1996; Derby et al., 2001a) and was used to describe the effectiveness of CTLm-mediated killing, encompassing TCR affinity and signaling, as well as the efficiency of MHC-peptide binding. Since functional avidity is dependent on the peptide concentration used for stimulation (Alexander-Miller et al., 1996), and because CTLm were stimulated with peptide prior to functional avidity determination, each epitope-specific CTLm line was stimulated with several different peptide concentrations as described above. This resulted in a range of functional avidities for each epitope-specific CTLm line. Overall functional avidity was calculated for each epitope-specific CTLm line, by determining the geometric mean of the functional avidity values of each of the corresponding individual sublines generated by stimulation with decreasing concentrations of peptide. The functional avidity of an individual subline was not determined if the maximal specific lysis of peptide-pulsed target cells was less than 10% above the specific lysis of target cells not pulsed with peptide. For graphical comparisons between CTLm of different peptide epitope specificities, CTLm functional avidity curves were generated for each CTLm line by plotting the functional avidity values against the corresponding peptide concentrations used for CTLm stimulation. A CTLm line was considered to be of high avidity if its overall avidity value was at least 10,000 times lower than that of the CTLm line with the lowest overall avidity.
Plasma neutralizing antibody activity in A2140
Neutralizing antibody activity in frozen heparinized plasma from A2140 was determined using a virus reduction method (Mealey et al., 2001). Briefly, 300 μl heparinized plasma was heat inactivated by incubation at 56°C for 30 min, then mixed with 300 μl EIAVWSU5 (approximate titer 104 TCID50/ml) stock, and incubated for 1 h at 37°C. The same was performed using negative control plasma from four horses not infected with EIAV. For each plasma–virus mixture, threefold serial dilutions were made, and titration was performed in EK cell culture (O’Rourke et al., 1988). Percentage virus reduction was calculated using the following formula: % virus reduction = [(mean negative control plasma titer − test plasma titer)/(mean negative control plasma titer)] ×100.
Viral RNA purification
Viral RNA was isolated from 140 μl frozen EDTA plasma using a QIAamp Viral RNA kit (Qiagen Inc., Chats-worth, CA) (Leutenegger et al., 2001) and DNase I treated on the spin column (DNase 1 set; Qiagen), eluted in 60 μl nuclease-free water, and frozen at −20°C. Only heparinized plasma was available for horse A2140 on several of the days assayed. For these samples, viral RNA was extracted as above, and then the eluates were treated with heparinase as described (Lin et al., 1997).
Real-time quantitative RT-PCR
Real-time RT-PCR was used to determine plasma viral load in each of the four horses by amplification of a 127-bp segment (nt 2984–3110) of the EIAVWSU5 (GenBank Accession No. AF247394) reverse transcriptase gene. The following oligonucleotides were used: forward primer, 5′-AGACATGGTAAAGAATCCAACCCTTAATG-3′; reverse primer, 5′-TCTAAACATCCCTTAGTATGAGCT-GCTATGTG-3′; and TaqMan (Applied Biosystems, Foster, CA) probe, 5′-TACTGTCAACCCTGGGACCCCTGAG-CTCAT-3′ (nt 3045–3074). The fluorescence reporter dye at the 5′ end of the TaqMan probe was FAM (6-carboxyfluorescein) and the quencher dye at the 3′ end was 6-carboxytetramethyl-rhodamine (TAMRA). An RNA standard template was made by cloning EIAVWSU5 nt 2078–3152 (which included the reverse transcriptase gene) into pCR2.1 (designated pWRT8; Invitrogen, Carlsbad, CA), followed by in vitro transcription of the HindIII-linearized pWRT8 using the RiboMax Large-Scale RNA Production System-T7 (Promega, Madison, WI). After transcription, RNA was DNase I treated and purified using an RNeasy Mini Kit (Qiagen). To quantitate the transcribed RNA template, an aliquot of the transcript eluate, along with serial dilutions of a known RNA standard (RNA Quantitation Kit; Sigma, St. Louis, MO), were analyzed by electrophoresis through a 1% agarose gel containing 0.15 μg/ml ethidium bromide. Before loading the gel, the RNA in each of the standard dilutions was denatured by heating at 65°C for 10 min in a mixture of 1.3 M formaldehyde, 11.8 M deionized formamide, 21.4 mM HEPES, and 1.1 mM EDTA disodium. The integrated density value of the RNA