Abstract
Heritable skin diseases represent a broad spectrum of clinical manifestations due to mutations in ~500 different genes. A number of model systems have been developed to advance our understanding of the pathomechanisms of genodermatoses. Zebrafish (Danio rerio), a freshwater vertebrate, has a well-characterized genome, the expression of which can be easily manipulated. The larvae develop rapidly, with all major organs having developed by 5–6 days post fertilization, including the skin, consisting of the epidermis comprising two cell layers and separated from the dermal collagenous matrix by a basement membrane. This perspective highlights the morphologic and ultrastructural features of zebrafish skin, in the context of cutaneous gene expression. These observations suggest that zebrafish provides a useful model system to study molecular aspects of skin development, as well as the pathogenesis and treatment of select heritable skin diseases.
The Spectrum of Heritable Skin Diseases
Genodermatoses manifest with a broad spectrum of clinical conditions where at one end of the spectrum the cutaneous findings can be relatively minor, limited to the skin, hair and nails, while at the other extreme of the spectrum the cutaneous manifestations can be part of multi-system pathology with significant morbidity and mortality (for reviews, see Pulkkinen et al, 2002; Feramisco et al, 2009; Uitto, 2009). With the advent of technologies in molecular genetics in general and completion of the human genome database, an increasingly large number of gene defects has been linked to specific heritable disorders with cutaneous manifestations. In fact, mutations are now known to occur in as many as 500 different genes in a manner that these genetic variations explain the phenotypic manifestations characteristic of a heritable disease with skin involvement (Feramisco et al, 2009).
Mutation Databases of the Candidate Genes
The genodermatosis mutation databases have revealed both obvious candidate genes and a number of surprises. For example, approximately 20 years ago, based on advances in the knowledge of the cutaneous basement membrane zone (BMZ), we made a prediction that mutations in the structural components of the basement membrane at the epidermal-dermal interface could explain the fragility of skin in different forms of epidermolysis bullosa (EB) (Uitto and Christiano, 1992). This prediction has since been proven correct by the demonstration of mutations in as many as 14 distinct genes encoding the structural components of the skin in different variants of EB (Uitto et al, 2010a).
In contrast to the apparent candidate genes, many of the mutated genes have turned out to be surprisingly unpredictable, with their role in skin biology being largely unrecognized prior to the identification of these specific mutations. For example, pseudoxanthoma elasticum (PXE), which was initially classified as a prototypic connective tissue disorder affecting the elastic fiber network in the skin, the eyes and the cardiovascular system, was subsequently shown to result from mutations in the ABCC6 gene, expressed primarily in the liver and the kidneys and at very low level, if at all, in tissues affected with mineralization in PXE (Li et al, 2009; Uitto et al, 2010b). This condition is now thought to be a metabolic disorder in which the absence of circulating anti-mineralization factors allows ectopic calcification of the connective tissues to ensue (Jiang et al, 2009).
Model Systems to Study Heritable Skin Diseases
In order to gain insight into the pathomechanistic details of heritable skin diseases and to provide model systems for testing of treatment modalities, a number of animal models that recapitulate features of a specific disease have been developed. Traditionally, mice have provided the preferred platform to develop models of human diseases, often through the development of “knock-out” (KO) animals by targeted ablation of the corresponding genes. While often the KO mice show remarkable similarity to the human phenotype both at the genetic, gross morphologic, histopathologic, and ultrastructural levels, mice as a model system can have considerable limitations (Lieschke and Currie, 2007). The drawback of the mouse model is its relatively long lifespan, and it may take several years to develop a KO mouse. In some cases, development of the KO mouse as a model system of the corresponding human disease is not feasible due to the absence of the corresponding gene in the mouse genome, as in case of the SAMD9 gene underlying normophosphatemic familial tumoral calcinosis in humans (Li et al, 2007; Sprecher, 2010). These considerations, together with cost containment issues, have prompted the search for alternative model systems to study heritable skin diseases (Vanchieri, 2001).
