Abstract
Tubulin tyrosine ligase (TTL) catalyzes the post-translational C-terminal tyrosination of α–tubulin. Tyrosination regulates recruitment of microtubule interacting proteins. TTL is essential. Its loss causes morphogenic abnormalities and is associated with cancers of poor prognosis. We present the first crystal structure of TTL (from Xenopus tropicalis), defining the structural scaffold upon which the diverse TTL-like family of tubulin-modifying enzymes is built. TTL recognizes tubulin using a bipartite strategy. It engages the tubulin tail through low-affinity, high-specificity interactions, and co-opts what is otherwise a homo-oligomerization interface in structurally related ATP-grasp fold enzymes to form a tight hetero-oligomeric complex with the tubulin body. Small-angle X-ray scattering and functional analyses reveal that TTL forms an elongated complex with the tubulin dimer and prevents its incorporation into microtubules by capping the tubulin longitudinal interface, possibly modulating the partition of tubulin between monomeric and polymeric forms.
Tubulin tyrosine ligase (TTL) adds a C-terminal Tyr to α–tubulin as part of a tyrosination/detyrosination cycle present in most eukaryotic cells. α–tubulin is synthesized with a C-terminal Tyr that can be removed by α–tubulin Tyr carboxypeptidase, resulting in detyrosinated tubulin with a C-terminal glutamate (“Glu-tubulin”). Acting on the αβ-tubulin heterodimer, TTL restores the C-terminal Tyr, producing tyrosinated “Tyr-tubulin”. TTL is essential for development and cellular function. TTL-null mice die within an hour after birth because of disorganized neuronal networks1,2 and suppression of TTL protein levels provides a marked selective advantage for tumor growth3. TTL suppression leads to formation of microtentacles rich in Glu-tubulin that penetrate endothelial layers to facilitate reattachment of circulating tumor cells during metastasis4. Not surprising, TTL suppression correlates with poor prognosis in neuroblastoma, breast and prostate cancer patients5.
The tyrosination/detyrosination cycle affects microtubule (MT) dynamics in cells through differential recruitment of cytoskeletal regulators. The unstructured α–tubulin tail lies on the outside of the MT, in close proximity to binding sites of motors and MT-associated proteins6. Studies using Tyr- or Glu-tubulin specific antibodies revealed that the cellular distribution as well as stability of these MT populations is markedly different7. Interphase MTs are mostly tyrosinated, spindle MTs are detyrosinated; MTs in dendrites are enriched in Tyr-tubulin, while MTs in axons are enriched in Glu-tubulin8–12. Detyrosinated MTs persist as long as 16 hours, while tyrosinated MTs turnover in only 3–5 minutes7. Mitotic centromere-associated kinesin (MCAK) preferentially binds and depolymerizes Tyr-MTs over Glu-MTs, thus contributing to their differential stability in the cell13. Tyrosination also affects MT end dynamics by acting as a localization signal for MT plus-end binding proteins cytoplasmic linker protein-170 (CLIP-170) and p150Glued14. These proteins use their cytoskeleton-associated protein Gly-rich (Cap-Gly) domains to recognize a composite binding site formed by the tyrosinated α–tubulin tail and the end-binding protein EB115,16. Consistent with the importance of tyrosination in recruitment of MT plus-end tracking proteins, TTL is needed in migrating fibroblasts for robust polarization of the MT cytoskeleton14. Tyrosination can also act as a negative cue. In neurons, the kinesin-1 KIF5 targets only to the axon (rich in Glu-MTs) and not dendrites (rich in Tyr-MTs)17,18 by sensing the presence of the C-terminal Tyr on tubulin. Mutation of KIF5 or TTL depletion breaks the asymmetric distribution of this kinesin, allowing it to enter both axon and dendrites17.
Tubulin tyrosination was the first tubulin-specific post-translational modification reported19,20, with a plethora of evolutionarily conserved tubulin modifications subsequently discovered21. TTL was first isolated from brain extracts in 1977 (ref. 22–24). The tyrosination/detyrosination cycle has been characterized in a wide range of eukaryotes including trypanosomes, nematodes and humans11,25–27 and TTL shows a striking degree of sequence conservation from echinoderms to humans (Supplementary Fig. 1), displaying more than 96% identity among mammalian orthologs.
Tubulin is also subject to polyglutamylation and polyglycylation. These abundant and evolutionarily conserved post-translational modifications result in addition of glutamate or glycine chains of variable lengths to the α– or β–tubulin C-terminal tails. Although the modifications themselves have been known for decades, only recently has it become apparent that the enzymes responsible are evolutionarily related to TTL28,29, most likely sharing a similar structure and enzymatic mechanism. The polyglutamylases and polyglycylases now form the TTL-like (TTLL) family30–33.
While the cell biological role of TTL and the tyrosination/detyrosination cycle have been extensively studied, the molecular mechanism of TTL action remains to be resolved. To this end, we have now determined the first crystal structure of TTL, in its apo, ATP- and ADP-bound states, thus defining the scaffold upon which the diversity of recognition for the TTLL family evolved. Using analytical ultracentrifugation (AUC), small-angle X-ray scattering (SAXS), and functional analyses, we show that TTL uses a conserved, positively-charged surface to recognize the α-tubulin tail and forms an elongated complex with the αβ–tubulin heterodimer. TTL caps the tubulin heterodimer on the interface where tubulin would make longitudinal interactions within the MT lattice, thereby preventing its incorporation into MTs. Consistent with this, TTL inhibits tubulin polymerization in vitro, and TTL overexpression decreases microtubule growth in vivo, suggesting that in addition to modifying the α–tubulin tail, TTL could possibly also influence the partition of tubulin between the monomeric and polymeric forms in cells.
RESULTS
Structural overview
Xenopus tropicalis TTL was expressed in Escherichia coli and purified to homogeneity. X. tropicalis TTL is 81 % identical to human TTL (Supplementary Fig. 1) and it tyrosinates bovine brain tubulin in vitro with activity comparable to that of the murine enzyme (data not shown). 95% of 386 TTL crystals tested were severely split and did not provide useful data. The structure of apo TTL was determined by combining low-resolution phases from Ta6Br12 soaks with phases from a two-wavelength SeMet multiple anomalous dispersion experiment. The 2.5 Å refined model of apo TTL has Rfree=29.6% (Table 1 and Methods). The structures of TTL bound to the slowly-hydrolyzable ATP analog AMPPNP (at 2.5 Å), and ADP (at 2.9 Å) were solved by molecular replacement and refined to Rfree=28.5% and 31.4%, respectively (Table 1, Methods, Supplementary Fig. 2 showing experimental map and superposition of apo, ADP and AMPPNP structures).
Table 1.
Crystallographic data and refinement statistics
| Native Apo TTLa | Apo TTL (SeMet)a | TTL:AMPPNPa | TTL:ADPa | ||
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | C2 | C2 | P21 | C2 | |
| Cell dimensions | |||||
| a, b, c (Å) | 116.6, 76.2, 44.2 | 116.6, 76.2, 44.2 | 116.6, 76.2, 44.2 | 44.4, 74.6, 117.3 | 117.0, 75.7, 44.2 |
| α, β, γ (°) | 90, 90.6, 90 | 90, 90.6, 90 | 90, 90.6, 90 | 90, 90.1, 90 | 90, 90.8, 90 |
| Inflection | Remote | ||||
| Wavelength | 0.9806 | 0.9637 | |||
| Resolution (Å) | 50.0-2.5 (2.54–2.5) | 50-2.6 (2.6–2.5) | 50.0-2.24 (2.28–2.24) | 30-2.5 (2.54–2.50) | 30-2.9 (2.95–2.9) |
| Rsym | 6.1 (33.3) | 5.3 (32.5) | 5.0 (32.9) | 7.4 (38.0) | 5.8 (22.6) |
| I / σI | 52 (5) | 23 (2) | 28 (2) | 37.3 (4.5) | 29 (5) |
| Completeness (%) | 99.9 (99.8) | 98.7 (88.7) | 92.2 (51.0) | 99.1 (98.5) | 98.7 (95.9) |
| Redundancy | 7.0 (5.8) | 2.3 (2) | 3.6 (2.4) | 7.4 (6.9) | 3.7 (3.5) |
| Refinement | |||||
| Resolution (Å) | 50.0–2.5 | 30–2.5 | 30–2.9 | ||
| No. reflections | 12946 | 26356 | 8195 | ||
| Rwork / Rfree | 24.9/29.8 | 25.4/28.5 | 23.2/31.3 | ||
| No. atoms | |||||
| Protein | 2395 | 4850 | 2442 | ||
| Ligand/ion | 0 | 62/7 | 27/3 | ||
| Water | 36 | 30 | 2 | ||
| B-factors | |||||
| Protein | 73.5 | 65.9 | 71.3 | ||
| Ligand/ion | 51 | 88.9/42.8 | 115.9/44.6 | ||
| Water | 63 | 49.8 | 43.8 | ||
| R.m.s deviations | |||||
| Bond lengths (Å) | 0.0084 | 0.0078 | 0.0085 | ||
| Bond angles (°) | 1.32 | 1.31 | 1.35 | ||
Data were collected on a single crystal for each dataset.
Values in parentheses are for highest-resolution shell.