bands in the gel was determined using an AlphaImager 2000 digital imaging system (Alpha Innotech Corp., San Leandro, CA). The equation of the best-fit regression line for the integrated density values of the known RNA standard dilutions was used to calculate the RNA concentration in the transcript eluate. The RNA transcripts were serially diluted to yield 106, 105, 104, 103, 102, 101, and 100 RNA copies/10 μl in nuclease-free water and stored in 25 μl aliquots at −80°C. Real-time RT-PCR was performed using the Platinum Quantitative RT-PCR ThermoScript One-Step System (Life Technologies, Gaithersburg, MD) in 25 μl reactions containing 12.5 μl 2× ThermoScript Reaction Mix, 0.75 μl forward primer (25 μM), 0.75 μl reverse primer (25 μM), 0.5 μl TaqMan probe (10 μM), 0.5 μl ThermoScript Plus RT/Platinum Taq Enzyme Mix, and 10 μl RNA sample or standard. Cycling conditions were 15 min at 50°C, 3.5 min at 95°C, followed by 50 cycles at 95°C for 15 s and 60°C for 1 min. Reactions were carried out in duplicate in an iCycler 96 Thermal Cycler using the iCycler iQ Real-Time PCR Detection System and software (Bio-Rad Laboratories, Hercules, CA). Real-time fluorescence measurements were performed and a threshold cycle (CT) value for each sample was calculated by determining the point at which the fluorescence exceeded a threshold limit (10 times the standard deviation of the baseline as determined between cycles 3 and 15). The CT value was proportional to the log of the starting amount of nucleic acid and reflected the earliest point in the amplification curve that could be used for quantitation of the template input (Heid et al., 1996; Leutenegger et al., 2001). The sensitivity of the real-time RT-PCR was determined using a dilution series of the standard RNA transcripts as above. The dilution containing 1 RNA copy was positive in 40% (4/10) of the replicates, while the dilutions containing 10 RNA copies or more were positive in 100% (10/10) of the replicates. The reliable detection limit, therefore, was somewhere between 43 and 428 RNA copies/ml when 140 μl plasma for the RNA extraction and 10 μl of the 60 μl eluate were used.
RT-PCR amplification of plasma EIAV genome regions containing CTLm epitopes
RT-PCR was performed using a SuperScript One-Step RT-PCR kit (Life Technologies). Initial RT-PCR was performed using 5 μl template RNA eluate in 50 μl total reaction volume. When nested PCR was performed, 5 μl from the initial reaction was used in a 100 μl total reaction volume. The epitope Env-RW12 recognized by CTLm from A2140 was contained within nt 5868–6164 and the following primers and reaction conditions were used: outside forward primer (5322F), 5′-ATGGTCAGCATCGCATTC-3′; inside forward primer (5616F), 5′-CAACATTATAT-AGGGTTGGTAGC-3′; reverse primer (6164R), 5′-AG-CAATCCCTTTCTCCTGT-3′; initial RT-PCR, 30 min at 45°C, 2 min at 94°C, followed by 40 cycles of 30 s at 94°C, 30 s at 59°C, 1 min at 72°C, and then 7 min at 72°C; nested PCR, 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 59°C, 45 s at 72°C, and then 7 min at 72°. The epitope Gag-GW12 recognized by CTLm from A2140 was contained within nt 406–694 and the following primers and reaction conditions were used: forward primer (406F), 5′-GACAGCAGAGGAGAACTTAC-3′; reverse primer (694R), 5′-CCTCTCTTTCTTGTCCTG-3′; 30 min at 45°C, 2 min at 94°C, followed by 40 cycles of 30 s at 94°C, 30 s at 54°C, 30 s at 72°C, and then 7 min at 72°C. The epitope Gag-FK10 recognized by CTLm from A2147 was contained within nt 1141–1471 and the following primers and reaction conditions were used: forward primer (1141F), 5′-CCTAT-TCCCATGACAGCAA-3′; reverse primer (1471R), 5′-CCTCTGGTCTTAAATGTCTCATA-3′; 40 min at 45°C, 2 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 58°C, 30 s at 72°C, and then 7 min at 72°C. Finally, the Rev epitopes recognized by CTLm from A2150 (Rev-QW11 and Rev-PR12) and A2153 (Rev-IR10 and Rev-GR9) were contained within nt 7143–7728 and the following primers and reaction conditions were used: outside forward primer (7143F), 5′-GGAGTCATATTGGAAATTGGA-3′; inside forward primer (7175F), 5′-GGGAGCTTCCATTATA-AAATATAT-3′; reverse primer (7728R), 5′-CTAGTC-CCCAAAATAGCCAT-3′; initial RT-PCR, 40 min at 45°C, 2 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 58°C, 45 s at 72°C, and then 7 min at 72°C; nested PCR, 2 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 58°C, 45 s at 72°C, and then 7 min at 72°C.
