Abstract
Glioblastoma multiforme (GBM) is a histopathologically heterogeneous disease with few treatment options. Therapy based on genomic alterations is rapidly gaining popularity because of the high response rate and high specificity. DNA copy number and exon sequencing studies of GBM samples have revealed recurrent genomic alterations in genes such as TP53, EGFR and IDH1 but to date this has not resulted in novel GBM therapies. Identification of expression subtypes have resulted in new insights such as the association between genomic abnormalities and expression signatures. This review describes the types of genomic studies that have been performed and that are underway, the most prominent results and the implications of genomic research for development of clinical treatment modalities.
Keywords: Glioblastoma, genomics, sequencing, personalized medicine, expression profiling, molecular subtypes
Background
Glioblastoma multiforme (GBM) is the most common primary brain tumor in adults. Currently, less than 3% of GBM patients in the United States and Europe will survive more than 5 years1. Care of GBM patients is in dire need of better treatment options. The current standard of care for GBM therapy is based on treatment with the alkylating agent temozolomide and ionizing radiation, which is aimed at causing DNA damage in the fast growing tumor cells. Methylation of the promoter of methyltransferase gene MGMT is a predictor of temozolomide response, but lack of alternative treatment options renders this biomarker clinically ineffective2. In the current era wherein whole genome sequencing technology is a reality, using targeted agents based on tumor specific genetic alterations has become feasible and provides a hopeful avenue towards new GBM treatment modalities3, 4.
High throughput genomic experiments have been broadly applied to catalogue somatic alterations that affect this fatal disease. Genomic characteristics, such as gene expression, somatic copy number alterations and somatic gene mutations have been interrogated and correlated to clinical factors5–26. Because of its dim prognosis, GBM was selected as the first cancer type studied by The Cancer Genome Atlas (TCGA) project, initially launched in 2005. TCGA aims to generate a comprehensive catalog of genomic abnormalities through application of whole genome copy number arrays, expression arrays, methylation arrays, exome sequencing, microRNA arrays on a large cohort of patient tumor samples, combined with high quality clinical data. TCGA has qualified over 500 GBM tumor samples with matching normal tissue samples for multidimensional genomic interrogation, which is aimed to be completed in 2012 (http://cancergenome.nih.gov).
In this review paper, we discuss the recent advances of our understanding of GBM, inspired by TCGA and other studies that resulted from the analysis of high dimensional genomic data.
Pathology and intratumoral heterogeneity of GBM
The histopathological diagnosis of glioblastoma, also known as stage IV glioma, requires presence of anaplastic glial cells, brisk mitotic activity, and vascular proliferation and/or necrosis16, 27, 28. Inter-observer variation in pathology review is rare for the diagnosis GBM but has been reported for lower grade gliomas29–31. However, significant intratumoral heterogeneity is found which may have a profound impact on the accuracy of genome alteration identification. A subset of GBMs shows intratumoral variation in the level of MGMT promoter methylation, suggesting the presence of a mixture of different clonal populations32. Investigation of gene expression profiles revealed differences between different samples of the same tumor, which were unrelated to differences in histology6. Comparison of genome wide copy number of different areas of the same tumor showed that glioblastomas may harbor genetic alterations common to all areas analyzed, but also area specific alterations33, 34. The same GBM may harbor different TP53 mutations in different tumor locations35.
These data suggest that GBM can contain both ancestral genomic abnormalities as well as genomic alterations unique to different areas of the same tumor. Such a tumor population structure can be the result of a clonal evolution process, in which selective pressure continuously results in growth advantages for the better adapted clones36. Not every GBM appears to contain regional heterogeneity, allowing the possibility for alternative theories, such as the cancer stem cell theory. In this model, each cell develops from a cancer stem cell and is therefore genomically homogeneous accept for sets of mutations acquired during the differentiation process from stem cell to bulk tumor cell37. It seems plausible that both models are not mutually exclusive and can be found in GBM.
The fact that GBMs can be polygenomic tumors that maintain multiple clonal populations has important implications for the design of studies that genomically interrogate GBM samples and clinical trials38. Ultra deep sequencing or assaying multiple tumor sections may be a requirement to adequately characterize GBM samples. Importantly, although targeted molecular cancer therapies such as gefitinib treatment of EGFR kinase domain mutations in lung adenocarcinoma are highly effective, acquired therapy resistance remains an important clinical problem and may be the result of tumor polyclonality4, 39.
Gene expression profiling identifies prognostic and subtype GBM signatures
Several groups have turned to gene expression profiling to classify morphologically indistinguishable gliomas into disease relevant subtypes9, 13, 14, 16, 18, 23, 40. Supervised expression profiling studies aim to establish signatures that recognize specific subsets of tumor samples, and this study design has been applied to improve the prediction of survival over histopathology, to predict response to temozolomide therapy, and to measure EGFR activation9, 14, 16, 40. Unsupervised studies have been aimed at grouping all tumor samples into naturally occurring expression subtypes13, 18, 23. Philips et al. used a cohort of 107 grade III and grade IV astrocytoma samples to define a 35 gene signature that defined subclasses named Proneural, Mesenchymal and Proliferative (Figure 1A)18. Importantly, the Proneural subclass, containing the majority of grade III astrocytomas, showed prolonged survival compared to the Mesenchymal and Proliferative subclasses even when grade III cases were excluded.
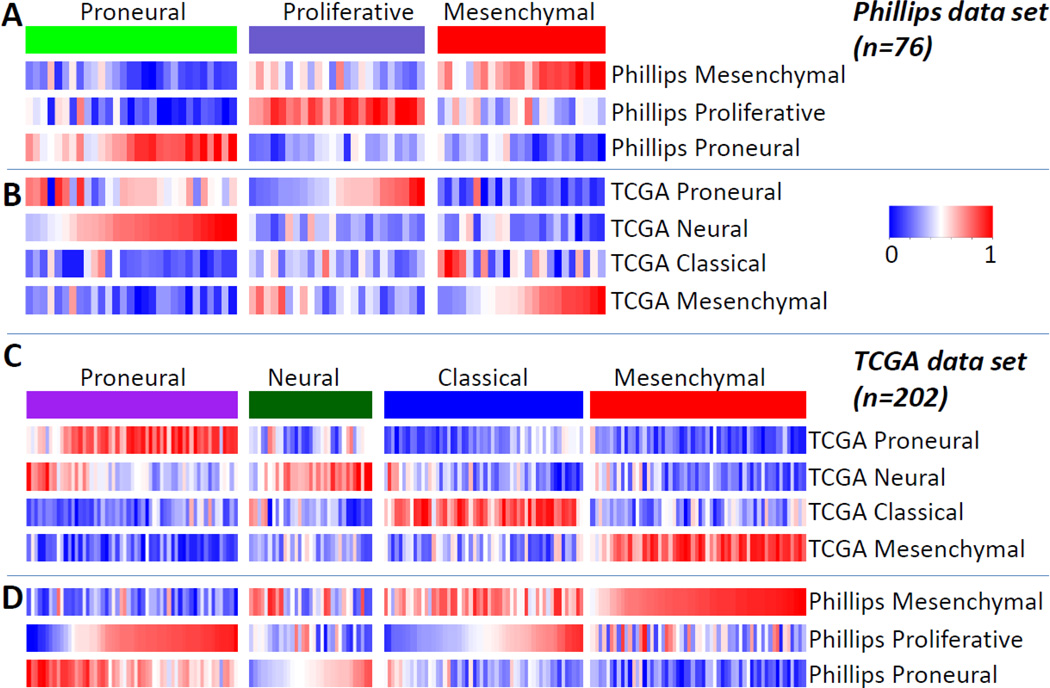
Figure 1.