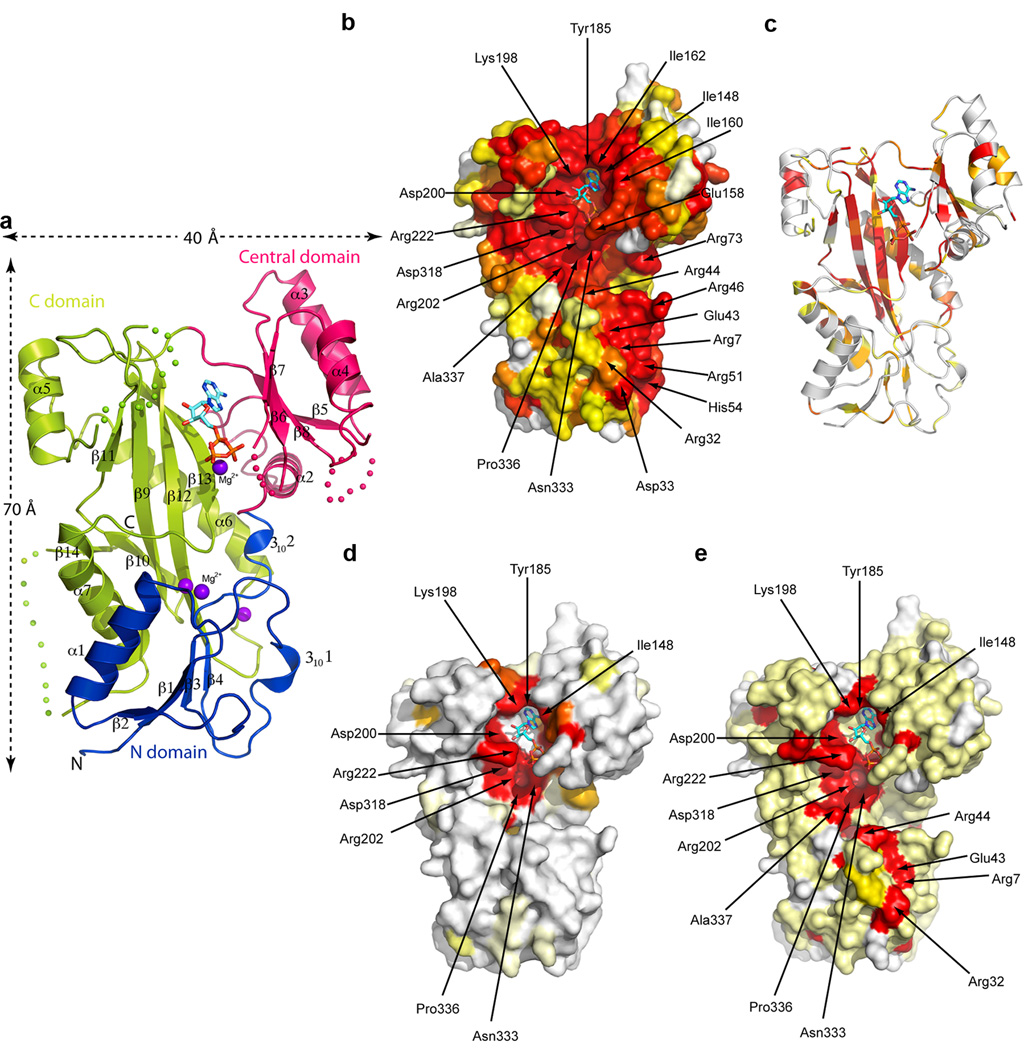
TTL is elongated (maximum dimensions 70 × 40 Å2; Fig. 1a) and comprises three domains: an N-domain (residues 1–71), a central domain (residues 72–188) and a C-domain (residues 189–377). The N-domain adopts an α/β fold comprised of a four-stranded parallel β-sheet flanked on one side by helix α1, and on the other by irregular segments that include two 310 helices. The central domain is formed by a four-stranded anti-parallel β-sheet flanked by helices α3 and α4 on one face and the C-domain on the other. Helix α2 lies on the β-sheet edge, contacting both the N and C-domains. The C-domain is formed by an elongated anti-parallel β-sheet (strands β11, 9, 12, 13). The long strand β12 reaches into the N-domain and bridges the β-sheets from the N- and C-domains to form a continuous curved β-sheet (Fig. 1 and Supplementary Fig. 2b). The C-domain β-sheet underlies the active site and is surrounded by three helices, with long helix α6 serving as a “brace” that makes stabilizing interactions across strands β11, 12 and 13. Both the central and C-domains contribute to nucleotide coordination, cradling it at their interface. There are no major conformational changes between the apo and nucleotide bound structures. The central domain shows some conformational plasticity, rotating ~3° between the apo (or ADP) and AMPPNP crystal forms (Supplementary Fig. 2). Structure-based sequence alignments of TTLs ranging from the sea urchin Strongylocentrotus purpuratus to human demonstrate the high degree of conservation of these enzymes, both within their hydrophobic cores as well as surface-exposed residues (Figs. 1b,c and Supplementary Fig. 1). Thus, the structure of X. tropicalis TTL is representative of that of all known TTLs.
Figure 1.
X-ray structure of the X. tropicalis TTL, conservation and specialization across the TTL and TTLL tubulin modifying enzyme families. (a) Ribbon representation of the crystal structure of TTL bound to AMPPNP. The bound nucleotide is shown as a stick atomic model. Colored dots represent regions of the polypeptide chain that are disordered in the crystal structure (b) TTL molecular surface, color-coded for conservation of TTL sequences ranging from S. purpuratum to human (Supplementary Fig. 1), colored on a gradient from white to red with red representing 100% identity (c) Ribbon representation of the TTL structure, color-coded according to sequence similarity scores (Protskin54) with TTLL family enzymes (TTLL 3, 4, 5, 6, 7 and 10); gradient from white to red with red representing 100% similarity. (d) TTL molecular surface color-coded based on sequence conservation with TTLL family enzymes (TTLL 3, 4, 5, 6, 7 and 10), colored as in panel b. (e) TTL molecular surface color-coded for conservation among the α–tubulin specific TTL and TTLL5 enzymes from X. tropicalis, M. musculus and human, colored as in panel b.
Despite having only 9% overall sequence identity, the fold of TTL closely resembles that of D-Ala:D-Ala ligase [PDBeFold Z score = 6.1 (ref. 34,35)], a member of the ATP-grasp superfamily which consists of enzymes of diverse specificities that catalyze the ATP-dependent ligation of a carboxylate-containing molecule to an amine or a thiol36. Their fold is characterized by two α/β domains (the central and C-domains in TTL) that “grasp” the ATP between them. While the overall fold of the TTL N-domain is similar to the corresponding domain in D-Ala:D-Ala ligase, the structural equivalencies are strongest for the central and C-domains (r.m.s.d. = 3.1 Å over 219 residues). The TTL fold also resembles that of glutathione S-transferase (GST), another ATP-grasp enzyme, as proposed previously29. However, our crystal structure reveals that TTL and GST are sufficiently divergent that the secondary structure prediction of TTL based on the latter has several inaccuracies29,30. Unique among known ATP-grasp family structures is the continuation of hydrogen bonding between strands β4 and β12 (from the N- and C-domains, respectively) which gives rise to a continuous 9–stranded β-sheet in TTL (Fig. 1 and Supplementary Fig. 2b). In other ATP-grasp superfamily members, the N- and C-domain β-sheets are disjoint.
A versatile common scaffold for TTL and TTLLs
Mapping of sequence conservation from a multiple sequence alignment of TTL and TTLL polyglutamylases and polyglycylases onto the TTL crystal structure reveals how the TTL scaffold supported the expansion of the repertoire of tubulin post-translational modification enzymes. The core secondary structural elements in the three domains are conserved, with the TTL-specific central elongated β-sheet underlying the active site being most highly conserved (Fig. 1c). This indicates that TTL and TTLL family members share a similar core architecture.
The conservation pattern of surface residues among members of the TTL and TTLL families with divergent specificities (TTL, TTLL3, TTLL4, TTLL5, TTLL6, TTLL7 and TTLL10) indicates that only residues immediately adjacent to the ATP binding site are invariant, while the rest of the molecular surface evolved to accommodate different recognition specificities (Fig. 1d). Interestingly, when comparing family members with similar specificities, putative common platforms of substrate recognition emerge. For example, TTL and TTLL5, both enzymes specific for the α–tubulin tail30, share an elongated conserved patch extending from the ATP binding site towards the N-domain (Fig. 1e). Mutations of residues in this patch impair α–tubulin tail recognition by TTL (below), suggesting a shared α–tubulin tail binding surface.