Cloning and sequencing of RT-PCR products
Amplified products from multiple RT-PCR reactions were purified from agarose gels using a QIAquick Gel Purification Kit (Qiagen) and cloned into the pCR2.1 TA cloning vector (Invitrogen) (Zhang et al., 1999). A QIAprep Spin Miniprep Kit (Qiagen) was used to extract plasmid DNA from each of the clones, and the nucleotide sequences determined by the Laboratory for Biotechnology and Bio-analysis (Washington State University, Pullman, WA) using an ABI Prism 377 (Applied Biosystems) automated DNA sequencer. Deduced aa sequences from each of the EIAV clones obtained were compared to the EIAVWSU5 consensus sequence using ClustalW and Boxshade programs.
Acknowledgments
The authors appreciate the technical assistance of Emma Karel, Andi Parsons, and Sarah Pownder. This research was supported by U.S. Public Health Service, National Institutes of Health Grants AI01575, AI24291, and Morris Animal Foundation Grant D01EQ-09.
References
- Alexander-Miller MA. Differential expansion and survival of high and low avidity cytotoxic T cell populations during the immune response to a viral infection. Cell Immunol. 2000;201:58– 62. doi: 10.1006/cimm.1999.1632. [DOI] [PubMed] [Google Scholar]
- Alexander-Miller MA, Leggatt GR, Berzofsky JA. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc Natl Acad Sci USA. 1996;93:4102–4107. doi: 10.1073/pnas.93.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersen S, Carpenter S. Characterization of variable regions in the envelope and S3 open reading frame of equine infectious anemia virus. J Virol. 1991;65:4255– 4262. doi: 10.1128/jvi.65.8.4255-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, O’Connor DH, Jing P, Dzuris JL, Mothe BR, Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, Kunstman KJ, Wang X, Allison DB, Hughes AL, Desrosiers RC, Altman JD, Wolinsky SM, Sette A, Watkins DI. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- Andrieu M, Chassin D, Desoutter JF, Bouchaert I, Baillet M, Hanau D, Guillet JG, Hosmalin A. Short communication: down-regulation of major histocompatibility class I on human dendritic cells by HIV Nef impairs antigen presentation to HIV-specific CD8+ T lymphocytes. AIDS Res Hum Retroviruses. 2001;17:1365–1370. doi: 10.1089/08892220152596623. [DOI] [PubMed] [Google Scholar]
- Antczak DF, Bailey E, Barger B, Guerin G, Lazary S, McClure J, Mottironi VD, Symons R, Templeton J, Varewyck H. Joint report of the Third International Workshop on Lymphocyte Alloantigens of the Horse, Kennett Square, Pennsylvania, 25–27 April 1984. Anim Genet. 1986;17:363–373. doi: 10.1111/j.1365-2052.1986.tb00730.x. [DOI] [PubMed] [Google Scholar]
- Bailey E. Identification and genetics of horse lymphocyte alloantigens. Immunogenetics. 1980;11:499–506. doi: 10.1007/BF01567818. [DOI] [PubMed] [Google Scholar]
- Ball JM, Rushlow KE, Issel CJ, Montelaro RC. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J Virol. 1992;66:732–742. doi: 10.1128/jvi.66.2.732-742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Craiu A, Santra S, Egan MA, Schmitz JE, Kuroda MJ, Fu TM, Nam JH, Wyatt LS, Lifton MA, Krivulka GR, Nickerson CE, Lord CI, Moss B, Lewis MG, Hirsch VM, Shiver JW, Letvin NL. Elicitation of high-frequency cytotoxic T-lymphocyte responses against both dominant and subdominant simian-human immunodeficiency virus epitopes by DNA vaccination of rhesus monkeys. J Virol. 2001a;75:2462–2467. doi: 10.1128/JVI.75.5.2462-2467.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Santra S, Kuroda MJ, Schmitz JE, Plishka R, Buckler-White A, Gaitan AE, Zin R, Nam JH, Wyatt LS, Lifton MA, Nickerson CE, Moss B, Montefiori DC, Hirsch VM, Letvin NL. Reduction of simian-human immunodeficiency virus 89.6P viremia in rhesus monkeys by recombinant modified vaccinia virus Ankara vaccination. J Virol. 