Single sample Gene Set Enrichment Analysis of the TCGA and Phillips et al expression data sets. Single sample Gene Set Enrichment Analysis (ssGSEA) assesses gene set activation levels based on gene expression levels of individual samples. The figure shows ssGSEA scores for the TCGA/Verhaak et al expression profiles (n = 202) and Phillips et al expression profiles (n = 76). Gene set scores were row-normalized to the same scale for each gene set.
A. ssGSEA scores of the three Phillips et al subtype signatures and Phillips expression profiles
B. ssGSEA scores of the four TCGA subtype signatures and Phillips et al expression profiles
C. ssGSEA scores of the four TCGA subtype signatures and TCGA expression profiles
D. ssGSEA scores of the three Phillips et al subtype signatures and TCGA expression profiles
We analyzed 202 expression profiles available through TCGA to establish four GBM subtypes which we termed Proneural, Neural, Classical and Mesenchymal (Figure 1C)23. Our approach differed from the results reported by Phillips et al in two important ways: 1) only GBMs were included and 2) 1,740 variably expressed genes, not specifically related to survival, were used to define the clusters. The patterns of these subtypes were well recapitulated by an independent validation data set consisting of 260 samples, including those analyzed by Phillips et al9, 10, 18, 41.
Single sample gene set enrichment analysis (ssGSEA) is a method that measures the activation level of a gene signature in an individual expression profile, independent of other profiles in the same data set. Combined analysis of the Phillips et al and TCGA data sets using both sets of gene signatures and ssGSEA shows that the Phillips et al cohort contains few samples characterized by the Classical signature, potentially prohibiting the identification of the Classical subgroup on the basis of this data set. Interestingly, the Phillips et al Proneural subset resembles a mixture of TCGA Proneural and Neural classes. The Phillips Proliferative class contains samples from both the TCGA Proneural and Classical samples. Both studies identified a Mesenchymal subtype which recapitulates a very similar set of samples.
Currently, gene expression subtypes have no direct clinical relevance due to the lack of alternative treatment options. However, the existence of transcriptomic classes suggests that each GBM cannot be treated similarly and that characterization of patient samples prior to inclusion into clinical trials may provide important new insights into why certain patients are more or less responsive to certain therapies. Examples of these are the efficacy of angiogenesis inhibitors specifically in the mesenchymal class, which signature suggests strong activation of angiogenesis gene sets, and the application of inhibitors of the mammalian target of rapamycin (mTOR) protein in the proneural group, whose expression phenotype suggests that PI(3)K/AKT pathway plays an important role.
Methylation profiling defines a G-CIMP phenotype
Genome wide DNA methylation profiling identified a subset of 9% of GBMs with a hypermethylated phenotype, known as the G-CIMP15. G-CIMP GBMs are younger at time of diagnosis, show significantly prolonged survival and typically express the Proneural expression signature. Interestingly, the G-CIMP subclass entirely explains the increased survival associated with the Proneural subtype. This observation holds for both Verhaak classification23 and Philips classification18, suggesting that this methylation defined subtype is more robust in predicting prognosis than the gene expression based subtypes.
Genomic analysis identifies new oncogenes and tumor suppressor genes in GBM
Copy number gains and activating mutations of proto-oncogenes provide proliferative advantage to tumor cells whereas deletions and disrupting mutations targeting tumor suppressor genes remove barriers that suppress cell growth. An overview of genes that have been repeatedly reported to be genomically altered in GBMs shows that similar numbers of amplified and deleted genes have been identified so far, suggesting that no specific emphasis on activation of oncogenes versus deactivation of tumor suppressors genes exists (Table 1). Early on, cytogenetic studies have shown the nearly uniform co-occurence of heterozygous loss of chromosome 10, leading to reduced transcript levels of cell cycle regulator PTEN, and low level gain of chromosome 7, which harbors receptor tyrosine kinase (RTK) signaling gene EGFR and the proto-oncogene MET10, 20, 42. Whole genome copy number analysis by TCGA and others allowed the study of these abnormalities in much higher detail, as well as the pinpointing of new genomic alterations. Interestingly, these analyses showed that genes and proteins can be activated or de-activated through multiple mechanisms. For instance, the gene EGFR is focally amplified at high levels in 30–40% of GBM. A subset of EGFR-amplified tumors contain EGFR point mutations, whereas another subset maintains activating deletions of exons 2–7 (EGFR variant III or EGFRvIII). Both the deletion and point mutations are found in the extracellular domain. Deletion of the p16 locus which contains the genes CDKN2A and CDKN2B often co-occurs with focal high level EGFR amplification, but few mutations in these genes have been found. Germline single nucleotide variants near the p16 locus have been identified that increase the risk of glioma43, 44.
TABLE 1.