Active Site Architecture
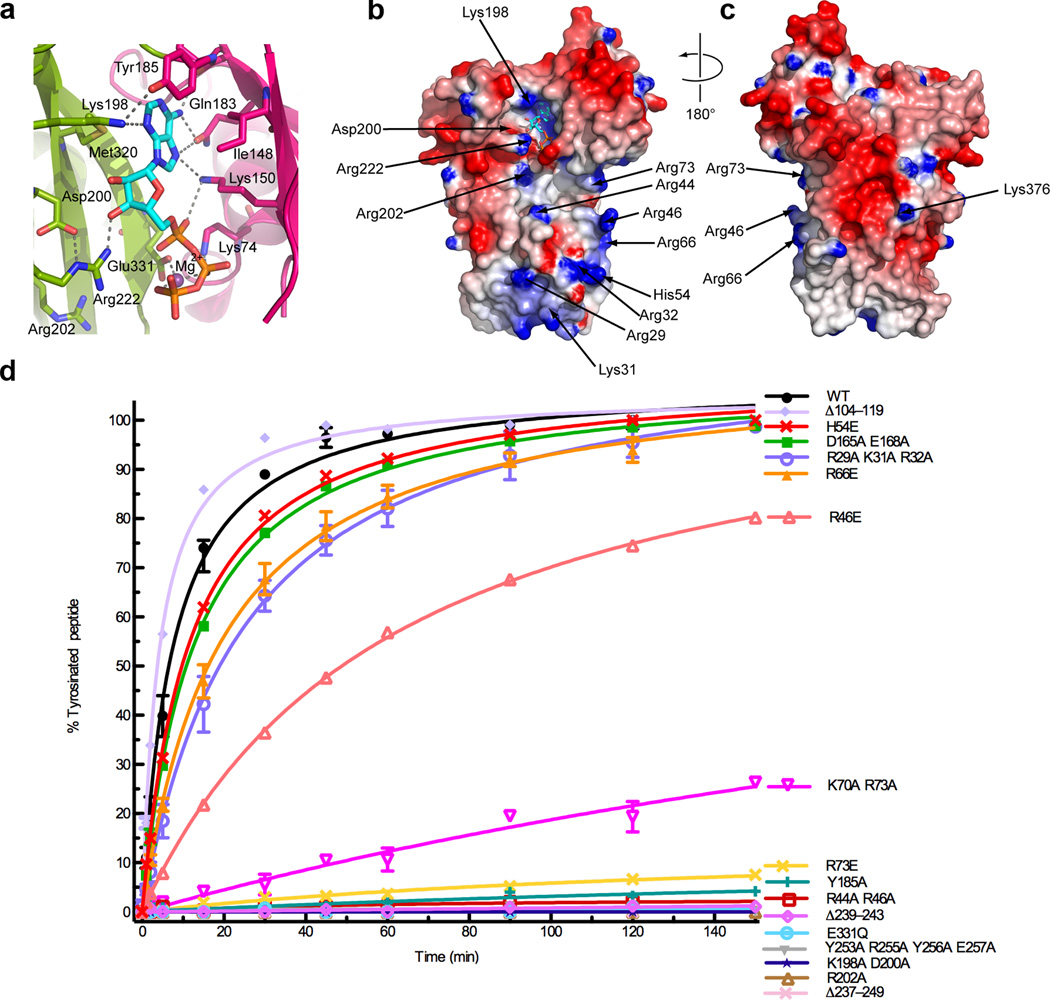
ATP is wedged against a loop connecting the central and C-domains (residues 184–198; Fig. 2a and Supplementary Fig. 2c). The phenyl group of Tyr185 interacts edge-on with the base of ATP, which is sandwiched between conserved hydrophobic residues (Met320 and Ile148) and hydrogen bonds with conserved functional groups in the central domain (Lys198, Lys184, Lys150; Figs. 1 and 2a–c). The α-, β-, and γ-phosphates make electrostatic interactions with the side-chains of invariant Lys150, Lys74 and Arg220, respectively, and the β- and γ-phosphates are bridged by a magnesium ion also coordinated by Glu331 (Fig. 2a). Consistent with the essential role of ATP for TTL activity22, mutation of residues that are involved in nucleotide binding (Y185A and K198A D200A) abolishes tyrosination activity (assayed with an α-tail peptide substrate; Fig. 2d and Supplementary Fig. 3). Glu331 is invariant in all TTLs and TTLLs (as well as ATP-grasp enzymes30); its mutation inactivates TTL (Fig. 2d). The conservation of active site architectures between TTL and ATP-dependent ligases like D-Ala:D-Ala ligase, strongly suggests that TTL and TTLLs employ a catalytic mechanism similar to that of these ligases (Supplementary Fig. 4).
Figure 2.
Active site architecture and α–tubulin C-terminal peptide recognition. (a) Conserved interactions in the TTL active site, colored as in Fig. 1a (b) Nucleotide (ventral) and (c) dorsal views of the TTL molecular surface color-coded for electrostatic potential55 (red, negative; blue, positive, ranging from −7 kBT to 7 kBT) (d) Tyrosination of a 14-residue α–tubulin C-terminal peptide by TTL and structure-guided TTL mutants (N≥2; Supplementary Fig. 3). The 14-residue peptide (VDSVEGEGEEEGEE) serves as an optimal TTL peptide substrate42. Error bars indicate s.e.m. and are frequently smaller than the symbols.
Molecular determinants of α–tubulin tail recognition
The TTL electrostatic surface exhibits a conserved, positively charged stripe running from the ATP binding site to the N-domain (Fig. 2b). This is likely a region for interaction with the negatively charged α–tubulin tail. In contrast, the dorsal surface of TTL is negatively charged and less conserved and thus unlikely to interact with the anionic tubulin tail (Fig. 2c). We carried out structure-guided mutagenesis of conserved surface and active site residues and performed tyrosination assays using an α-tail peptide as substrate for the purified mutant TTLs (Methods and Supplementary Fig. 3). The guanidinium group of Arg202 is situated 6.9 Å from the γ-phosphate and is part of a positively charged pocket ideal for binding the C-terminal glutamate of the α–tubulin tail (Figs. 2a,b). TTL and TTLLs catalyze the addition of an amino acid to a glutamate (Supplementary Fig. 4). Arg202 is invariant in all TTL and TTLLs (Fig. 1d and Supplementary Fig. 1) and its mutation inactivates TTL (Fig. 2d). The mobile loop connecting β11 and α5 (residues 230–260) is proximal to the active site and contains two highly conserved motifs (Fig. 3a and Supplementary Fig. 1). The first [HL(T or C)N] is common to all TTLs and TTLLs, while the second [(Y or F)G(R or K)YEE] is unique to the TTLs. The structurally equivalent loop is used in D-Ala:D-Ala for substrate recognition37. This loop displays no conservation within the broader ATP-grasp family, consistent with its specialization for substrate binding. Deletion of residues in this loop (Δ239–243 or Δ237–249) or mutation of residues in the TTL specific motif (Y253A R255A Y256A E257A) inactivate TTL (Fig. 2d), but do not abolish tubulin binding, supporting its essential role in recognizing the tubulin tail.
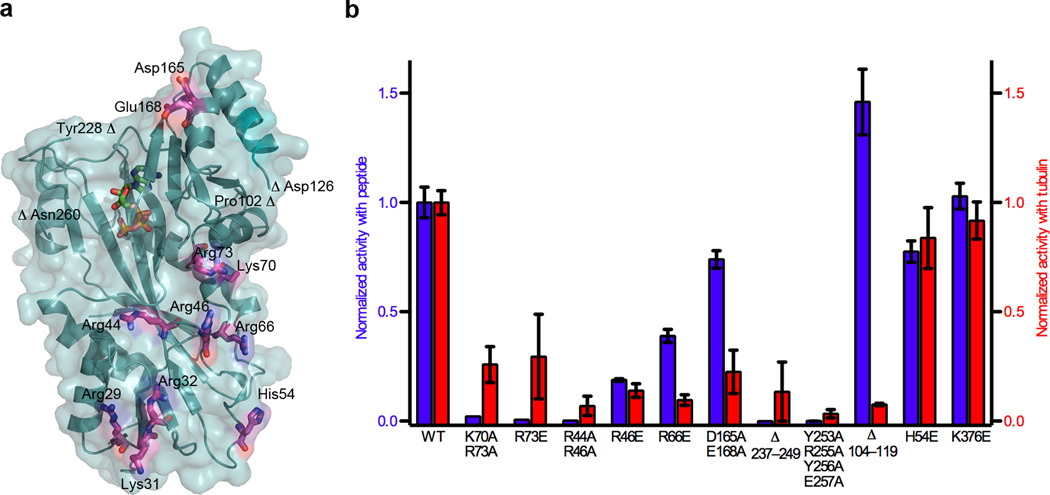
Figure 3.
Molecular determinants for tubulin tyrosination. (a) TTL molecular surface with conserved residues important for tubulin tyrosination shown in stick atomic representation (b) Normalized relative tyrosination activity with α-tubulin peptide (blue) or tubulin (red) substrates of structure-guided TTL mutants (N≥3). Error bars indicate s.e.m.