2001b;75:5151–5158. doi: 10.1128/JVI.75.11.5151-5158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Santra S, Schmitz JE, Kuroda MJ, Fu TM, Wagner W, Bilska M, Craiu A, Zheng XX, Krivulka GR, Beaudry K, Lifton MA, Nickerson CE, Trigona WL, Punt K, Freed DC, Guan L, Dubey S, Casimiro D, Simon A, Davies ME, Chastain M, Strom TB, Gelman RS, Montefiori DC, Lewis MG, Emini EA, Shiver JW, Letvin NL. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486– 492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- Belshan M, Baccam P, Oaks JL, Sponseller BA, Murphy SC, Cornette J, Carpenter S. Genetic and biological variation in equine infectious anemia virus Rev correlates with variable stages of clinical disease in an experimentally infected pony. Virology. 2001;279:185–200. doi: 10.1006/viro.2000.0696. [DOI] [PubMed] [Google Scholar]
- Belshan M, Harris ME, Shoemaker AE, Hope TJ, Carpenter S. Biological characterization of Rev variation in equine infectious anemia virus. J Virol. 1998;72:4421– 4426. doi: 10.1128/jvi.72.5.4421-4426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103– 6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- Brodie SJ, Lewinsohn DA, Patterson BK, Jiyamapa D, Krieger J, Corey L, Greenberg PD, Riddell SR. In vivo migration and function of transferred HIV-1-specific cytotoxic T cells. Nat Med. 1999;5:34–41. doi: 10.1038/4716. [DOI] [PubMed] [Google Scholar]
- Carpenter S, Evans LH, Sevoian M, Chesebro B. Role of the host immune response in selection of equine infectious anemia virus variants. J Virol. 1987;61:3783–3789. doi: 10.1128/jvi.61.12.3783-3789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheevers WP, McGuire TC. Equine infectious anemia virus: immunopathogenesis and persistence. Rev Infect Dis. 1985;7:83– 88. doi: 10.1093/clinids/7.1.83. [DOI] [PubMed] [Google Scholar]
- Chen ZW, Craiu A, Shen L, Kuroda MJ, Iroku UC, Watkins DI, Voss G, Letvin NL. Simian immunodeficiency virus evades a dominant epitope-specific cytotoxic T lymphocyte response through a mutation resulting in the accelerated dissociation of viral peptide and MHC class I. J Immunol. 2000;164:6474–6479. doi: 10.4049/jimmunol.164.12.6474. [DOI] [PubMed] [Google Scholar]
- Crawford TB, Wardrop KJ, Tornquist SJ, Reilich E, Meyers KM, McGuire TC. A primary production deficit in the thrombocytopenia of equine infectious anemia. J Virol. 1996;70:7842–7850. doi: 10.1128/jvi.70.11.7842-7850.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby M, Alexander-Miller M, Tse R, Berzofsky J. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J Immunol. 2001a;166:1690–1697. doi: 10.4049/jimmunol.166.3.1690. [DOI] [PubMed] [Google Scholar]
- Derby MA, Snyder JT, Tse R, Alexander-Miller MA, Berzofsky JA. An abrupt and concordant initiation of apoptosis: antigen-dependent death of CD8(+) CTL. Eur J Immunol. 2001b;31:2951–2959. doi: 10.1002/1521-4141(2001010)31:10<2951::aid-immu2951>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Ellis SA, Martin AJ, Holmes EC, Morrison WI. At least four MHC class I genes are transcribed in the horse: phylogenetic analysis suggests an unusual evolutionary history for the MHC in this species. Eur J Immunogenet. 1995;22:249–260. doi: 10.1111/j.1744-313x.1995.tb00239.x. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Partin KM, Carpenter S, Cullen BR. Identification of the activation domain of equine infectious anemia virus rev. J Virol. 1993;67:7317–7323. doi: 10.1128/jvi.67.12.7317-7323.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore A, Dumrese T, Hengartner H, Zinkernagel RM, Rammensee HG. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J Exp Med. 1998a;187:1647–1657. doi: 10.1084/jem.187.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore A, Hombach J, Dumrese T, Rammensee HG, Zinkernagel RM, Hengartner H. A protective cytotoxic T cell response to a subdominant epitope is influenced by the stability of the MHC class I/peptide complex and the overall spectrum of viral peptides generated within infected cells. Eur J Immunol. 1998b;28:3301–3311. doi: 10.1002/(SICI)1521-4141(199810)28:10<3301::AID-IMMU3301>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Brander C, Tang Y, Tremblay C, Colbert RA, Addo MM, Rosenberg ES, Nguyen T, Allen R, Trocha A, Altfeld M, He S, Bunce M, Funkhouser R, Pelton SI, Burchett SK, McIntosh K, Korber BT, Walker BD. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997a;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Sewell AK, Lalloo DG, Price DA, Whelan JA, Evans J, Taylor GP, Luzzi G, Giangrande P, Phillips RE, McMichael AJ. Patterns of immunodominance in HIV-1-specific cytotoxic T lymphocyte responses in two human histocompatibility leukocyte antigens (HLA)-identical siblings with HLA-A*0201 are influenced by epitope mutation. J Exp Med. 1997b;185:1423–1433. doi: 10.1084/jem.185.8.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SA, Cook SJ, Lichtenstein DL, Issel CJ, Montelaro RC. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrer T, Harrer E, Barbosa P, Kaufmann F, Wagner R, Bruggemann S, Kalden JR, Feinberg M, Johnson RP, Buchbinder S, Walker BD. Recognition of two overlapping CTL epitopes in HIV-1 p17 by CTL from a long-term nonprogressing HIV-1-infected individual. J Immunol. 1998;161:4875– 4881. [PubMed] [Google Scholar]
- Harrer T, Harrer E, Kalams SA, Barbosa P, Trocha A, Johnson RP, Elbeik T, Feinberg MB, Buchbinder SP, Walker BD. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection. Breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. J Immunol. 1996;156:2616–2623. [PubMed] [Google Scholar]
- Harris ME, Gontarek RR, Derse D, Hope TJ. Differential requirements for alternative splicing and nuclear export functions of equine infectious anemia virus Rev protein. Mol Cell Biol. 1998;18:3889–3899. doi: 10.1128/mcb.18.7.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Hussain KA, Issel CJ, Schnorr KL, Rwambo PM, Montelaro RC. Antigenic analysis of equine infectious anemia virus (EIAV) variants by using monoclonal antibodies: epitopes of glycoprotein gp90 of EIAV stimulate neutralizing antibodies. J Virol. 1987;61:2956–2961. doi: 10.1128/jvi.61.10.2956-2961.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain KA, Issel CJ, Schnorr KL, Rwambo PM, West M, Montelaro RC. Antigenic mapping of the envelope proteins of equine infectious anemia virus: Identification of a neutralization domain and a conserved region on glycoprotein 90. Arch Virol. 1988;98:213–224. doi: 10.1007/BF01322170. [DOI] [PubMed] [Google Scholar]
- Iversen AK, Shpaer EG, Rodrigo AG, Hirsch MS, Walker BD, Sheppard HW, Merigan TC, Mullins JI. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J Virol. 1995;69:5743–5753. doi: 10.1128/jvi.69.9.5743-5753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig S, Conley AJ, Brewah YA, Jones GM, Leath S, Boots LJ, Davey V, Pantaleo G, Demarest JF, Carter C, et al. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat Med. 1995;1:330–336. doi: 10.1038/nm0495-330. [DOI] [PubMed] [Google Scholar]
- Kono Y, Kobayashi K, Fukunaga Y. Antigenic drift of equine infectious anemia virus in chronically infected horses. Arch Gesamte Virusforsch. 1973;41:1–10. doi: 10.1007/BF01249923. [DOI] [PubMed] [Google Scholar]