Gene Abnormalities in GBM.
| Gene | Location | Mutation | Amplification | Deletion | Intragenic deletion |
Methylation | Pathway | Reference |
|---|---|---|---|---|---|---|---|---|
| CDKN2C | 1p32 | hom 2%20 | Cell cycle | 20, 42, 54 | ||||
| ERRFI1 | 1p36 | hom 3%20 | RTK signaling | 20, 43 | ||||
| IGFBP2 | 1p36.22 | Y44 | Cell migration and invasion | 44 | ||||
| CHD5 | 1p36.31 | het 30%94 | Chromatin structure regulation | 20, 45, 94 | ||||
| MDM4 | 1q32 | 7%20 | p53 signaling | 20, 80 | ||||
| AKT3 | 1q44 | 2%20 | RTK signaling | 20, 94 | ||||
| MYCN | 2p24.3 | 2%20 | Transcription regulation | 7, 20 | ||||
| LRP1B | 2q21.2 | het 7%47 | 7%47 | Signal transduction | 20, 47, 95 | |||
| IDH1 | 2q33.3 | 12%8 | Cellular respiration and metabolism | 8, 48 | ||||
| PIK3CA | 3q26.3 | 7%20 | 2%20 | RTK signaling | 20, 49 | |||
| SOX2 | 3q26.3-q27 | 1496 | Cell differentiation | 20, 96, 97 | ||||
| PDGFRA | 4q12 | 3%23 | 13%20 | 3%17 | RTK signaling | 20, 17, 23 | ||
| PIK3R1 | 5q13.1 | 10%20 | RTK signaling | 8, 20 | ||||
| PARK2 | 6q25.2-q27 | 9%50 | het 22%50 hom 2%50 |
2%50 | Signal transduction | 20, 50 | ||
| QKI | 6q26 | het 2547 hom 4%47 |
mRNA splicing, transportation/cell differentiation | 20, 47 | ||||
| EGFR | 7p12 | 18%20 | 42%20 | 20–30%98 | RTK signaling | 20, 51, 98 | ||
| CDK6 | 7q21-q22 | 1%20 | Cell cycle | 20, 52 | ||||
| MET | 7q31 | 2%20 | 4%20 | RTK signaling | 20, 53 | |||
| CDKN2A/2B | 9p21 | hom 50%20 | Cell cycle | 20, 55 | ||||
| PTPRD | 9p23-p24.3 | 6%22 | 40%22 | Y22 | 37%22 | Signal transduction | 20, 22 | |
| PTEN | 10q23.3 | 31%20 | hom 8%20 | RTK signaling | 20, 56 | |||
| CDK4 | 12q14 | 18%20 | Cell cycle | 20, 55, 57 | ||||
| MDM2 | 12q14.3-q15 | 14%20 | p53 signaling | 20, 58 | ||||
| RB1 | 13q14.2 | 10%20 | hom 3%20 | Cell cycle | 20, 59 | |||
| NFKBIA | 14q13 | het 23%11 | NF-kappa-B signaling | 11 | ||||
| TP53 | 17p13.1 | 34%20 | hom 1%20 | Cell cycle/apoptosis | 20, 60 | |||
| NF1 | 17q11.2 | 14%20 | hom 2%20 | RTK signaling | 20 | |||
| CCNE1 | 19q12 | Y20 | Cell cycle | 20 | ||||
| BCL2L12 | 19q13.3 | Y61 | p53 signaling | 61 | ||||
| IL6 | 7p21 | 42%100 | Growth arrest and differentiation | 62, 99, 100 | ||||
| MGMT | 10q26 | 21%20 | DNA repair | 2, 20 | ||||
| CCND2 | 12p13 | 2%20 | Cell cycle | 20, 63 |
hom, homozygous; het, heterozygous; Y, reported but no specific percentage available.
Exon sequencing analysis allows identification of different types of mutations, such as single nucleotide variants (SNVs) that lead to altered amino acids, alternative stop codons, alternative mRNA splice sites, alternative transcription start sites and small insertions and deletions. This methodology was applied by TCGA, who used Sanger sequencing to analyze 91 samples and 601 genes which found both known mutations such as in TP53 and EGFR, as was well as novel mutations such as in PIK3R1 and NF120. At the same time, a group at Johns Hopkins University performed exome wide sequencing on 22 samples, similarly recognizing known mutations as well novel mutations, most notably in the isocytrate dehydrogenase gene IDH18. TCGA is currently performing genome wide characterization of its entire cohort of approximately 500 samples, scheduled for completion in 2012. Identification of sequence variants is dependent on the tumor purity of a sample, as well as the allele frequency of a mutation. For instance, if a gene is amplified and a mutation is present in only one out of four alleles, and the tumor purity is 80%, the fraction of sequence reads of that position that covers the mutation is 22%. SNV discovery is further affected by mutations that may only be present in a subset of the cells in a tumor. Effective mutation discovery thus requires sufficient sequencing depth to account for these factors, but may aid to assess the clonality of a tumor sample through comparison of allele frequencies.
Genome wide sequencing can additionally identify intra- and interchromosomal rearrangements. This methodology was applied to the U87 glioma cellline and showed that there was only a limited overlap in comparison to TCGA results, suggesting an intrinsic difference in the genetic context of this cell line and human primary GBM samples45.
Integrated analysis of genomic data
The TCGA data set presented a unique opportunity to characterize gene expression subtypes by genomic abnormalities23. A number of significant associations between subtype and gene alterations were found, which suggests that the expression pattern is an important determinant of the growth enhancing ability of specific genomic alterations. An example of this is the preference of PDGFRA amplifications and mutations to occur within the Proneural expression group. The Proneural signature resembles the signature of normal oligodendrocyte progenitor cells, whereas PDGFRα protein expression is an important marker of normal oligodendrocyte development46. Moreover, a recent study suggests the oligodendrocyte progenitor as a cell of origin for Proneural GBMs47. PDGFRα activation may thus be an important step in the ability to maintain a progenitor like cell state for tumors arising from the oligodendrocyte lineage46.
EGFR signaling is an important regulator of astrocyte reactivity and proliferation48. Interestingly, the Classical subtype shows high activation of normal astrocyte progenitor signatures, and is hallmarked by high level amplifications of EGFR and EGFR abnormalities such as the vIII deletion, point mutations and c-terminal deletions. An association between the Classical subtype and a cell of origin is yet to be reported but may reveal the importance of the transcriptomic context for EGFR abnormalities.
Heterozygous deletion of putative tumor suppressor NFKBIA rarely occurs within tumor samples expressing the Classical signature but is frequently found in non-Classical tumors, again suggesting the importance of the expression pattern in which the NFKBIA abnormalities are found11.
The availability of multidimensional genomic data sets facilitated identification of the striking co-occurrence of the G-CIMP methylation phenotype with mutations in the IDH115. This finding raises important questions on the mechanism of action of IDH1 associated gliomagenesis and whether the G-CIMP profile is the result of the IDH1 mutation, vice versa, or whether they are otherwise related. The G-CIMP phenotype/IDH1 mutation is found at high frequency amongst lower grade gliomas which may explain the significant improvement in outcome that this phenotype confers, even within GBMs. Like the majority of lower grade tumors, IDH1 mutated/G-CIMP tumors predominantly express the Proneural gene signature.