Continuing towards the interface between the central and N-domains, invariant Arg73 as well as conserved Lys70 are important in α–tubulin peptide recognition, as the mutation R73E or the double mutation K70A R73A leads to 99.4% and 98% reductions in tyrosination rate, respectively (Fig. 2d). It is noteworthy that TTL contains a predicted cAMP-dependent kinase serine phosphorylation site (RKAS)28 at Ser76 and adjacent to Arg73, suggesting a possible mode of TTL regulation. In the N-domain, mutation of conserved residues Arg44 and Arg46 in the loop connecting β3 and 3101 leads to more than 99.8% reduction in activity (Fig. 2d). The charge reversal mutant R46E has a more modest reduction in activity, underscoring that Arg44 is the more critical residue in α-tail recognition. Interestingly, Arg44 is conserved in TTLL5 (an α–tubulin-specific glutamyl-ligase) but not in other TTLLs (Figs. 1d,e), suggesting that it is important for α–tubulin tail recognition in that case also. The structurally equivalent loop in D-Ala:D-Ala ligase and other ATP-grasp proteins partially occludes the active site and is also used for substrate binding37,38; however, the loop in TTL is considerably shorter than in other ATP-grasp proteins, consistent with TTL binding to a larger ligand at the interface between the N- and C-domains, compared to the small substrates typical for ATP-grasp enzymes (i.e. D-Ala, glutathione). Mutation of the conserved positive patch residues at the tip of the N-domain (R29A K31A R32A or H54E; Figs. 1b and 2b,c) has a small effect on tyrosination, indicating that they are not a determinant for α–tail recognition.
Tubulin recognition by a coöpted oligomerization interface
We next examined the effects of our structure-guided mutations on tubulin tyrosination. Not surprisingly, mutations that strongly impair activity with the α-tail peptide (K70A R73A, R73E, R44A R46A, Y253A R255A Y256A E257A) also do so with tubulin (Fig. 3a,b). Deletion of the mobile loop connecting β11 and α5 (TTL Δ237–249) inactivates TTL both with peptide as well as tubulin substrate. This mutant also shows a modest loss in tubulin binding affinity as assayed by gel filtration (Figs. 4a–c). Nearby conserved Asp165 and Glu168 in the central domain are also important for tubulin recognition, as their mutation reduces tubulin tyrosination by 75%, while the effect on peptide tyrosination is small (Fig. 3b). The central domain has the highest B-factors in the TTL structures, suggesting that its plasticity might be important for accommodating tubulin. The charge reversal mutant of invariant Arg66, which has robust activity with the α–tail peptide (Fig. 2d), displays very low activity with tubulin (~90% reduction; Fig. 3). Size-exclusion chromatography shows that this mutant is defective in tubulin binding (Fig. 4d), suggesting that the tubulin binding interface lies close to the junction of the central and N-domains. Interestingly, a TTL mutant missing the disordered loop between β5 and α3 (Δ104–119) in the central domain shows a 30% enhancement in activity with the peptide, but has a more than 90% decrease in activity with tubulin compared to wild-type (Fig. 3b). Unlike the R66E mutant, the Δ104–119 mutant still binds tubulin (Fig. 4e), suggesting that residues connecting the β5 and α3 structural elements might possibly play a role in the release of tubulin from the TTL–tubulin complex. Consistent with the non-involvement of the dorsal face in α-tail or tubulin recognition, the charge reversal mutation of conserved Lys376 (Fig. 2c) has no effect on tyrosination activity (Fig 3b).
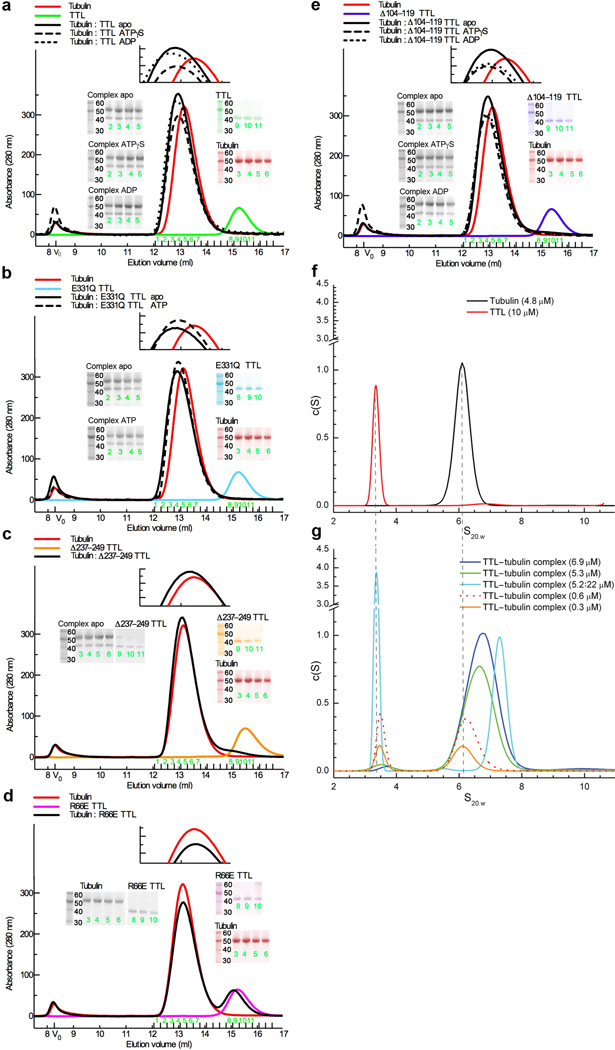
Figure 4.
Gel filtration studies and sedimentation velocity ultracentrifugation analysis of TTL binding to tubulin. (a–e) Complex formation between TTL or TTL mutants and tubulin in various nucleotide conditions monitored by gel filtration chromatography. Binding is monitored by the disappearance of the slower mobility peak (corresponding to uncomplexed TTL) when TTL or TTL mutants are incubated with excess tubulin (68 µM tubulin and 34 µM TTL or TTL mutant). The gels correspond to peak fractions indicated in green. (f) Sedimentation coefficient distributions (c(s)) obtained for 10 µM TTL and 4.8 µM tubulin (g) c(s) for TTL–tubulin complexes at varying concentrations. All complexes are at 1:1 stoichiometry, except that shown in the cyan trace, which denotes a 1:4 tubulin:TTL ratio. The sedimentation coefficient distributions obtained with mixtures of 1:1 tubulin:TTL ratio at increasing total concentrations show two peaks, a small s value peak corresponding to free TTL, with the area of the peak decreasing with increasing protein concentration, and the high s value peak representing the position of the TTL–tubulin complex reaction boundary. At lower concentrations, the position of the higher s value peak corresponds to that of the tubulin dimer alone and increases in s value with increasing concentration. By analyzing the shift in position of the complex peak with increasing protein concentration the affinity constant of TTL for tubulin was determined to be ~1 µM (method described in56; Methods). When TTL is in excess so that all tubulin is in complex with TTL, a narrow symmetrical boundary of TTL–tubulin is observed.
Mutations on the TTL molecular surface that affect tubulin tyrosination, but not peptide tyrosination, cluster on the upper quadrant of the molecule, in the C- and central domains or at the N- and central-domain junction (Fig. 3a). Remarkably, all hitherto characterized ATP-grasp superfamily members (e.g. D-Ala:D-Ala ligase37, GST38, synapsin39, Lys biosynthesis enzyme LysX40), are either dimers or tetramers and use their equivalent surfaces for oligomerization. TTL is monomeric in our crystals, and AUC demonstrates that it is monomeric in solution. The sedimentation coefficient distribution [c(s)] of TTL shows a single peak representing the TTL monomer with an s value of 3.36 (Fig. 4f) and a corresponding molecular mass of 42,696 Da, in excellent agreement with the mass predicted for monomeric TTL (43,686Da). Thus, the homo-oligomerization interface of ATP-grasp enzymes has evolved in TTL to recognize the tubulin dimer, a substrate substantially larger than those of typical ATP-grasp ligases. TTL, which has uniquely evolved to be a monomer, is unstable in the absence of tubulin22,41, which completes its oligomerization interface.