Biology of Zebrafish Skin Development
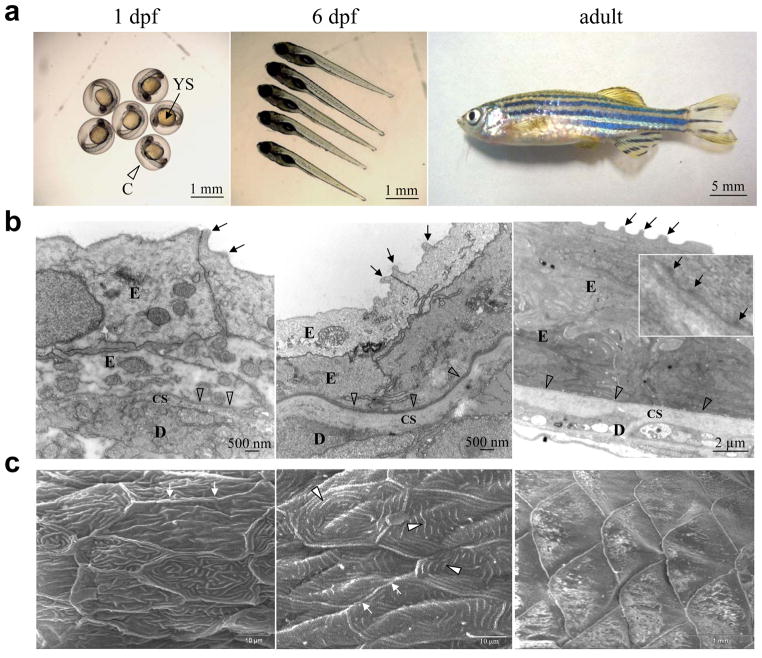
There are several characteristics that favor choosing zebrafish (Dario rerio) for genetic testing in developmental studies (Table 1) (Lieschke and Currie, 2007; Brittijn et al, 2009; Rakers et al, 2010). This small freshwater fish is easily maintained in the laboratory setting with a rapid rate of maturation from embryos to fully developed fish (Fig. 1a). A single female can lay 50–100 eggs per laying. The development of various organs is easy to visualize in vivo because the embryos are optically transparent during the first several days. By day 5–6, all important internal organs as well as skin compartments are largely formed, as can be visualized by transmission electron microscopy (Fig. 1b). At 1 day post-fertilization (dpf) different skin layers representing the epidermis and the dermis can be recognized, although the cutaneous BMZ at the dermal-epidermal junction is not yet developed. However, at 6 dpf the epidermis, composed of two cell layers, is readily noticeable and clearly separated from the underlying connective tissue stroma by a basement membrane. At the surface of the epidermal contour, there are spicule-like protrusions which correspond to microridges of the epidermal cells. On the dermal side there is a well developed collagenous stroma with adjacent fibroblastic cells with well developed rough endoplasmic reticulum (Le Guellec et al, 2004). In fully developed adult zebrafish skin, there is a multi-layer epidermis separated from the underlying collagenous stroma by the BMZ and at high magnification, hemidesmosomal structures can be visualized (Fig. 1b), (Sonawane et al, 2005). Thus, zebrafish skin has a clearly demarcated dermal-epidermal BMZ, separating epidermis from the underlying dermis. Previous studies have shown that the basement membrane may be formed as early as 32 hpf, and lamina lucida and lamina densa can be identified by transmission electron microscopy at 48 hpf (Le Guellec et al, 2004).
Table 1.
Comparison of selected attributes for the use of zebrafish or mouse as a model system to study human diseases1)
| Attribute | Zebrafish | Mouse |
|---|---|---|
| Practical Issues | ||
| • Husbandry infrastructure | $ | $$$ |
| • Cost per animal per year | $ | $$ |
| • Characterized inbred strains | + | ++++ |
| • Outbred laboratory strains | +++ | ++ |
| • Anatomical similarity2) | + | ++ |
| • Genetic similarity2) | ++ | +++ |
| • Pathological similarity2) | ++ | +++ |
| Molecular Biology Tools | ||
| • Transgenesis3) | ++ | ++ |
| • Targeted gene modification3) | − | ++++ |
| • Transient in vivo assays3) | ++++ | + |
| • Feasibility of large-scale screens | +++ | ++ |
| • Affordability of large-scale screens | +++ | + |
| • Genome sequencing progress | ++ | +++ |
| • Genome annotation progress | ++ | ++++ |
| Cell Biology Tools | ||
| • Cell lines and tissue cultures | + | +++ |
| • Antibody reagents | + | ++++ |
Designations: $ and $$$; and −, +, ++ and +++ refer to the relative cost and strength of the model system in each category; ++++ signifies outstanding strength of the model. (Adapted with modifications from Lieschke and Currie, 2007).