Examination of co-occurring and mutually exclusively occurring genomic abnormalities showed that glioblastomas nearly universally (1) circumvent cell cycle inhibition through genetic alterations to the RB1 pathway, (2) evade apoptosis by blocking the TP53 pathway and (3) maintain enhanced proliferation properties through activation of the RTK/phosphatidylinositol-3-OH kinase (PI(3)K) pathway20, 24. Although mutations, deletions and amplifications in the individual members of these pathways had been identified to occur at significantly higher rates than random chance, thereby suggesting their importance for GBM, integrated analysis of combined copy number and mutation patterns resulted in the important new insight that GBMs require at least one alteration in each of these three pathways, and the striking mutual exclusivity of genomic abnormalities within the RTK, TP53 and RB1 pathways20, 24.
The emphasis on these core pathways has resulted in the identification of novel pathway members and therapeutic opportunities. Co-deletion of CDKN2A and CDKN2C sensitizes glioma cells to treatment with RB1 pathway inhibitors targeting CDK4 and CDK649. Integrated network modeling of DNA copy number and transcriptome data allowed the identification of new TP53 pathway related genes such as NDN50. A similar analysis specific to the Mesenchymal subtype revealed STAT3 and C/EBPβ as master network regulators of mesenchymal transformation12.
Clinical developments
Personalized therapy based on the genetic characterization of a given patient's tumor offers great potential in cancer treatment. These therapies that target the molecular drivers of cancers offer high efficacy and more palatable side effect profiles compared to standard chemotherapies. Successes include imatinib which treats chronic myeloid leukemia with its characteristic translocations causing the fusion protein BCR-ABL and gefitinib for non small cell lung cancers with point mutations in the intracellular kinase domain of EGFR3, 4, 39, 51. Activating mutations and amplifications are easier to block, than it is to reinstall the activity of disrupting mutations and deletions.
In GBM, developing targeted therapies is complicated: we need to (1) identify true driver mutations and alterations while steering clear of the hundreds of non-driver "passenger" mutations that result from massive genomic instability of GBM; (2) effectively deliver drug to the tumor despite the blood-brain barrier; and (3) completely perfuse the heterogeneous tumor with high enough drug levels to affect the target. Additionally, successful targeted therapies for GBM might be particularly challenging to develop because of co-activation of multiple, independent, signaling cascades that might require hitting multiple targets in order to effectively kill the tumor20, 52. Targeting multiple pathways with multiple agents in this way makes clinical trial design more difficult and trials take longer, since the dose limiting toxicities of each drug must be demonstrated, and there might be synergistic effects between toxicities from multiple targeted agents combined. The field is further muddled by clinical trials that do not require evidence of given pathway activation as an inclusion criterion for study entry for the given targeted therapy. Such trials fail to account for the fact that GBM is a heterogeneous disease in which targeted therapies will have minimal effects when the target is not the driver for the treated patient's tumor. While such data can still be used to study dose limiting toxicity, it is not helpful in extrapolating the potential utility of the drug. The last limitation to interpreting results of targeted therapies is the failure to select agents that truly cause cell death -- and not just senescence or decreased cell proliferation -- in their targeted cells in vitro. Although cell death might not be required for the utility of targeted agents when used in combination with other drugs, it should be a distinguishing feature of drugs that are moved to high priority for further clinical testing. On a practical level, assessing successful target shut down in brain tumor patients is complicated: in order to select the appropriate patients, tissue is required. However, to assess successful target shut down requires more tissue after a given time of treatment. This therefore requires a minimum of two brain surgeries. While the first surgery might only require a relatively low risk biopsy for tissue, sample bias as a result of intratumoral heterogeneity is further complicating when it comes to improving treatment of GBM. The success of targeted therapies depends on patient selection, either by histological and staining criteria that define the susceptible pathway targeted, or tumor signatures that accurately predict response. For example, one study sponsored by North Central Cancer Treatment Group and NCI is collecting protein expression, immunohistochemistry staining, and gene expression data on patients to identify the best candidates for successful EGFR inhibition.
Of the 579 GBM clinical trials in the US and Europe presently listed on http://www.clinicaltrials.org, a number aim to bring targeted therapies to GBM. Although far from an exhaustive, this list serves as an example of the depth of what is being tried, such as various targeted agents at different levels of action from the cell membrane down to cyclin dependent kinases.
Since EGFR and other RTK pathways are aberrant at the genomic level of a high fraction of tumors, these offer suitable targets for GBM therapies. Early trials of RTK-targeting were hampered by inadequate drug levels in the tumor or not assessing the tumor after treatment for target activity53, 54. Erlotinib, which blocks internal tyrosine kinase activation of EGFR, cetuximab and nimotuxumab, which block ligand binding to EGFR, have been or are in trial55–57. These might be a hopeful strategy for EGFR-amplified tumors, but not necessarily tumors with constitutively activated EGFRvIII. Gefitinib, which effectively blocks EGFR with intracellular point mutations in lung adenocarcinoma, is under study although GBM is different from lung cancer in that the GBM EGFR mutations are typically extracellular and therefore the promise of gefitinib for GBM is less clear. PF00299804 is an irreversible pan ERBB family inhibitor that has shown efficacy in non small cell lung cancer models and is being tried in GBM given its EGFR inhibition58. Other RTKs are being targeted as well, for example, XL184 which targets MET in addition to VEGFR2, also known as KDR, and MEDI-575, an antibody that targets the platelet-derived growth factor-receptor PDGFRα59. Sunitinib targets PDGFRβ as well as KDR60, 61. Tyrosine kinases are the targets in a trial with E7050 in combination with E7080 in addition to KDR61, 62. A trial of pazopanib, which blocks PDGFR, FLT1 (VEGFR1), KDR, (VEGFR2), FLT4 (VEGFR3), and c-KIT, did not prolong progression free survival in patients with recurrent GBM63
While RAS genes are not found to be frequently mutated in GBM, tipifarnib is a RAS farnesyltransferase inhibitor that is under investigation, after it failed to gain approval for several other cancers64. Lonafarnib is a farnesyltransferase that inhibits post-translational modification of H-RAS, but not N-RAS or K-RAS65.
Downstream of RTK and RAS signaling, there are multiple agents that target the PI3K pathway such as pan PI3K inhibitor BKM12066. Multiple studies include sirolimus or rapamycin, which target mTOR67. There is an ongoing trial that includes nelfinavir, a protease inhibitor used in the treatment of HIV that also interferes with Akt activity68. Sorafenib is a RAF inhibitor that also targets PDGFR as well as VEGFR69. An example of targeting the cell cycle alterations found in GBM genomes is the CDK4/CDK6 inhibitor PD0332991; patients must have activated Rb in order to be enrolled70. There are agents in trial that affect less commonly aberrant pathways in GBM, like an attempt to target TGFB2 with an antisense oligonucleotide, AP1200971. Lastly, a large number of trials have been focusing on targeting members of pathways that enable vascularization, like VEGF receptors. While in studies there might be improvements in imaging characteristics (decreased gadolinium enhancement on MRI) and the ability for patients to decrease their steroid dose (less brain edema with VEGF-targeting agents), so far, there has been minimal benefit in survival72. VEGFR status on the tumor cells is typically not used to decide whether a patient receives these agents and it possible that some subclasses of patients will be found to do better, such as those that express the angiogenesis marker. Interestingly, the Mesenchymal expression subtype signature contains several angiogenesis markers. An ongoing study of bevacizumab and temozolomide in older patients with newly diagnosed GBM, sponsored by UCLA and Genentech, is collecting tumor microarray data. Such studies are then able to ask questions like whether the Mesenchymal subclass signature predicts response to bevacizumab.