The TTL–tubulin complex in solution
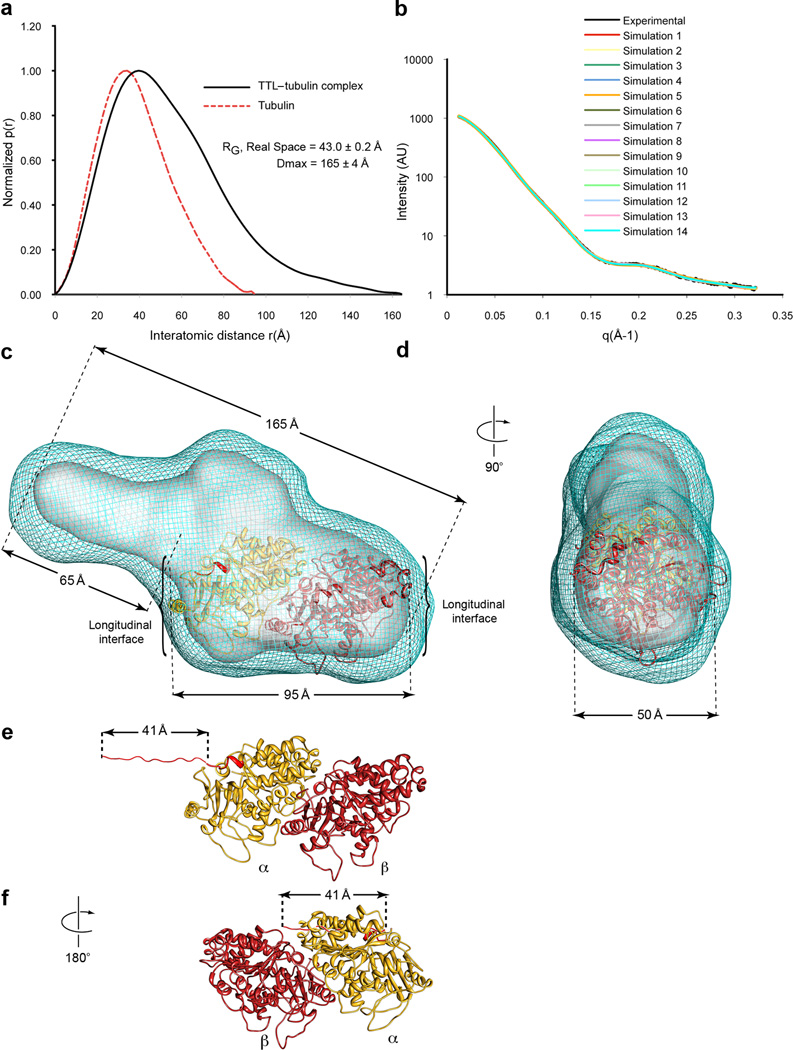
Our ultracentrifugation experiments demonstrate that purified TTL forms a 1:1 complex with tubulin (the calculated molecular mass of 141,974 Da is in good agreement with the predicted mass of 143,584 Da; Fig. 4g and Methods), consistent with sucrose gradient analysis of endogenous TTL–tubulin complexes24. Isolated α–tubulin tail peptide binds to TTL with low affinity (Kd~144 µM; Methods) when compared to tubulin (Kd~1 µM, Fig. 4g, Methods,) and the specific activity of TTL with this peptide is 98% lower than with tubulin42, indicating that the enzyme makes important contacts with the tubulin dimer in addition to the α–tail. The low binding affinity for the α–tail peptide explains the puzzling earlier observation that TTL is inefficient at modifying α-tubulin in MTs43 despite the fact that the flexible tail is exposed on the MT surface. To further characterize the TTL–tubulin complex, we carried out SAXS experiments. The SAXS data show that the TTL–tubulin complex has a considerably larger maximal dimension (Dmax) than either TTL or tubulin alone: 165±3Å versus 72±5Å or 95Å, respectively (Fig. 5a and Supplementary Fig. 5; in the case of tubulin, the Dmax was determined from the known electron crystal structure; 1JFF.pdb44). The elongated nature of the complex is consistent with our sedimentation velocity data, which indicates that the shape of the TTL–tubulin complex can be approximated by a prolate ellipsoid with an aspect ratio of 3.8. We benchmarked our analysis by determining through AUC the aspect ratio for the tubulin heterodimer alone. This resulted in an aspect ratio of 2.2, in excellent agreement with the known tubulin structure44. Comparison of the calculated pair-distribution [P(r)] function for tubulin and the experimental P(r) for the TTL–tubulin complex also shows that in addition to being more elongated, the complex is asymmetric as the function is relatively skewed (Fig. 5a).
Figure 5.
Model of the TTL–tubulin complex from small-angle X-ray scattering. (a) Pair-distance probability distributions, [P(r)], computed from the experimental SAXS data for the TTL–tubulin complex and the crystal structure of the tubulin dimer (1JFF.pdb44) (b) Agreement of the experimental scattering for the TTL–tubulin complex with the scattering profiles of the models obtained from 14 ab initio simulations using Gasbor57 (χavg= 1.49 ± 0.09). The average normalized spatial discrepancy (NSD) for 14 models is 1.314, indicating good agreement (c,d) Composite (cyan mesh) and filtered (solid grey surface) SAXS envelopes for the TTL–tubulin complex. The composite structure consists of the aligned, superimposed and summed models from 14 independent simulations, whereas the filtered model corresponds to the most probable density map. The docked αβ-tubulin dimer is shown in ribbon representation, α–tubulin, yellow, β-tubulin, dark red. The last 2 residues of the α-tubulin atomic model are colored red and mark the beginning of the α-tail. The resolution of the reconstruction does not allow the unambiguous determination of the relative orientation of the tubulin dimer along its long axis. (e,f) Ribbon structure of the αβ-tubulin dimer in two different possible orientations in the SAXS reconstruction. The unstructured region of the α–tubulin tail absent from the tubulin crystal structure was modeled as a fully extended strand to illustrate its maximal possible span, and is colored red. Panel f shows the α-tubulin tail is unlikely to reach the active site of TTL if the interface of TTL with the globular core of the tubulin heterodimer is through β-tubulin. For all panels mean ±sd
SAXS data were used to generate low-resolution ab initio models of three-dimensional arrangements of scattering centers that provide the shape of the molecular envelope for the TTL–tubulin complex (Fig. 5b and Methods). We performed fifteen ab initio simulations and the resulting models were aligned, averaged and filtered on the basis of occupancy (Figs. 5c,d and Methods). The filtered structure of the TTL–tubulin complex is a deformed prolate ellipsoid with a narrower side. The wider section has dimensions consistent with those of the αβ-tubulin heterodimer (95 × 50 Å2). The remaining volume fits well the TTL molecular envelope with the N-domain lying at the edge of the complex and the central and C-domains contacting the tubulin body. This proposed model is consistent with our mutagenesis data showing that mutations at the tip of the N-domain do not affect tyrosination, but mutations in the central and C-domains as well as at the N- and central domains junction do (Figs. 3 and 4).
Given the symmetric nature of the tubulin dimer and the limited resolution of the SAXS reconstruction, it is not possible to unequivocally position the tubulin heterodimer in the reconstruction and establish which tubulin protomer is recognized by TTL. However, the length of the α–tubulin tail and the substrate requirements of TTL support a model in which the major recognition interface is on α–tubulin. The last eleven residues in the α-tubulin tail are disordered in the tubulin structure44. Assuming that these residues are in a completely extended conformation (resulting in a maximum length of 41 Å, Fig. 5e), the distance the α-tail would have to span if TTL were to recognize mainly the β-tubulin protomer is too long for it to reach the active site at the center of the TTL volume (Fig. 5f). Moreover, solution NMR and molecular dynamics studies show that the α-tail is not fully extended, but adopts a slightly helical conformation with a ~29 Å45 span. Thus, in the absence of large conformational changes upon binding, it is likely that TTL recognizes α-tubulin on the surface that would form the longitudinal interface in the context of the MT.
We tested this model by performing tubulin tyrosination assays in the presence of antibodies that recognize various αβ-tubulin dimer epitopes (Supplementary Fig. 6). Antibodies that recognize an epitope in β-tubulin at the longitudinal interface or in the H11-H12 C-terminal helices do not inhibit tyrosination, while an antibody that recognizes the α–tubulin tail inhibits the reaction robustly. Moreover, an antibody that recognizes an epitope on α-tubulin (residues 65–97) proximal to the αβ protomer interface that lies close to the interface between TTL and tubulin in our model also inhibited tyrosination (Fig. 5c and Supplementary Fig. 6). The engagement of the tubulin dimer through α-tubulin with minimal β-tubulin contacts is unexpected as TTL was proposed to have a β-tubulin binding site based on crosslinking studies and that this interaction was the determining factor for TTLs high specificity for the tubulin heterodimer46. Our data do not exclude an interaction between TTL and β-tubulin; however, they support a model in which the major interaction surface is not through β-tubulin. The extensive interaction surface with the tubulin dimer explains the high degree of specificity TTL has for tubulin as well as the earlier observation that TTL interacts only with monomeric tubulin and not MTs20,43 as the TTL binding surface is partially buried upon MT assembly.
TTL inhibits tubulin polymerization in vitro and in vivo
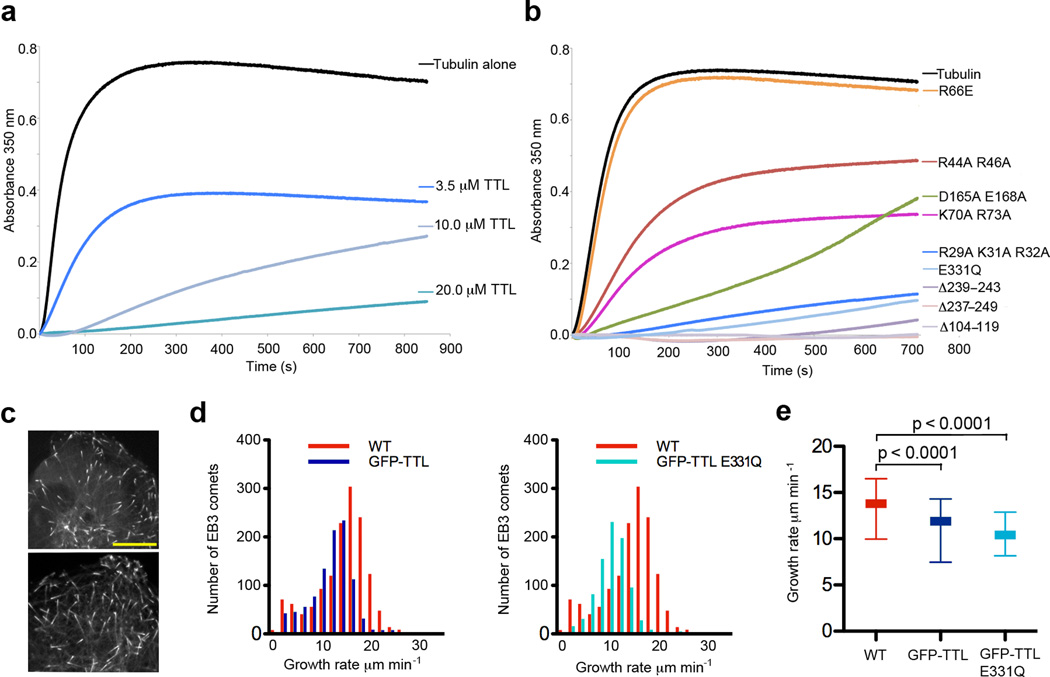
Based on our SAXS reconstruction, TTL should inhibit tubulin polymerization since it caps the longitudinal MT interface of tubulin. We tested this by examining the effects of TTL on the spontaneous polymerization of tubulin in vitro (Fig. 6a; Supplementary Methods). Addition of purified TTL to a solution containing 20 µM tubulin results in dose-dependent reduction of polymerization. When added in 1:1 molar ratio to tubulin, TTL effectively prevents MT assembly, consistent with its ability to bind and prevent the incorporation of tubulin into the MT lattice (Fig. 6a).