In comparison to human.
Reverse-genetics approach.
Figure 1.
Cutaneous biology of the developing zebrafish. (a) The figure illustrates the growth of zebrafish from 1 dpf embryos, which are surrounded by a transparent chorion (C) and display a prominent yolk sac (YS), to an adult fish. At 6 dpf pigmentation becomes apparent on the skin. (b) Transmission electron microscopy reveals at 1 and 6 dpf an epidermis (E) consisting of two cell layers, and at 6 dpf the epidermis is separated from the underlying collagenous stroma (CS) and dermis (D) by a clearly demarcated basement membrane (open arrowheads). In adult fish, there is a multi-layered epidermis, and higher magnification of the basement membrane zone reveals the presence of hemidesmosomes (arrows in the inset). The spicule-like extensions of the surface of the skin (arrows) correspond to microridges. (c) Scanning electron microscopy reveals well demarcated keratinocytes with distinct cell-cell borders (small arrows). In the middle of the keratinocyte surface, there are developing microridges which at 6 dpf become well organized (open arrowheads). In an adult fish, the epidermis is covered by scales.
Examination of the developing zebrafish skin surface at 1 dpf by scanning electron microscopy reveals well demarcated keratinocytes with a surface contour containing microridges, which are well organized by 6 dpf (Fig. 1c). In adult zebrafish the epidermis is covered by scales, which form under the control of a genetic cascade, including sonic hedgehog expression, at around 30 dpf (Sire and Akimendo, 2004). However, early on, at least up to 6 dpf as shown here, the developing zebrafish epidermis has characteristic landmark features that can be altered by perturbed keratinocyte gene expression.
In addition to the structural elements of the epidermis, BMZ, and the collagenous dermis, zebrafish skin has a neural-crest-derived pigment cell system, including the presence of melanocytes, that can serve as a target to study developmental biology and pathology of pigmentation (O’Reilly-Pol and Johnson, 2009; Lee et al, 2010). Zebrafish skin also has structures that are specialized for the aquatic environment, such as scales, presence of mucous secreting cells, and the lateral line. The latter organ contains 54 neuromasts that are topographically highly conserved neural elements consisting of hair cells, serving as a sensory organ regarding the rheological movements of the fish (Froehlicher et al, 2009). Of interest is our recent finding that the gene product of col17a1b is localized to neuromasts, and “knockdown” of the corresponding gene by a morpholino abolishes the formation of functional neuromasts (Kim et al, 2010). A key difference between the zebrafish and human skin is, however, the lack of mammalian appendages, including hair follicles and sebaceous glands.
The Zebrafish Genome
Significant progress has recently been made in sequencing the zebrafish genome encompassing 25 chromosomes. This diploid genome essentially contains the full repertoire of vertebrate genes, and the most recent gene-set comprises 24,147 protein-encoding genes based on the integrated whole genome shotgun assembly at 6.5–7-fold coverage (Ensembl, Zebrafish Zv8; http://uswest.ensembl.org/Danio_rerio/Info/Index). In spite of the extensive coverage, this assembly is still considered preliminary because there are contigs that are of lower sequence quality, and the assembly contains misjoins, misassemblies and artificial duplications.