Lessons learned and future directions
Genomic experiments have accelerated our knowledge of GBM in the past decade. Microarray expression data allowed researchers to identify prognostic signatures and to stratify patients into molecular subtypes. Copy number and sequencing data analysis have identified the most frequent genomic alterations such as amplification of chromosome 7, heterozygous deletion of chromosome 10, TP53 mutation, EGFR focal amplification and p16 homozygous deletion which may serve as potential drug targets. Methylation profiling surprisingly revealed the presence of a G-CIMP hypermethylation phenotype and co-occurrence with IDH1 mutations. Combined, these results improved our understanding of the mechanistic characterizations of GBM.
However, the fact that the clinical outcome has not been much improved shows we are still in the curve. One problem we are facing now is how to combine the many outputs from genomic data analysis to generate novel therapeutic options, and to distinguish the real drivers behind gliomagenesis.
Large multi-institutional research projects like TCGA aim to shed light on this problem, by providing high quality multi-faceted genomic data on a curated set of 500 GBMs. The number of samples interrogated allows not only to identify the most abnormalities in GBM as a whole, but to perform analysis such as network modeling and mutual exclusivity analysis on expression, methylation based subsets of GBM.
Even though GBM patients today still face a dim prognosis, the prospects for improving the outcome to this diagnosis are hopeful. The door has been opened for translational research in which clinical trials are directly connected to genomic characteristics. It is up to the current generation of researchers and clinicians to self organize into teams of clinical, biological and computational investigators as it clear that each expertise is required to change the bleak future of GBM patients.
Acknowledgements
MCG is supported by National Institutes of Health grant number 5K08NS062907. SZ and RGWV acknowledge support by National Cancer Institute grant number U24CA143883. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109(1):93–108. doi: 10.1007/s00401-005-0991-y. Available from http://www.ncbi.nlm.nih.gov/pubmed/15685439. [DOI] [PubMed] [Google Scholar]
- 2.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. Available from http://www.ncbi.nlm.nih.gov/pubmed/15758010. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. Available from http://www.ncbi.nlm.nih.gov/pubmed/15118073. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. Available from http://www.ncbi.nlm.nih.gov/pubmed/15118125. [DOI] [PubMed] [Google Scholar]
- 5.Rich JN, Hans C, Jones B, Iversen ES, McLendon RE, Rasheed BK, et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65(10):4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. Available from http://www.ncbi.nlm.nih.gov/pubmed/15899794. [DOI] [PubMed] [Google Scholar]
- 6.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102(16):5814–5819. doi: 10.1073/pnas.0402870102. Available from http://www.ncbi.nlm.nih.gov/pubmed/15827123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui AB, Lo KW, Yin XL, Poon WS, Ng HK. Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest. 2001;81(5):717–723. doi: 10.1038/labinvest.3780280. Available from http://www.ncbi.nlm.nih.gov/pubmed/11351043. [DOI] [PubMed] [Google Scholar]
- 8.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. Available from http://www.ncbi.nlm.nih.gov/pubmed/18772396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. Available from http://www.ncbi.nlm.nih.gov/pubmed/18565887. [DOI] [PubMed] [Google Scholar]
- 10.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. Available from http://www.ncbi.nlm.nih.gov/pubmed/18077431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364(7):627–637. doi: 10.1056/NEJMoa1006312. Available from http://www.ncbi.nlm.nih.gov/pubmed/21175304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. Available from http://www.ncbi.nlm.nih.gov/pubmed/20032975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li A, Walling J, Ahn S, Kotliarov Y, Su Q, Quezado M, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69(5):2091–2099. doi: 10.1158/0008-5472.CAN-08-2100. Available from http://www.ncbi.nlm.nih.gov/pubmed/19244127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22(15):2361–2373. doi: 10.1038/sj.onc.1206344. Available from http://www.ncbi.nlm.nih.gov/pubmed/12700671. [DOI] [PubMed] [Google Scholar]
- 15.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. Available from http://www.ncbi.nlm.nih.gov/pubmed/20399149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–1607. Available from http://www.ncbi.nlm.nih.gov/pubmed/12670911. [PubMed] [Google Scholar]
- 17.Ozawa T, Brennan CW, Wang L, Squatrito M, Sasayama T, Nakada M, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24(19):2205–2218. doi: 10.1101/gad.1972310. Available from http://www.ncbi.nlm.nih.gov/pubmed/20889717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. Available from http://www.ncbi.nlm.nih.gov/pubmed/16530701. [DOI] [PubMed] [Google Scholar]
- 19.Rothenberg SM, Mohapatra G, Rivera MN, Winokur D, Greninger P, Nitta M, et al. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res. 2010;70(6):2158–2164. doi: 10.1158/0008-5472.CAN-09-3458. Available from http://www.ncbi.nlm.nih.gov/pubmed/20215515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. Available from http://www.ncbi.nlm.nih.gov/pubmed/18772890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tso CL, Freije WA, Day A, Chen Z, Merriman B, Perlina A, et al. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66(1):159–167. doi: 10.1158/0008-5472.CAN-05-0077. Available from http://www.ncbi.nlm.nih.gov/pubmed/16397228. [DOI] [PubMed] [Google Scholar]