Figure 6.
TTL inhibits spontaneous polymerization of purified tubulin in vitro and attenuates MT growth rates in vivo. (a) Tubulin polymerization determined via turbidity (at 350 nm) in the absence or presence of TTL at various concentrations. For each experiment tubulin concentration was 20 µM. (b) Tubulin polymerization determined via turbidity (at 350 nm) in the absence or presence of various TTL mutants. For each experiment tubulin concentration was 20 µM and the concentration of the indicated TTL mutants was 20 µM. (c) Spinning disc confocal image of a U2OS cell expressing GFP-TTL (top panel) and TTL E331Q mutant (bottom panel) and the plus-end tracker EB3 fused to the fluorescent protein mKusabira-Orange. Scale bar corresponds to 10 µm. (d) Histograms of growth velocities of all tracked MTs in a wild-type and a GFP-TTL expressing cell (left panel) and a wild-type and a GFP-TTL E331Q expressing cell (right panel). Distributions comprise 1427, 983 and 858 measurements for the wild-type, GFP-TTL and GFP-TTL E331Q cell, respectively. (e) MT growth rates in wild-type, GFP-TTL and TTL E331Q expressing cells: 25th percentile (bottom line), median (middle thick line), 75th percentile (top line). The average growth rate is 13.04 ± 0.07 µm min−1 (mean±s.d.; N= 4225 tracks from 4 cells) for wild-type, 10.86 ± 0.08 µm min−1 for GFP-TTL expressing cells (N=3787 from 4 cells) and 10.24 ± 0.12 µm min−1 for GFP-TTL E331Q expressing cells (N=5222 from 5 cells). Statistical significance of the difference was determined by a permutation t-test.
To better understand the nature of the TTL-tubulin interaction, we examined the effects of our structure-guided surface mutants on tubulin polymerization (Fig. 6b). Not surprising, TTL mutations that affect α–tail recognition and/or tubulin heterodimer recognition show impairment in inhibiting tubulin polymerization. Importantly, mutation of Arg66 at the junction between the central and N-domains fails to inhibit MT polymerization, consistent with its loss in tubulin binding despite its ability to recognize the α–tail peptide (Figs. 2d and 4d). Consistent with our proposed TTL–tubulin model, mutants of conserved residues positioned at the edge of the N-domain (R29A K31A R32A) inhibit polymerization similarly to wild-type TTL, indicating they are competent in tubulin binding. The ability to modify the tubulin tail does not seem to affect tubulin sequestration activity, as the catalytic E331Q mutant has comparable activity to the wild-type protein, consistent with our gel-filtration studies showing this mutant is unimpaired in tubulin binding regardless of nucleotide occupancy (Fig. 4b). Consistent with the tubulin sequestration activity observed in vitro, TTL displays strong product inhibition42,47 and our equilibrium sedimentation studies using substrate and product α-tubulin peptides show that they bind with similar affinities (144 µM for substrate versus 96 µM for product peptide; Supplementary Methods).
We supplemented our in vitro results with studies in living cells. We analyzed microtubule growth rates in human osteosarcoma cells (U2OS line) while manipulating TTL levels. We detected and tracked48 the position of fluorescently tagged plus end-binding protein EB3 that marks the ends of growing MTs (Fig. 6c) to determine the growth rates of MTs from time-lapse image series. Overexpression of GFP-TTL at low levels reduced the speed of MT growth (Figs. 6d,e), consistent with our in vitro results. Since tyrosination can influence the recruitment of Cap-Gly domain containing plus end binding proteins (e.g. CLIP-170, p150Glued14) that affect MT dynamics, we also investigated the effects of a TTL mutant (E331Q) that is defective in tubulin tyrosination (Fig. 2d), but not tubulin binding (Fig. 4b). Live cell imaging with this mutant shows that its effect on MT growth rates are similar to wild-type TTL (Figs. 6d,e), indicating that the reduction in MT growth rates is not due to increased tubulin tyrosination.
DISCUSSION
TTL uses a unique bipartite substrate recognition strategy. It makes canonical use of its ATP-grasp ligase active site, which has been exquisitely tuned evolutionarily to recognize small molecule substrates (i.e. the end of the α-tubulin tail) and catalyze peptide bond formation. However, in addition, and uniquely among ATP-grasp enzymes, TTL has co-opted what is otherwise a homo-oligomerization interface to form a hetero-oligomeric complex with the tubulin body. TTL caps the longitudinal tubulin interface and forms an elongated 1:1 complex with the tubulin heterodimer (Fig. 5), rendering it unable to incorporate into the MT lattice. Consistent with this, TTL inhibits spontaneous tubulin polymerization in vitro and its overexpression decreases MT growth rates in vivo (Fig. 6). This mode of tubulin heterodimer recognition by a protein that is able to sequester tubulin is novel. Tubulin sequestering protein stathmin binds two tubulin dimers and caps α–tubulin49, while centrosomal P4.1.-associated protein sequesters a single heterodimer by capping the β-subunit longitudinal interface50. Unlike TTL, both these proteins are intrinsically disordered in solution in the absence of tubulin49,50. A better understanding of how TTL recognizes the tubulin dimer will require a higher-resolution TTL–tubulin complex structure.
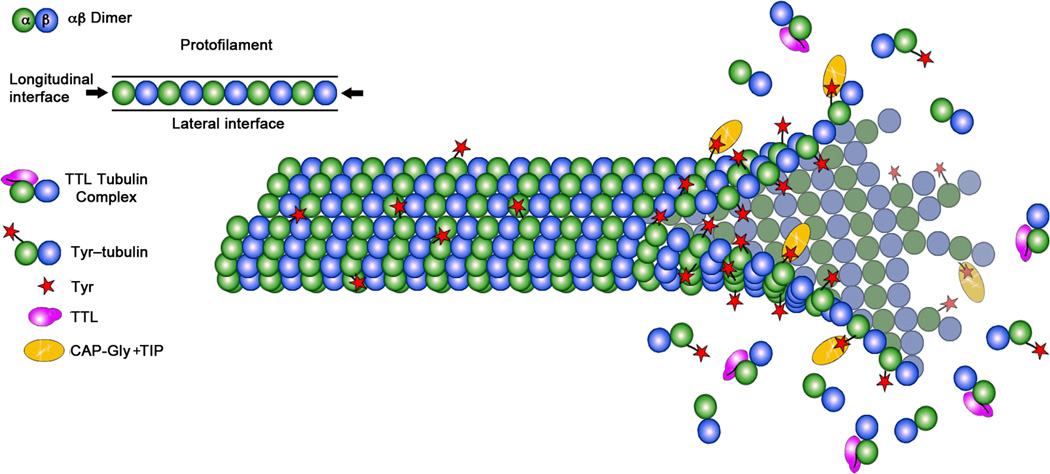
The intracellular concentration of monomeric tubulin is 5–10 µM51. Since TTL binds to tubulin with 1 µM Kd, this implies that TTL is bound to tubulin most of the time in the cell, consistent with earlier observations that TTL isolated from brain extracts copurified with tubulin22,41. TTL acts preferentially on soluble tubulin and the detryosination reaction preferentially takes place on MTs13. This creates an asymmetry in the distribution of Tyr-tubulin within the MT, with tyrosinated tubulin enriched at the growing plus end (Fig. 7). The degree of asymmetry depends on the relative rates of tyrosination and detyrosination, and thus has the potential to tune the recruitment to the MT of proteins sensitive to the tyrosination status, such as CLIP-170 or MCAK7,15,16.
Figure 7.
Model for TTL action. TTL binds to monomeric tubulin and prevents its incorporation into the MT lattice by occluding the longitudinal interface of the tubulin heterodimer (left panel). TTL acts preferentially on the soluble tubulin pool and detyrosination takes place on MTs. This results in an asymmetry in the distribution of Tyr-tubulin within the MT, with tyrosinated tubulin enriched at the growing plus end where it can aid in the recruitment of proteins sensitive to the tubulin tyrosination status such as plus-end tracking proteins (+TIPS) (right panel).
