Highlights
► Current reporter systems for translational readthrough are gene-specific and cannot be used in living cells. ► We developed a readthrough reporter based on green fluorescent protein. ► The reporter is gene-independent and can be used to sort living cells based on fluorescence.
Keywords: Aminoglycoside, Readthrough reporter, Premature termination codon
Abbreviations: PTC, premature termination codon; GFP, green fluorescent protein; DMD, Duchenne muscular dystrophy; CF, cystic fibrosis; NMD, nonsense mediated decay
Abstract
Agents to induce readthrough of premature termination codons (PTCs) are useful research tools and potential therapeutics. Reporters used to detect PTC readthrough are gene-specific and thus are not suited to for general assessment of readthrough activity or in cases where PTC-inactivated genes are unknown. Here we describe a GFP-based reporter construct pMHG-W57∗ which is capable of detecting dose-dependent drug-induced PTC readthrough both by fluorescence microscopy and flow cytometry. pMHG-W57∗ may be used as a general indicator of PTC readthrough in living cells and obviates the need for gene-specific recoding sequences in reporter constructs.
1. Introduction
Almost one third of all heritable disorders result from premature termination codons (PTCs) mutations [1]. PTCs may result from nonsense or frameshift mutations, as well as exon skipping or splice site intron inclusions. Heritable disorders where premature stop codon mutations (TAA, TAG or TGA) commonly occur include Duchenne muscular dystrophy (DMD) (10–20% of patients) and cystic fibrosis (CF) (2–5% of patients) [2], Furthermore, PTCs may occur in tumor suppressor genes. In general, these mutations result in the activation of nonsense mediated decay (NMD), whereby PTC-containing mRNAs are targeted for rapid degradation. Inhibition of NMD to uncover PTCs, called gene identification by NMD Inhibition (GINI), was first described in colon cancer and used to confirm a known PTC in MLH1, among others [3]. Subsequent studies have successfully used this strategy to uncover inactivating tumor suppressor mutations in colon and prostate cancer and in melanoma [4–6].
Strategies for correction of PTCs have focused on the identification of compounds which allow the translational machinery to overcome aberrant termination codons. One such class of compounds are aminoglycosides, which have been widely used as antibiotics for several decades (e.g. neomycin, kanamycin, paromomycin) [7]. Aminoglycosides bind with high specificity to the 16S ribosome in prokaryotes, resulting in altered ribosomal conformation and loss of codon–anticodon fidelity. Loss of fidelity causes incorporation of incorrect amino acids at sense codons or failure to recognize stop codons. Aminoglycoside specificity in prokaryotes is mediated through adenine 1408 in the 16S ribosome. In eukaryotic ribosomes, the corresponding nucleotide is a guanine, resulting in lower binding affinity. Consequently, higher doses of aminoglycosides are required to affect protein translation in eukaryotes (>10 times therapeutic dose in prokaryotes).
Many groups have demonstrated the ability of aminoglycoside compounds to circumvent PTCs and restore synthesis of full-size proteins. Both G-418 (geneticin) and gentamicin promote readthrough of nonsense mutations (R553X and G542X) in the cystic fibrosis transmembrane conductance regulator (CFTR), resulting in full length protein expression and restoration of cyclic-AMP activated chloride ion channel activity [8]. Significant readthrough was achieved with aminoglycoside concentrations of 0.05–0.2 mg/ml. The first in vivo demonstration of aminoglycoside-induced readthrough was carried out in the DMD mouse model (mdx mouse) [9]. Gentamicin (0.1–0.4 mg/ml) provided restoration of functional dystrophin with increases in myotube size and number [9]. Other genetic diseases in which aminoglycoside therapy has been applied successfully include hemophilia and ataxia–talangiectasia [10,11]. More recently, the novel synthetic readthrough agent PTC124 (Ataluren®) showed promising results in mdx mice with low levels of toxicity [12].
The identification and characterization of PTC readthrough-inducing compounds necessitates the use of reporter systems. The most commonly used system is based on the dual luciferase reporter assay, and consists of a recoding sequence inserted between firefly and renilla luciferase genes [13]. The ratio of the two luciferase activities provides a measure of translational efficiency associated with the test sequence. When a recoding event occurs (e.g. readthrough of a stop codon), an increase in firefly luciferase activity is observed, but in the absence of such an event no activity is observed. This system was successfully used to assess −1 and +1 frameshifting, as well as readthrough events [13]. Others have adapted the system by modifying the recoding sequence according to the gene of interest, e.g. APC [14] or DMD [15].
Here we describe the generation and characterization of a green fluorescent protein (GFP) reporter for PTC readthrough assessment (pMHG-W57∗). Unlike, gene-specific reporters the recoding event in pMHG-W57∗ occurs in the reporter protein (i.e. GFP). Thus, the reporter may be used as a general indicator of PTC readthrough in cell culture models. pMHG-W57∗ can be useful for optimizing readthrough conditions when many types of recoding events are possible, e.g. nonsense and frameshift mutations in cancer cells.