- 22.Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, Solit DB, et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci U S A. 2009;106(23):9435–9440. doi: 10.1073/pnas.0900571106. Available from http://www.ncbi.nlm.nih.gov/pubmed/19478061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. Available from http://www.ncbi.nlm.nih.gov/pubmed/20129251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciriello G, Cerami EG, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. doi: 10.1101/gr.125567.111. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21908773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bredel M, Scholtens DM, Harsh GR, Bredel C, Chandler JP, Renfrow JJ, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302(3):261–275. doi: 10.1001/jama.2009.997. Available from http://www.ncbi.nlm.nih.gov/pubmed/19602686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 2010;5(2):e8918. doi: 10.1371/journal.pone.0008918. Available from http://www.ncbi.nlm.nih.gov/pubmed/20169195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. Available from http://www.ncbi.nlm.nih.gov/pubmed/20414201. [DOI] [PubMed] [Google Scholar]
- 28.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. Available from http://www.ncbi.nlm.nih.gov/pubmed/17618441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician's perspective. Acta Neuropathol. 2010;120(3):297–304. doi: 10.1007/s00401-010-0725-7. Available from http://www.ncbi.nlm.nih.gov/pubmed/20644945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coons SW, Johnson PC, Scheithauer BW, Yates AJ, Pearl DK. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79(7):1381–1393. doi: 10.1002/(sici)1097-0142(19970401)79:7<1381::aid-cncr16>3.0.co;2-w. Available from http://www.ncbi.nlm.nih.gov/pubmed/9083161. [DOI] [PubMed] [Google Scholar]
- 31.Giannini C, Scheithauer BW, Weaver AL, Burger PC, Kros JM, Mork S, et al. Oligodendrogliomas: reproducibility and prognostic value of histologic diagnosis and grading. J Neuropathol Exp Neurol. 2001;60(3):248–262. doi: 10.1093/jnen/60.3.248. Available from http://www.ncbi.nlm.nih.gov/pubmed/11245209. [DOI] [PubMed] [Google Scholar]
- 32.Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101(1):124–131. doi: 10.1038/sj.bjc.6605127. Available from http://www.ncbi.nlm.nih.gov/pubmed/19536096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nobusawa S, Lachuer J, Wierinckx A, Kim YH, Huang J, Legras C, et al. Intratumoral patterns of genomic imbalance in glioblastomas. Brain Pathol. 20(5):936–944. doi: 10.1111/j.1750-3639.2010.00395.x. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20406234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vital AL, Tabernero MD, Crespo I, Rebelo O, Tao H, Gomes F, et al. Intratumoral patterns of clonal evolution in gliomas. Neurogenetics. 11(2):227–239. doi: 10.1007/s10048-009-0217-x. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19760258. [DOI] [PubMed] [Google Scholar]
- 35.Ren ZP, Olofsson T, Qu M, Hesselager G, Soussi T, Kalimo H, et al. Molecular genetic analysis of p53 intratumoral heterogeneity in human astrocytic brain tumors. J Neuropathol Exp Neurol. 2007;66(10):944–954. doi: 10.1097/nen.0b013e318156bc05. Available from http://www.ncbi.nlm.nih.gov/pubmed/17917588. [DOI] [PubMed] [Google Scholar]
- 36.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 71(12):4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21628493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 17(4):362–375. doi: 10.1016/j.ccr.2009.12.049. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&listuids=20385361. [DOI] [PubMed] [Google Scholar]
- 38.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 472(7341):90–94. doi: 10.1038/nature09807. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21399628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. Available from http://www.ncbi.nlm.nih.gov/pubmed/15737014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64(18):6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. Available from http://www.ncbi.nlm.nih.gov/pubmed/15374961. [DOI] [PubMed] [Google Scholar]
- 41.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9(4):287–300. doi: 10.1016/j.ccr.2006.03.003. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16616334. [DOI] [PubMed] [Google Scholar]
- 42.Bigner SH, Mark J, Burger PC, Mahaley MS, Jr, Bullard DE, Muhlbaier LH, et al. Specific chromosomal abnormalities in malignant human gliomas. Cancer Res. 1988;48(2):405–411. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=3335011. [PubMed] [Google Scholar]
- 43.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. Available from http://www.ncbi.nlm.nih.gov/pubmed/19578366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. Available from http://www.ncbi.nlm.nih.gov/pubmed/19578367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark MJ, Homer N, O'Connor BD, Chen Z, Eskin A, Lee H, et al. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6(1) doi: 10.1371/journal.pgen.1000832. e1000832. Available from http://www.ncbi.nlm.nih.gov/pubmed/20126413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, et al. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51(2):187–199. doi: 10.1016/j.neuron.2006.06.012. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16846854. [DOI] [PubMed] [Google Scholar]
- 47.Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 146(2):209–221. doi: 10.1016/j.cell.2011.06.014. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21737130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaufmann K, Bach K, Thiel G. The extracellular signal-regulated protein kinases Erk1/Erk2 stimulate expression and biological activity of the transcriptional regulator Egr-1. Biol Chem. 2001;382(7):1077–1081. doi: 10.1515/BC.2001.135. Available from http://www.ncbi.nlm.nih.gov/pubmed/11530939. [DOI] [PubMed] [Google Scholar]
- 49.Wiedemeyer WR, Dunn IF, Quayle SN, Zhang J, Chheda MG, Dunn GP, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci U S A. 107(25):11501–11506. doi: 10.1073/pnas.1001613107. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20534551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jornsten R, Abenius T, Kling T, Schmidt L, Johansson E, Nordling TE, et al. Network modeling of the transcriptional effects of copy number aberrations in glioblastoma. Mol Syst Biol. 2011;7:486. doi: 10.1038/msb.2011.17. Available from http://www.ncbi.nlm.nih.gov/pubmed/21525872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1031–1037. doi: 10.1056/NEJM200104053441401. Available from http://www.ncbi.nlm.nih.gov/pubmed/11287972. [DOI] [PubMed] [Google Scholar]
- 52.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–290. doi: 10.1126/science.1142946. Available from http://www.ncbi.nlm.nih.gov/pubmed/17872411. [DOI] [PubMed] [Google Scholar]