- Leroux C, Craigo JK, Issel CJ, Montelaro RC. Equine infectious anemia virus genomic evolution in progressor and nonprogressor ponies. J Virol. 2001;75:4570– 4583. doi: 10.1128/JVI.75.10.4570-4583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux C, Issel CJ, Montelaro RC. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J Virol. 1997;71:9627–9639. doi: 10.1128/jvi.71.12.9627-9639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutenegger CM, Higgins J, Matthews TB, Tarantal AF, Luciw PA, Pedersen NC, North TW. Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res Hum Retroviruses. 2001;17:243–251. doi: 10.1089/088922201750063160. [DOI] [PubMed] [Google Scholar]
- Lin HJ, Tanwandee T, Hollinger FB. Improved methods for quantification of human immunodeficiency virus type 1 RNA and hepatitis C virus RNA in blood using spin column technology and chemiluminescent assays of PCR products. J Med Virol. 1997;51:56– 63. [PubMed] [Google Scholar]
- Lonning SM, Zhang W, Leib SR, McGuire TC. Detection and induction of equine infectious anemia virus-specific cytotoxic T-lymphocyte responses by use of recombinant retroviral vectors. J Virol. 1999;73:2762–2769. doi: 10.1128/jvi.73.4.2762-2769.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire TC, Leib SR, Lonning SM, Zhang W, Byrne KM, Mealey RH. Equine infectious anaemia virus proteins with epitopes most frequently recognized by cytotoxic T lymphocytes from infected horses. J Gen Virol. 2000;81:2735–2739. doi: 10.1099/0022-1317-81-11-2735. [DOI] [PubMed] [Google Scholar]
- McGuire TC, O’Rourke KI, Perryman LE. Immunopathogenesis of equine infectious anemia lentivirus disease. Dev Biol Stand. 1990;72:31–37. [PubMed] [Google Scholar]
- McGuire TC, Tumas DB, Byrne KM, Hines MT, Leib SR, Brassfield AL, O’Rourke KI, Perryman LE. Major histocompatibility complex-restricted CD8+ cytotoxic T lymphocytes from horses with equine infectious anemia virus recognize Env and Gag/PR proteins. J Virol. 1994;68:1459–1467. doi: 10.1128/jvi.68.3.1459-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire TC, Zhang W, Hines MT, Henney PJ, Byrne KM. Frequency of memory cytotoxic T lymphocytes to equine infectious anemia virus proteins in blood from carrier horses. Virology. 1997;238:85–93. doi: 10.1006/viro.1997.8795. [DOI] [PubMed] [Google Scholar]
- McKinney DM, Lewinsohn DA, Riddell SR, Greenberg PD, Mosier DE. The antiviral activity of HIV-specific CD8+ CTL clones is limited by elimination due to encounter with HIV-infected targets. J Immunol. 1999;163:861– 867. [PubMed] [Google Scholar]
- Mealey RH, Fraser DG, Oaks JL, Cantor GH, McGuire TC. Immune reconstitution prevents continuous equine infectious anemia virus replication in an arabian foal with severe combined immunodeficiency: lessons for control of lentiviruses. Clin Immunol. 2001;101:237–247. doi: 10.1006/clim.2001.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelaro RC, Ball JM, Rushlow KE. Equine Retroviruses. In: Levy JA, editor. The Retroviridae. Vol. 2. Plenum Press; New York/London: 1993. pp. 257–360. [Google Scholar]
- Montelaro RC, Parekh B, Orrego A, Issel CJ. Antigenic variation during persistent infection by equine infectious anemia virus, a retrovirus. J Biol Chem. 1984;259:10539–10544. [PubMed] [Google Scholar]
- Newman MJ, Issel CJ, Truax RE, Powell MD, Horohov DW, Montelaro RC. Transient suppression of equine immune responses by equine infectious anemia virus (EIAV) Virology. 1991;184:55–66. doi: 10.1016/0042-6822(91)90821-r. [DOI] [PubMed] [Google Scholar]