2. Materials and methods
2.1. Generation of pMHG-W57∗ reporter
A GFP (S65T redshifted variant)-containing plasmid, pMONO-hygro-gfp (pMHG) (Invivogen, San Diego, CA) was used for the generation of the readthrough reporter. The plasmid contains a ferritin heavy chain (FerH) promoter fused to the SV40 enhancer at its 5′ end and the mouse elongation factor 1 alpha gene at its 3′ end. Hygromycin B resistance is provided by the Escherichia coli phosphotransferase gene hph. Site-directed mutagenesis of GFP tryptophan 57 codon to a stop codon (W57∗) (G171A, mRNA) was carried out using the Quikchange® XL mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s guidelines. An opal stop codon (TGA) was introduced using the following primers (forward, G CTG CCA GTG CCT TGA CCA ACT CTG GTG AC; reverse, GTC ACC AGA GTT GGT CAA AGG CAC TGG CAG C). The W57∗ mutation in pMHG-W57∗ was confirmed by direct sequencing.
2.2. Cell culture and transient transfections
HCT-116 colorectal cancer cells were obtained from the American Tissue Type Collection and maintained in McCoy’s 5A media supplemented with 10% fetal bovine serum. Cells were passaged every 2–3 days. Cells were seeded in 96-wells plates (3 × 104/well) or 12-well plates (2 × 105/well) 24 h prior to transfection. Cells were transfected using lipofectamine 2000 (Invitrogen, Carlsbad, CA) reagent with pMHG or pMHG-W57∗ (320 ng/well DNA for 96-well plate and 1.6 μg/well DNA for 12-well plate). After 24 h, cell media was removed and cells were treated with Geneticin® (G-418) (0–2000 μg/ml) for 24 h. GFP in transfected cells was imaged using a Zeiss Axiovert 25 inverted microscope (Carl Zeiss, Thornwood, NY) with a fluorescein isothiocyanate (FITC) filter at 50× magnification. A Zeiss Axiocam MR camera was used to capture fluorescent cell images.
2.3. Flow cytometry
After transfection and treatment cells were cells were detached from plates using trypsin and resuspended in McCoy’s 5A media. Green fluorescent cells in pMHG/pMHG-W57∗ transfected cells were quantified using a LSRII flow cytometer (BD Biosciences, San Hose, CA), running FACSDiva software (v 6.1.2). A negative control (non-transfected, non-treated cells) and a positive control (pMHG transfected cells) were used to optimize voltages and detection thresholds for GFP fluorescence. Two biological replicates were analyzed for each condition.
3. Results
3.1. pMHG-W57∗ reporter generation
Assessment of chemically-induced PTC readthrough is enabled by the use of reporter systems. Most PTC readthrough reporter constructs contain a recoding sequence which lies upstream of a reporter enzyme such as luciferase. This approach requires the generation of a new construct containing a recoding sequence specific to each gene of interest, as well as each recoding event in that gene (e.g. DMD, CFTR). However, if multiple recoding events occur in a single biological system, it may be more advantageous to use a more general readthrough reporter. Therefore, we generated a PTC readthrough reporter, pMHG-W57∗, which can be used as a general indicator of readthrough efficiency. A PTC mutation has been introduced into the coding sequence of GFP (W57∗). In the absence of translational readthrough a truncated non-functional protein is generated, while readthrough results in fully functional, fluorescent GFP (Fig. 1). Therefore, global readthrough can be tested by parallel transient transfection of pMHG-W57∗ and pMHG (wildtype GFP). GFP fluorescence in pMHG-W57∗ treated cells can be normalized to pMHG fluorescence in order to calculate readthrough efficiency.
Fig. 1.
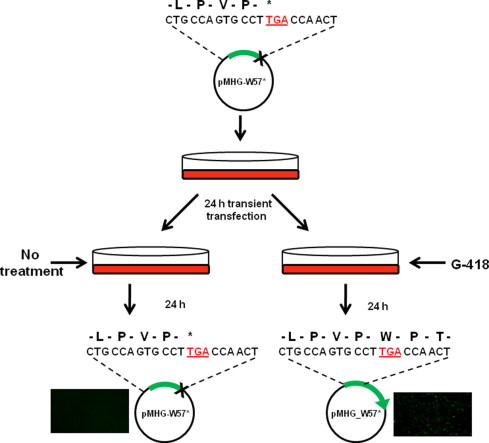
Schematic representation of pMHG-W57∗ reporter for premature termination codon (PTC) readthrough.