- 53.De Witt Hamer PC. Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies. Neuro Oncol. 2010;12(3):304–316. doi: 10.1093/neuonc/nop068. Available from http://www.ncbi.nlm.nih.gov/pubmed/20167819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solomon DA, Kim JS, Jenkins S, et al. Identification of p18 INK4c as a tumor suppressor gene in glioblastoma multiforme. Cancer Res. 2008;68(8):2564–2569. doi: 10.1158/0008-5472.CAN-07-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belda-Iniesta C, Carpeno Jde C, Saenz EC, Gutierrez M, Perona R, Baron MG. Long term responses with cetuximab therapy in glioblastoma multiforme. Cancer Biol Ther. 2006;5(8):912–914. doi: 10.4161/cbt.5.8.3118. Available from http://www.ncbi.nlm.nih.gov/pubmed/16929166. [DOI] [PubMed] [Google Scholar]
- 56.Combs SE, Heeger S, Haselmann R, Edler L, Debus J, Schulz-Ertner D. Treatment of primary glioblastoma multiforme with cetuximab, radiotherapy and temozolomide (GERT)--phase I/II trial: study protocol. BMC Cancer. 2006;6:133. doi: 10.1186/1471-2407-6-133. Available from http://www.ncbi.nlm.nih.gov/pubmed/16709245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diaz Miqueli A, Rolff J, Lemm M, Fichtner I, Perez R, Montero E. Radiosensitisation of U87MG brain tumours by anti-epidermal growth factor receptor monoclonal antibodies. Br J Cancer. 2009;100(6):950–958. doi: 10.1038/sj.bjc.6604943. Available from http://www.ncbi.nlm.nih.gov/pubmed/19293809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67(24):11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. Available from http://www.ncbi.nlm.nih.gov/pubmed/18089823. [DOI] [PubMed] [Google Scholar]
- 59.Wen PY. American Society of Clinical Oncology 2010: report of selected studies from the CNS tumors section. Expert Rev Anticancer Ther. 2010;10(9):1367–1369. doi: 10.1586/era.10.117. Available from http://www.ncbi.nlm.nih.gov/pubmed/20836670. [DOI] [PubMed] [Google Scholar]
- 60.George S. Sunitinib, a multitargeted tyrosine kinase inhibitor, in the management of gastrointestinal stromal tumor. Curr Oncol Rep. 2007;9(4):323–327. doi: 10.1007/s11912-007-0040-1. Available from http://www.ncbi.nlm.nih.gov/pubmed/17588358. [DOI] [PubMed] [Google Scholar]
- 61.Nakagawa T, Tohyama O, Yamaguchi A, Matsushima T, Takahashi K, Funasaka S, et al. E7050: a dual c-Met and VEGFR-2 tyrosine kinase inhibitor promotes tumor regression and prolongs survival in mouse xenograft models. Cancer Sci. 2010;101(1):210–215. doi: 10.1111/j.1349-7006.2009.01343.x. Available from http://www.ncbi.nlm.nih.gov/pubmed/19832844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsui J, Yamamoto Y, Funahashi Y, Tsuruoka A, Watanabe T, Wakabayashi T, et al. E7080, a novel inhibitor that targets multiple kinases, has potent antitumor activities against stem cell factor producing human small cell lung cancer H146, based on angiogenesis inhibition. Int J Cancer. 2008;122(3):664–771. doi: 10.1002/ijc.23131. Available from http://www.ncbi.nlm.nih.gov/pubmed/17943726. [DOI] [PubMed] [Google Scholar]
- 63.Iwamoto FM, Lamborn KR, Robins HI, Mehta MP, Chang SM, Butowski NA, et al. Phase II trial of pazopanib (GW786034), an oral multi-targeted angiogenesis inhibitor, for adults with recurrent glioblastoma (North American Brain Tumor Consortium Study 06-02) Neuro Oncol. 2010;12(8):855–861. doi: 10.1093/neuonc/noq025. Available from http://www.ncbi.nlm.nih.gov/pubmed/20200024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reid TS, Beese LS. Crystal structures of the anticancer clinical candidates R115777 (Tipifarnib) and BMS-214662 complexed with protein farnesyltransferase suggest a mechanism of FTI selectivity. Biochemistry. 2004;43(22):6877–6884. doi: 10.1021/bi049723b. Available from http://www.ncbi.nlm.nih.gov/pubmed/15170324. [DOI] [PubMed] [Google Scholar]
- 65.Awada A, Eskens FA, Piccart M, Cutler DL, van der Gaast A, Bleiberg H, et al. Phase I and pharmacological study of the oral farnesyltransferase inhibitor SCH 66336 given once daily to patients with advanced solid tumours. Eur J Cancer. 2002;38(17):2272–2278. doi: 10.1016/s0959-8049(02)00379-9. Available from http://www.ncbi.nlm.nih.gov/pubmed/12441264. [DOI] [PubMed] [Google Scholar]
- 66.Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2(51) doi: 10.1126/scitranslmed.3001599. 51ra70. Available from http://www.ncbi.nlm.nih.gov/pubmed/20881279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5(1):e8. doi: 10.1371/journal.pmed.0050008. Available from http://www.ncbi.nlm.nih.gov/pubmed/18215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plastaras JP, Vapiwala N, Ahmed MS, Gudonis D, Cerniglia GJ, Feldman MD, et al. Validation and toxicity of PI3K/Akt pathway inhibition by HIV protease inhibitors in humans. Cancer Biol Ther. 2008;7(5):628–635. doi: 10.4161/cbt.7.5.5728. Available from http://www.ncbi.nlm.nih.gov/pubmed/18285707. [DOI] [PubMed] [Google Scholar]
- 69.Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66(24):11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. Available from http://www.ncbi.nlm.nih.gov/pubmed/17178882. [DOI] [PubMed] [Google Scholar]
- 70.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3(11):1427–1438. Available from http://www.ncbi.nlm.nih.gov/pubmed/15542782. [PubMed] [Google Scholar]
- 71.Jaschinski F, Rothhammer T, Jachimczak P, Seitz C, Schneider A, Schlingensiepen KH. The Antisense Oligonucleotide Trabedersen (AP 12009) for the Targeted Inhibition of TGF-beta2. Curr Pharm Biotechnol. 2011 doi: 10.2174/138920111798808266. Available from http://www.ncbi.nlm.nih.gov/pubmed/21619536. [DOI] [PubMed] [Google Scholar]
- 72.Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27(28):4733–4740. doi: 10.1200/JCO.2008.19.8721. Available from http://www.ncbi.nlm.nih.gov/pubmed/19720927. [DOI] [PubMed] [Google Scholar]
- 73.Wiedemeyer R, Brennan C, Heffernan TP, Xiao Y, Mahoney J, Protopopov A, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13(4):355–364. doi: 10.1016/j.ccr.2008.02.010. Available from http://www.ncbi.nlm.nih.gov/pubmed/18394558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duncan CG, Killela PJ, Payne CA, Lampson B, Chen WC, Liu J, et al. Integrated genomic analyses identify ERRFI1 and TACC3 as glioblastoma-targeted genes. Oncotarget. 2010;1(4):265–277. doi: 10.18632/oncotarget.137. Available from http://www.ncbi.nlm.nih.gov/pubmed/21113414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song SW, Fuller GN, Zheng H, Zhang W. Inactivation of the invasion inhibitory gene IIp45 by alternative splicing in gliomas. Cancer Res. 2005;65(9):3562–3567. doi: 10.1158/0008-5472.CAN-04-3392. Available from http://www.ncbi.nlm.nih.gov/pubmed/15867349. [DOI] [PubMed] [Google Scholar]