- O’Connor DH, Allen TM, Vogel TU, Jing P, DeSouza IP, Dodds E, Dunphy EJ, Melsaether C, Mothe B, Yamamoto H, Horton H, Wilson N, Hughes AL, Watkins DI. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat Med. 2002;8:493– 499. doi: 10.1038/nm0502-493. [DOI] [PubMed] [Google Scholar]
- O’Rourke K, Perryman LE, McGuire TC. Antiviral, antiglycoprotein and neutralizing antibodies in foals with equine infectious anaemia virus. J Gen Virol. 1988;69:667– 674. doi: 10.1099/0022-1317-69-3-667. [DOI] [PubMed] [Google Scholar]
- Pantaleo G, Fauci AS. New concepts in the immunopathogenesis of HIV infection. Annu Rev Immunol. 1995;13:487–512. doi: 10.1146/annurev.iy.13.040195.002415. [DOI] [PubMed] [Google Scholar]
- Payne SL, Fang FD, Liu CP, Dhruva BR, Rwambo P, Issel CJ, Montelaro RC. Antigenic variation and lentivirus persistence: variations in envelope gene sequences during EIAV infection resemble changes reported for sequential isolates of HIV. Virology. 1987;161:321–331. doi: 10.1016/0042-6822(87)90124-3. [DOI] [PubMed] [Google Scholar]
- Perryman LE, O’Rourke KI, McGuire TC. Immune responses are required to terminate viremia in equine infectious anemia lentivirus infection. J Virol. 1988;62:3073–3076. doi: 10.1128/jvi.62.8.3073-3076.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo C, Huang XL, Fan ZF, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M, et al. High levels of anti-human immunodeficiency virus type 1 (HIV-1) memory cytotoxic T-lymphocyte activity and low viral load are associated with lack of disease in HIV-1-infected long-term nonprogressors. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rwambo PM, Issel CJ, Adams WVJ, Hussain KA, Miller M, Montelaro RC. Equine infectious anemia virus (EIAV) humoral responses of recipient ponies and antigenic variation during persistent infection. Arch Virol. 1990;111:199–212. doi: 10.1007/BF01311054. [DOI] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857– 860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Sellon DC, Fuller FJ, McGuire TC. The immunopathogenesis of equine infectious anemia virus. Virus Res. 1994;32:111–138. doi: 10.1016/0168-1702(94)90038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifka MK, Whitton JL. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat Immunol. 2001;2:711–717. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]
- Van Baalen CA, Pontesilli O, Huisman RC, Geretti AM, Klein MR, de WF, Miedema F, Gruters RA, Osterhaus AD. Human immunodeficiency virus type 1 Rev- and Tat-specific cytotoxic T lymphocyte frequencies inversely correlate with rapid progression to AIDS. J Gen Virol. 1997;78:1913–1918. doi: 10.1099/0022-1317-78-8-1913. [DOI] [PubMed] [Google Scholar]
- Van Baalen CA, Schutten M, Huisman RC, Boers PH, Gruters RA, Osterhaus AD. Kinetics of antiviral activity by human immunodeficiency virus type 1-specific cytotoxic T lymphocytes (CTL) and rapid selection of CTL escape virus in vitro. J Virol. 1998;72:6851– 6857. doi: 10.1128/jvi.72.8.6851-6857.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz D, Nowak MA. CD8 memory, immunodominance, and antigenic escape. Eur J Immunol. 2000;30:2704–2712. doi: 10.1002/1521-4141(200009)30:9<2704::AID-IMMU2704>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Zhang W, Auyong DB, Oaks JL, McGuire TC. Natural variation of equine infectious anemia virus Gag protein cytotoxic T lymphocyte epitopes. Virology. 1999;261:242–252. doi: 10.1006/viro.1999.9862. [DOI] [PubMed] [Google Scholar]
- Zhang W, Lonning SM, McGuire TC. Gag protein epitopes recognized by ELA-A-restricted cytotoxic T lymphocytes from horses with long-term equine infectious anemia virus infection. J Virol. 1998;72:9612–9620. doi: 10.1128/jvi.72.12.9612-9620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]