3.2. Fluorescence imaging of pMHG-W57∗ transfected cells
HCT-116 colorectal cancer cells were transiently transfected with either pMHG or pMHG-W57∗ and subsequently treated with increasing concentrations of G-418 (0–2000 μg/ml). Cells were examined by fluorescence microscopy. No fluorescent cells were observed in untreated pMHG-W57∗ transfected cells. In G-418-treated, pMHG-W57∗-transfected cells a dose-dependent increase in fluorescent cells was observed (Fig. 2). The highest G-418 concentration (2000 μg/ml) generated the largest number of fluorescent cells, but less than observed in pMHG-transfected cells, indicating that chemically-induced readthrough is relatively inefficient.
Fig. 2.
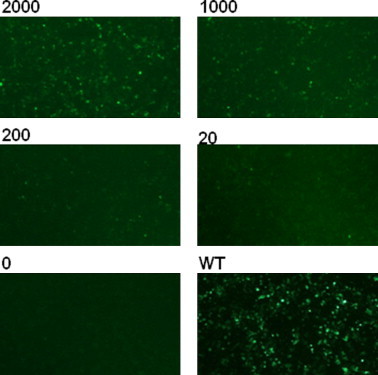
Assessment of chemically-induced readthrough by G-418 in pMHG-W57∗-transfected cells by fluorescence microscopy. HCT-116 cells were transiently transfected with pMHG-W57∗ or pMHG (wildtype) for 24 h and then treated with G-418 (20–2000 μg/ml) for a further 24 h.
3.3. Quantitative assessment of readthrough by flow cytometry
In order to provide a relative quantitative assessment of readthrough efficiency in G-418-treated, pMHG-W57∗ transfected cells, GFP-positive cells were quantified by flow cytometry. As before, a dose-dependent increase in GFP-positive cells was seen in G-418-treated cell cultures. In pMHG-W57∗-transfected cells, maximum readthrough (5.5% GFP positive) was achieved at the 2000 μg/ml G-418 level (Fig. 3A). Significantly higher levels of GFP-positive cells were observed in pMHG-transfected cells (26.5%). When readthrough levels in G-418-treated cells are normalized to pMHG levels, readthrough efficiencies ranged between 1% (20 μg/ml) and 20% (2000 μg/ml) (Fig. 3B). This result is in general agreement with previously reported efficiencies for chemically induced readthrough in cultured cells. The data demonstrate that pMHG-W57∗ can serve as a sensitive, quantitative reporter for PTC readthrough and is capable of detecting dose-dependent recoding of PTCs in cultured cells.
Fig. 3.
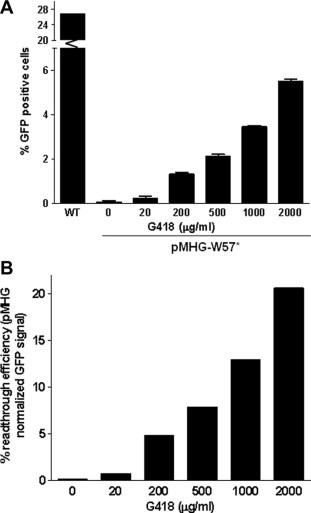
Quantitative evaluation of readthrough by flow cytometry. (A) HCT-116 cells were transiently transfected with pMHG-W57∗ or pMHG (wildtype) for 24 h and then treated with G-418 (20–2000 μg/ml) for a further 24 h. GFP positive cells were quantified by flow cytometry. (B) GFP positive cell numbers in G-418-treated, pMHG-W57∗-transfected cells were normalized to pMHG-transfected cell number in order to estimate readthrough efficiency values.
4. Discussion
Chemically induced readthrough of PTCs has become a widely adopted strategy for the restoration of normal protein expression in several human genetic disorders [7] and a tool to identify genes inactivated by PTCs [3]. Although most readthrough reagents result in relatively low levels of restoration, in many diseases this may be sufficient to cause significant clinical improvements. This is especially true of recessive disorders where the protein of interest is usually absent. Several studies have demonstrated the utility of aminoglycosides as readthrough inducers in conditions such as CF, DMD and hemophilia [9,10,13]. However, at high doses aminoglycosides may interfere with normal protein translation, resulting in significant toxicity. Therefore, establishing sub-toxic, therapeutic concentrations is critical for the development of readthrough reagents.
Here we describe a novel reporter system, pMHG-W57∗, for the assessment of PTC readthrough in cultured cells which can be used to aid in the characterization and investigation of readthrough reagents. Most reporter plasmids contain a recoding sequence specific to the gene under investigation, as well as a reporter enzyme whose expression is indicative of a readthrough event. In pMHG-W57∗, the recoding event occurs in the reporter protein (GFP), thus obviating the need for a gene-specific recoding sequence. We have successfully demonstrated by fluorescence microscopy and flow cytometry that pMHG-W57∗ detected readthrough over a range of G-418 concentrations (20–2000 μg/ml) and that GFP positive cells increased in a dose-dependent manner. Further experiments will be required to see how pMHG-W57∗ performs with readthrough drugs other than G-418.