TTL levels vary between wild-type and cancer cells52, as well as between different tissues and developmental stages19,53. It is possible that the changes in MT dynamics and morphology observed in cancer cells may be due not only to down-regulation of tubulin tyrosination and effects on the recruitment of MT tip binding proteins, but also an increase in the polymerization-competent tubulin pool. Studies of TTL levels in various cell types together with comprehensive MT dynamics analyses will be needed to examine TTL’s possible role in affecting the partition between the monomer and polymer pools of tubulin. Further elucidation of the mechanistic details of TTL interaction with tubulin and its effects on MT dynamics may provide insight into the origin of drug resistance of tumors with low TTL levels.
METHODS
Protein Production and X-ray Structure Determination
Xenopus tropicalis tubulin tyrosine ligase (residues 2–377) was expressed in Escherichia coli as a GST-fusion protein and purified by standard chromatography techniques (Supplementary Methods). Crystals of apo TTL (~ 8 mg ml−1) grew at room temperature by hanging drop vapor diffusion in 0.1 M Hepes pH 7.5, 8% (v/v) PEG8000, 8% (v/v) ethylene glycol, 0.05 M MgCl2 with the symmetry of space group C2 with one TTL copy per asymmetric unit (AU) (Unit Cell: a = 116.6 Å, b = 76.2 Å, c = 44.2 Å, β=90.6°; diffraction limit=2.8 Å). More than 90% of the crystals were split and/or weakly diffracting and not suitable for structure determination. Se-Met TTL crystals grew with symmetry C2 in 0.1 M Hepes pH 7.5, 8% (v/v) PEG8000, 12% (v/v) ethylene glycol, 0.05 M MgCl2. The crystals were flash frozen in liquid nitrogen and used for X-ray data collection [Advanced Light Source (ALS), Beamlines 8.2.1 and 8.2.2]. Se-Met MAD data were collected at two X-ray wavelengths, corresponding to the high-energy remote and inflection of the Se K-absorption edge (Table 1). Detailed procedures for structure determination and refinement can be found in Supplementary Methods. The current refinement model of TTL consists of 308 residues, two magnesium ions and 36 waters. Four regions of the polypeptide chain (residues 103–126; residues 155–158; residues 226–259; residues 364–373) are not well resolved in the electron density map and presumed disordered. The current apo TTL crystallographic model at 2.5Å resolution has working and free R factors of 24.7% and 29.6%, respectively (Table 1) with no unfavorable (ϕ,ψ) combinations.
Crystals of TTL–AMPPNP grew with space group symmetry P21 with two TTL copies per AU (Table 1 for unit cell parameters). The structure was solved by molecular replacement using the apo TTL structure as the search model (residues around the active site were excluded from the model). Detailed descriptions of data collection and structure determination for the AMPPNP bound structure can be found in Supplementary Methods. The current refinement model of TTL–AMPPNP consists of 622 residues, seven magnesium ions and 30 water molecules. Four regions of the polypeptide chain (residues 104–125; residues 154–158; residues 229–259; residues 363–371) are not well resolved in the electron density map in either of the molecules in the AU, and are presumed disordered. The current TTL–AMPPNP crystallographic model at 2.5Å resolution has working and free R factors of 25.4% and 28.5%, respectively (Table 1) with no unfavorable (ϕ,ψ) combinations and main chain and sidechain structural parameters consistently better than average. Crystals of TTL–ADP grew with space group symmetry C2 with one TTL copy per AU (see Table 1 for unit cell parameters). The structure was solved by molecular replacement using the apo TTL structure as the search model. Detailed descriptions of data collection and structure determination for the ADP bound structure can be found in Supplementary Methods. The current refinement model of TTL–ADP consists of 312 residues, three magnesium ions and two water molecules. Four regions of the polypeptide chain (residues 103–126; residues 155–158; residues 227–258; residues 364–371) are not well resolved in the electron density map, and are presumed disordered. The current TTL–ADP crystallographic model at 2.9Å resolution has working and free R factors of 23.1% and 31.4%, respectively (Table 1) with no unfavorable (ϕ,ψ) combinations. All structure figures were prepared with PyMOL (http://www.pymol.org/).
Tyrosination Assays
The tyrosination assay with an α–tubulin C-terminal peptide exploits the increased hydrophobicity of the tyrosinated (product) peptide relative to the substrate to separate them through reversed-phase high-performance liquid chromatography (Supplementary Fig. 3 and Supplementary Methods for detailed descriptions). The tubulin tyrosination assay using Tyr-specific antibodies is described in Supplementary Methods.
Analytical Ultracentrifugation
Sedimentation velocity experiments were performed on peak fractions of TTL, tubulin or TTL–tubulin after size exclusion chromatography. Data were analyzed in terms of continuous c(s) distributions using the SEDFIT program and with the discrete species model in the SEDPHAT program58. The c(s) analysis of SV data obtained with TTL and tubulin alone shows in both cases single peak distributions representing TTL monomer with s20,w of 3.36 S and the tubulin dimer with s20,w of 6.15 S, respectively (Fig. 4). The c(s) distributions obtained with protein mixtures containing 1:1 tubulin:TTL ratios at increasing concentrations show two peaks, a small s value peak representing free TTL, with the area of the peak decreasing with increasing protein concentration, and the high s value peak representing the position of the tubulin–TTL complex reaction boundary. A complete description of data collection and analyses for the sedimentation velocity experiments as well as the sedimentation equilibrium experiments with substrate and product α-tubulin peptides can be found in Supplementary Methods.
Small Angle X-ray Scattering
SAXS data for the reconstructions of the Xenopus tropicalis TTL–tubulin complex were collected at the SIBYLS beamline 12.3.1 at the ALS (see Supplementary Methods for details regarding sample preparation and data collection,). SAXS measurements were performed at concentrations of 5.2 mg ml−1, 2.6 mg ml−1, with no evidence of aggregation at the higher concentration. Individual scattering curves were visually inspected for radiation damage prior to averaging. The scattering curves collected at different concentrations were normalized and merged together in PRIMUS59. SAXS ab initio modeling was performed with GASBOR57 and minimization against the experimental scattering data rather than the pair-distribution function. The fourteen selected ab initio models agree well, yielding 1.31 ±0.05 (NSD ± SD). The crystallographic αβ-tubulin model (PDB ID 1JFF44) was fit as a rigid-body into the SAXS envelope manually, and then locally adjusted using the fit-in-map algorithms implemented in CHIMERA60. A complete description of data collection and analyses for the SAXS experiments can be found in Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Ralston for access to beamlines at the Advanced Light Source (Lawrence Berkeley Laboratories), L. Kizub for assistance with molecular biology, protein expression and purification, S. Abrams for making the X. tropicalis TTL and GFP-TTL clones, early imaging efforts and the initial observation with W. Shin that TTL expression affects MT growth rates, W. Shin for microscopy help, R. Sunyer for early live cell imaging and running the tip tracking program set-up with help from Y. Nishimura and K. Myers, and C. Waterman (National Institutes of Health) for mKusabira Orange-EB3 U2OS cells. We are grateful to N. Tjandra for temporary space while our laboratory was under renovation, C. Waterman for access to microscopes and her lab’s expertise, R. Levine and D.-Y. Lee for mass spectrometry analyses and access to their HPLC, and S. Buchanan for crystallization incubator space. A. R.-M. thanks H. Bourne, A. Ferré-D’ Amaré, S. Gottesman, E.D. Korn , N. Tjandra and C. Waterman for support and critical reading of the manuscript. The authors thank the reviewers for their helpful comments regarding the TTL effects on MT dynamics. A.R.-M. is a Searle Scholar and is supported by the intramural program of the National Institutes of Health.
Footnotes
Accession Codes
Atomic coordinates and structure factors for the TTL structure in the apo, ADP and AMPPNP state have been deposited in the Protein Data Bank, accession codes ###, ### and ### (to be provided at proof stage).
SUPPLEMENTARY METHODS
Includes Full Methods, seven figures and associated references.