- 76.Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128(3):459–475. doi: 10.1016/j.cell.2006.11.052. Available from http://www.ncbi.nlm.nih.gov/pubmed/17289567. [DOI] [PubMed] [Google Scholar]
- 77.Riemenschneider MJ, Buschges R, Wolter M, Reifenberger J, Bostrom J, Kraus JA, et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999;59(24):6091–6096. Available from http://www.ncbi.nlm.nih.gov/pubmed/10626796. [PubMed] [Google Scholar]
- 78.Yin D, Ogawa S, Kawamata N, Tunici P, Finocchiaro G, Eoli M, et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol Cancer Res. 2009;7(5):665–677. doi: 10.1158/1541-7786.MCR-08-0270. Available from http://www.ncbi.nlm.nih.gov/pubmed/19435819. [DOI] [PubMed] [Google Scholar]
- 79.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. Available from http://www.ncbi.nlm.nih.gov/pubmed/19228619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. Available from http://www.ncbi.nlm.nih.gov/pubmed/15016963. [DOI] [PubMed] [Google Scholar]
- 81.Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, et al. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42(1):77–82. doi: 10.1038/ng.491. Available from http://www.ncbi.nlm.nih.gov/pubmed/19946270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci U S A. 1992;89(10):4309–4313. doi: 10.1073/pnas.89.10.4309. Available from http://www.ncbi.nlm.nih.gov/pubmed/1584765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wiedemeyer WR, Dunn IF, Quayle SN, Zhang J, Chheda MG, Dunn GP, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci U S A. 2010;107(25):11501–11506. doi: 10.1073/pnas.1001613107. Available from http://www.ncbi.nlm.nih.gov/pubmed/20534551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fischer U, Muller HW, Sattler HP, Feiden K, Zang KD, Meese E. Amplification of the MET gene in glioma. Genes Chromosomes Cancer. 1995;12(1):63–65. doi: 10.1002/gcc.2870120111. Available from http://www.ncbi.nlm.nih.gov/pubmed/7534113. [DOI] [PubMed] [Google Scholar]
- 85.Basto D, Trovisco V, Lopes JM, Martins A, Pardal F, Soares P, et al. Mutation analysis of B-RAF gene in human gliomas. Acta Neuropathol. 2005;109(2):207–210. doi: 10.1007/s00401-004-0936-x. Available from http://www.ncbi.nlm.nih.gov/pubmed/15791479. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt EE, Ichimura K, Reifenberger G, Collins VP. CDKN2 (p16/MTS1) gene deletion or CDK4 amplification occurs in the majority of glioblastomas. Cancer Res. 1994;54(24):6321–6324. Available from http://www.ncbi.nlm.nih.gov/pubmed/7987821. [PubMed] [Google Scholar]
- 87.Wang SI, Puc J, Li J, Bruce JN, Cairns P, Sidransky D, et al. Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res. 1997;57(19):4183–4186. Available from http://www.ncbi.nlm.nih.gov/pubmed/9331071. [PubMed] [Google Scholar]
- 88.Rollbrocker B, Waha A, Louis DN, Wiestler OD, von Deimling A. Amplification of the cyclin-dependent kinase 4 (CDK4) gene is associated with high cdk4 protein levels in glioblastoma multiforme. Acta Neuropathol. 1996;92(1):70–74. doi: 10.1007/s004010050491. Available from http://www.ncbi.nlm.nih.gov/pubmed/8811128. [DOI] [PubMed] [Google Scholar]
- 89.Biernat W, Kleihues P, Yonekawa Y, Ohgaki H. Amplification and overexpression of MDM2 in primary (de novo) glioblastomas. J Neuropathol Exp Neurol. 1997;56(2):180–185. doi: 10.1097/00005072-199702000-00009. Available from http://www.ncbi.nlm.nih.gov/pubmed/9034372. [DOI] [PubMed] [Google Scholar]
- 90.Ichimura K, Schmidt EE, Goike HM, Collins VP. Human glioblastomas with no alterations of the CDKN2A (p16INK4A, MTS1) and CDK4 genes have frequent mutations of the retinoblastoma gene. Oncogene. 1996;13(5):1065–1072. Available from http://www.ncbi.nlm.nih.gov/pubmed/8806696. [PubMed] [Google Scholar]
- 91.Chung R, Whaley J, Kley N, Anderson K, Louis D, Menon A, et al. TP53 gene mutations and 17p deletions in human astrocytomas. Genes Chromosomes Cancer. 1991;3(5):323–331. doi: 10.1002/gcc.2870030502. Available from http://www.ncbi.nlm.nih.gov/pubmed/1686725. [DOI] [PubMed] [Google Scholar]
- 92.Stegh AH, Brennan C, Mahoney JA, Forloney KL, Jenq HT, Luciano JP, et al. Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev. 2010;24(19):2194–2204. doi: 10.1101/gad.1924710. Available from http://www.ncbi.nlm.nih.gov/pubmed/20837658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tchirkov A, Rolhion C, Bertrand S, Dore JF, Dubost JJ, Verrelle P. IL-6 gene amplification and expression in human glioblastomas. Br J Cancer. 2001;85(4):518–522. doi: 10.1054/bjoc.2001.1942. Available from http://www.ncbi.nlm.nih.gov/pubmed/11506489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ichimura K, Vogazianou AP, Liu L, et al. 1p36 is a preferential target of chromosome 1 deletions in astrocytic tumours and homozygously deleted in a subset of glioblastomas. Oncogene. 2008;27(14):2097–2108. doi: 10.1038/sj.onc.1210848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roversi G, Pfundt R, Moroni RF, et al. Identification of novel genomic markers related to progression to glioblastoma through genomic profiling of 25 primary glioma cell lines. Oncogene. 2006;25(10):1571–1583. doi: 10.1038/sj.onc.1209177. [DOI] [PubMed] [Google Scholar]
- 96.Annovazzi L, Mellai M, Caldera V, Valente G, Schiffer D. SOX2 expression and amplification in gliomas and glioma cell lines. Cancer Genomics Proteomics. 2011;8(3):139–147. [PubMed] [Google Scholar]
- 97.Schmitz M, Temme A, Senner V, et al. Identification of SOX2 as a novel glioma-associated antigen and potential target for T cell-based immunotherapy. Br J Cancer. 2007;96(8):1293–1301. doi: 10.1038/sj.bjc.6603696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60(5):1383–1387. [PubMed] [Google Scholar]
- 99.Van Meir E, Sawamura Y, Diserens AC, Hamou MF, de Tribolet N. Human glioblastoma cells release interleukin 6 in vivo and in vitro. Cancer Res. 1990;50(20):6683–6688. [PubMed] [Google Scholar]
- 100.Tchirkov A, Khalil T, Chautard E, et al. Interleukin-6 gene amplification and shortened survival in glioblastoma patients. Br J Cancer. 2007;96(3):474–476. doi: 10.1038/sj.bjc.6603586. [DOI] [PMC free article] [PubMed] [Google Scholar]