As a GFP-based reporter, pMHG-W57∗ offers several advantages over conventional luciferase-based readthrough reporters. Readthrough may be assessed directly in intact cells by fluorescence microscopy or flow cytometry. After readthrough measurement, cells can be sorted and collected, allowing for further experimental manipulation. Luciferase reporters require that cells be lysed and a substrate is needed to generate luminescence, thus preventing further analysis of the intact cells.
Because a gene-specific resequencing code is not required by pMHG-W57∗, it can be used as a general indicator to test compounds for NMD-inhibition and to compare cell lines for readthrough competency. For example, in cancer cells gene identification by NMD inhibition (GINI) has been used to detect genes containing inactivating mutations [3]. By preventing NMD-mediated degradation of PTC-containing transcripts, novel cancer-related mutations may be identified [4–6]. NMD suppression coupled with aminoglycoside-induced translational readthrough would allow novel mutants to be detected at the protein level. It should be noted that gene-specific reporters may be better suited for the assessment of readthrough when sequence context is a major factor in determining efficiency. Nonetheless, the use of a general readthrough indicator such as our GFP-based model could help in the screening of novel compounds and in optimization of treatment conditions.
In conclusion, we generated a reporter construct which is capable of detecting dose-dependent chemically-induced PTC readthrough in viable cultured cells. The use of a GFP-based reporter which does not require gene-specific recoding sequence may offer several advantages over luciferase-based reporters.
Acknowledgement
This work was supported by NIH Grant U24 CA126479.
References
- 1.OMIM – Online Mendelian Inheritance in Man. National Center for Biotechnology Information.
- 2.The Leiden DMD mutation database. Center for Human and Clinical Genetics, Leiden University Medical Center.
- 3.Noensie E.N., Dietz H.C. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol. 2001;19:434–439. doi: 10.1038/88099. [DOI] [PubMed] [Google Scholar]
- 4.Bloethner S., Mould A., Stark M., Hayward N.K. Identification of ARHGEF17, DENND2D, FGFR3, and RB1 mutations in melanoma by inhibition of nonsense-mediated mRNA decay. Genes Chromosom. Cancer. 2008;47:1076–1085. doi: 10.1002/gcc.20598. [DOI] [PubMed] [Google Scholar]
- 5.Huusko P. Nonsense-mediated decay microarray analysis identifies mutations of EPHB2 in human prostate cancer. Nat. Genet. 2004;36:979–983. doi: 10.1038/ng1408. [DOI] [PubMed] [Google Scholar]
- 6.Ionov Y., Nowak N., Perucho M., Markowitz S., Cowell J.K. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer cells with microsatellite instability. Oncogene. 2004;23:639–645. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- 7.Zingman L.V., Park S., Olson T.M., Alekseev A.E., Terzic A. Aminoglycoside-induced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy. Clin. Pharmacol. Ther. 2007;81:99–103. doi: 10.1038/sj.clpt.6100012. [DOI] [PubMed] [Google Scholar]
- 8.Howard M., Frizzell R.A., Bedwell D.M. Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations. Nat. Med. 1996;2:467–469. doi: 10.1038/nm0496-467. [DOI] [PubMed] [Google Scholar]
- 9.Barton-Davis E.R., Cordier L., Shoturma D.I., Leland S.E., Sweeney H.L. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J. Clin. Invest. 1999;104:375–381. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James P.D., Raut S., Rivard G.E., Poon M.C., Warner M., McKenna S., Leggo J., Lillicrap D. Aminoglycoside suppression of nonsense mutations in severe hemophilia. Blood. 2005;106:3043–3048. doi: 10.1182/blood-2005-03-1307. [DOI] [PubMed] [Google Scholar]
- 11.Lai C.H., Chun H.H., Nahas S.A., Mitui M., Gamo K.M., Du L., Gatti R.A. Correction of ATM gene function by aminoglycoside-induced read-through of premature termination codons. Proc. Natl. Acad. Sci. USA. 2004;101:15676–15681. doi: 10.1073/pnas.0405155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welch E.M. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 13.Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 14.Zilberberg A., Lahav L., Rosin-Arbesfeld R. Restoration of APC gene function in colorectal cancer cells by aminoglycoside- and macrolide-induced read-through of premature termination codons. Gut. 2010;59:496–507. doi: 10.1136/gut.2008.169805. [DOI] [PubMed] [Google Scholar]
- 15.Howard M.T., Shirts B.H., Petros L.M., Flanigan K.M., Gesteland R.F., Atkins J.F. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 2000;48:164–169. [PubMed] [Google Scholar]