AUTHOR CONTRIBUTIONS
A.S. purified TTL and TTL mutants, obtained TTL crystals, performed tyrosination and gel filtration assays and wrote the corresponding methods; A.D. processed SAXS data, obtained the reconstructions and wrote the corresponding methods; G.P. performed and analyzed all AUC experiments and wrote the corresponding methods; A.R.-M. grew and flash-froze crystals, collected crystallographic and SAXS data, solved and refined the crystal structures, performed in vitro polymerization assays, live cell imaging, and analyzed MT dynamics data. A.R.-M. conceived the project and planned experiments in consultation with all authors. All authors prepared figures and A.R.-M. wrote the manuscript, which was reviewed by all authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Erck C, et al. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc. Natl. Acad. Sci. U S A. 2005;102:7853–7858. doi: 10.1073/pnas.0409626102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcos S, et al. Tubulin tyrosination is required for the proper organization and pathfinding of the growth cone. PLoS One. 2009;4:e5405. doi: 10.1371/journal.pone.0005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lafanechère L, et al. Suppression of tubulin tyrosine ligase during tumor growth. J. Cell Sci. 1998;111(Pt 2):171–181. doi: 10.1242/jcs.111.2.171. [DOI] [PubMed] [Google Scholar]
- 4.Whipple RA, et al. Epithelial-to-mesenchymal transition promotes tubulin detyrosination and microtentacles that enhance endothelial engagement. Cancer Research. 2010;70:8127–8137. doi: 10.1158/0008-5472.CAN-09-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mialhe A, et al. Tubulin detyrosination is a frequent occurrence in breast cancers of poor prognosis. Cancer Research. 2001;61:5024–5027. [PubMed] [Google Scholar]
- 6.Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- 7.Webster DR, Gundersen GG, Bulinski JC, Borisy GG. Differential turnover of tyrosinated and detyrosinated microtubules. Proc. Natl. Acad. Sci. U S A. 1987;84:9040–9044. doi: 10.1073/pnas.84.24.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gundersen GG, Kalnoski MH, Bulinski JC. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. Cell. 1984;38:779–789. doi: 10.1016/0092-8674(84)90273-3. [DOI] [PubMed] [Google Scholar]
- 9.Bre MH, Pepperkok R, Kreis TE, Karsenti E. Cellular interactions and tubulin detyrosination in fibroblastic and epithelial cells. Biol. Cell. 1991;71:149–160. doi: 10.1016/0248-4900(91)90061-q. [DOI] [PubMed] [Google Scholar]
- 10.Kreis TE. Microtubules containing detyrosinated tubulin are less dynamic. EMBO J. 1987;6:2597–2606. doi: 10.1002/j.1460-2075.1987.tb02550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J. Cell Biol. 1987;104:439–446. doi: 10.1083/jcb.104.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wehland J, Weber K. Turnover of the carboxy-terminal tyrosine of alpha-tubulin and means of reaching elevated levels of detyrosination in living cells. J Cell Sci. 1987;88(Pt 2):185–203. doi: 10.1242/jcs.88.2.185. [DOI] [PubMed] [Google Scholar]
- 13.Peris L, et al. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 2009;185:1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peris L, et al. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J Cell Biol. 2006;174:839–849. doi: 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bieling P, et al. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 2008;183:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisbrich A, et al. Structure-function relationship of CAP-Gly domains. Nat. Struct. Mol. Biol. 2007;14:959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- 17.Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- 18.Liao G, Gundersen GG. Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J. Biol. Chem. 1998;273:9797–9803. doi: 10.1074/jbc.273.16.9797. [DOI] [PubMed] [Google Scholar]
- 19.Barra HS, Rodriguez JA, Arce CA, Caputto R. A soluble preparation from rat brain that incorporates into its own proteins (14 C)arginine by a ribonuclease-sensitive system and (14 C)tyrosine by a ribonuclease-insensitive system. J. Neurochem. 1973;20:97–108. doi: 10.1111/j.1471-4159.1973.tb12108.x. [DOI] [PubMed] [Google Scholar]
- 20.Arce CA, Rodriguez JA, Barra HS, Caputo R. Incorporation of L-tyrosine, L-phenylalanine and L-3,4-dihydroxyphenylalanine as single units into rat brain tubulin. Eur. J. Biochem. 1975;59:145–149. doi: 10.1111/j.1432-1033.1975.tb02435.x. [DOI] [PubMed] [Google Scholar]
- 21.Wloga D, Gaertig J. Post-translational modifications of microtubules. J. Cell Sci. 2010;123:3447–3455. doi: 10.1242/jcs.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raybin D, Flavin M. Enzyme which specifically adds tyrosine to the alpha chain of tubulin. Biochemistry. 1977;16:2189–2194. doi: 10.1021/bi00629a023. [DOI] [PubMed] [Google Scholar]
- 23.Murofushi H. Purification and characterization of tubulin-tyrosine ligase from porcine brain. J. Biochem. 1980;87:979–984. doi: 10.1093/oxfordjournals.jbchem.a132828. [DOI] [PubMed] [Google Scholar]
- 24.Schröder HC, Wehland J, Weber K. Purification of brain tubulin-tyrosine ligase by biochemical and immunological methods. J. Cell Biol. 1985;100:276–281. doi: 10.1083/jcb.100.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell DG, Miller D, Gull K. Tubulin heterogeneity in the trypanosome Crithidia fasciculata. Mol. Cell Biol. 1984;4:779–790. doi: 10.1128/mcb.4.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warn RM, Harrison A, Planques V, Robert-Nicoud N, Wehland J. Distribution of microtubules containing post-translationally modified alpha-tubulin during Drosophila embryogenesis. Cell Motil. Cytoskeleton. 1990;17:34–45. doi: 10.1002/cm.970170106. [DOI] [PubMed] [Google Scholar]
- 27.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat. Rev. Mol. Cell Biol. 2003;4:938–948. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 28.Ersfeld K, et al. Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 1993;120:725–732. doi: 10.1083/jcb.120.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janke C, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 30.van Dijk J, et al. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell. 2007;26:437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Ikegami K, et al. TTLL10 is a protein polyglycylase that can modify nucleosome assembly protein 1. FEBS Lett. 2008;582:1129–1134. doi: 10.1016/j.febslet.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogowski K, et al. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 2009;137:1076–1087. doi: 10.1016/j.cell.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 33.Wloga D, et al. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev. Cell. 2009;16:867–876. doi: 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 35.Fan C, Moews PC, Shi Y, Walsh CT, Knox JR. A common fold for peptide synthetases cleaving ATP to ADP: glutathione synthetase and D-alanine:D-alanine ligase of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1995;92:1172–1176. doi: 10.1073/pnas.92.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galperin MY, Koonin EV. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan C, Moews PC, Walsh CT, Knox JR. Vancomycin resistance: structure of D-alanine:D-alanine ligase at 2.3 Å resolution. Science. 1994;266:439–443. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- 38.Hara T, Kato H, Katsube Y, Oda J. A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 Å resolution. Biochemistry. 1996;35:11967–11974. doi: 10.1021/bi9605245. [DOI] [PubMed] [Google Scholar]
- 39.Esser L, et al. Synapsin I is structurally similar to ATP-utilizing enzymes. EMBO J. 1998;17:977–984. doi: 10.1093/emboj/17.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakai H, et al. Crystal structure of a lysine biosynthesis enzyme, LysX, from Thermus thermophilus HB8. J. Mol. Biol. 2003;332:729–740. doi: 10.1016/s0022-2836(03)00946-x. [DOI] [PubMed] [Google Scholar]
- 41.Wehland J, Schröder HC, Weber K. Isolation and purification of tubulin tyrosine ligase. Meth. Enzymol. 1986;134:170–179. doi: 10.1016/0076-6879(86)34086-2. [DOI] [PubMed] [Google Scholar]
- 42.Rüdiger M, Wehland J, Weber K. The carboxy-terminal peptide of detyrosinated alpha tubulin provides a minimal system to study the substrate specificity of tubulin-tyrosine ligase. Eur. J. Biochem. 1994;220:309–320. doi: 10.1111/j.1432-1033.1994.tb18627.x. [DOI] [PubMed] [Google Scholar]
- 43.Raybin D, Flavin M. An enzyme tyrosylating alpha-tubulin and its role in microtubule assembly. Biochem. Biophys. Res. Commun. 1975;65:1088–1095. doi: 10.1016/s0006-291x(75)80497-9. [DOI] [PubMed] [Google Scholar]
- 44.Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 Å resolution. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 45.Pal D, et al. Conformational properties of alpha-tubulin tail peptide: implications for tail-body interaction. Biochemistry. 2001;40:15512–15519. doi: 10.1021/bi015677t. [DOI] [PubMed] [Google Scholar]
- 46.Wehland J, Weber K. Tubulin-tyrosine ligase has a binding site on beta-tubulin: a two-domain structure of the enzyme. J. Cell Biol. 1987;104:1059–1067. doi: 10.1083/jcb.104.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deans NL, Allison RD, Purich DL. Steady-state kinetic mechanism of bovine brain tubulin: tyrosine ligase. Biochem. J. 1992;286(Pt 1):243–251. doi: 10.1042/bj2860243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matov A, et al. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods. 2010;7:761–768. doi: 10.1038/nmeth.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravelli RB, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 50.Cormier A, et al. The PN2-3 domain of centrosomal P4.1-associated protein implements a novel mechanism for tubulin sequestration. J. Biol. Chem. 2009;284:6909–6917. doi: 10.1074/jbc.M808249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiller G, Weber K. Radioimmunoassay for tubulin: a quantitative comparison of the tubulin content of different established tissue culture cells and tissues. Cell. 1978;14:795–804. doi: 10.1016/0092-8674(78)90335-5. [DOI] [PubMed] [Google Scholar]
- 52.Kato C, et al. Low expression of human tubulin tyrosine ligase and suppressed tubulin tyrosination/detyrosination cycle are associated with impaired neuronal differentiation in neuroblastomas with poor prognosis. Int. J. Cancer. 2004;112:365–375. doi: 10.1002/ijc.20431. [DOI] [PubMed] [Google Scholar]
- 53.Deanin GG, Gordon MW. The distribution of tyrosyltubulin ligase in brain and other tissues. Biochem. Biophys. Res. Commun. 1976;71:676–683. doi: 10.1016/0006-291x(76)90841-x. [DOI] [PubMed] [Google Scholar]
- 54.Deprez C, et al. Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J. Mol. Biol. 2005;346:1047–1057. doi: 10.1016/j.jmb.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 55.Rocchia W, et al. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J. Comput. Chem. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 56.Schuck P. Diffusion-deconvoluted sedimentation coefficient distributions for the analysis of interacting and non-interacting protein mixtures. Cambridge: The Royal Society of Chemistry; 2005. [Google Scholar]
- 57.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 60.Pettersen E, et al. UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.