One of the characteristic features of the zebrafish genome is that there are duplicate copies of a number of genes (see Postlethwait, 2007; Roch et al, 2009). The current hypotheses put forward suggest that the whole genome underwent two sequential rounds of duplication in the vertebrate stem well before the divergence of ray-finned and lobe-finned fish. Evidence suggests another further round of whole genome duplication occurring near the origin of teleost fish, i.e., about 350 million years ago. In many cases, one gene copy of the duplicate genes may have been lost or silenced, but if both copies survive in a functional state, these duplicates in some cases have evolved a novel function in distinct spatial distribution. For example, as indicated below, we have identified two copies of the zebrafish type XVII collagen genes, col17a1a and col17a1b, which by in situ hybridization and morpholino knockdown technologies were shown to have either epidermal or neural distribution, respectively (Kim et al, 2010). In higher vertebrates, including human and mouse, type XVII collagen can display either an epithelial or a neural isoform, which, however, are apparently products of the same gene (Seppänen et al, 2006; Has and Kern, 2010).
The overall conservation of different orthologous genes in different species, such as in human and zebrafish, can be determined by constructing phylogenetic trees based upon the nucleotide information on these genes. For example, the two zebrafish type XVII collagen genes are phylogenetically close, yet they show a high degree of divergence compared to other col17 orthologs in different species (Kim et al, 2010).
The conservation of individual genes between human and zebrafish can be variable, and certain protein domains may be well conserved. For example, the overall sequence homology at the amino acid level between the human and zebrafish ABCC6 genes is 47 percent, yet certain critical domains, such as the nucleotide binding folds, indispensable for the function of this transmembrane efflux transporter, are conserved by ~60 percent (Li et al, 2010).
Cutaneous Gene Expression Profiles in Zebrafish
Since ultrastructural survey of the developing zebrafish skin showed clearly recognizable epidermal structures separated from the underlying dermis by BMZ, we have profiled the zebrafish skin gene expression by RT-PCR for selected genes that are known to be expressed in human epidermis, BMZ or dermis, respectively (Table 2). RT-PCR using total RNA isolated from zebrafish as a template revealed expression of selective epidermal markers, including keratins (krt1 and krt5), and 230-kD bullous pemphigoid antigen (bpag1) at 6 dpf, and the expression of these genes continued into adulthood. The plectin gene (plec1) encoding an intracellular epidermal adhesion molecule, was expressed as early as 1 dpf, and its expression increased at later stages. Among the BMZ genes, mRNA expression corresponding to subunit polypeptides of type IV collagen, as well as collagens VII and XVII, was readily detected. Furthermore, expression of the genes encoding subunit polypeptides of the integrin α6β4 (itga6, itgb4) was noted at 1 or 6 dpf. A number of human collagen genes, as exemplified by collagens I, V and VI, the major collagens in human dermis, were also expressed at 6 dpf. These collagen mRNAs, as well as those corresponding to collagens XII, XIV, XV, XVI, XVIII, and XIX, could be temporally detected at different stages of zebrafish development (Table 2). It is important to note, however, that a search of the latest zebrafish genome database does not reveal the presence of the filaggrin, involucrin, envoplakin and trichohyalin genes. These observations may well reflect the fact that the zebrafish epidermis does not undergo terminal differentiation towards formation of stratum corneum with barrier function similar to humans. This difference limits the study of human epidermal disorders using zebrafish as a model system.
Table 2.
Expression of selected epidermal, BMZ and dermal genes in zebrafish at different stages of development 1)
| Gene | Developmental stage
|
||
|---|---|---|---|
| 1 dpf | 6 dpf | adult | |
| Epidermal | |||
| krt1 | − | + | + |
| krt5 | − | + | ++ |
| bpag1 | − | ++ | + |
| plec1 | + | + | ++ |
| BMZ | |||
| col4a1 | + | + | ++ |
| col4a2 | − | ++ | + |
| col4a3 | − | + | + |
| col4a4 | − | + | + |
| col4a5 | − | − | − |
| col4a6 | ++ | ++ | + |
| col7a1a | + | + | + |
| col7a1b | + | + | + |
| col17a1a | − | + | + |
| col17a1b | + | ++ | + |
| itga6 | − | + | ++ |
| itgb4 | + | ++ | +++ |
| Dermal | |||
| col1a1 | + | ++ | +++ |
| col1a2 | − | ++ | + |
| col1a3 | + | ++ | +++ |
| col5a1 | +++ | ++ | + |
| col5a2 | − | − | − |
| col6a1 | + | + | − |
| col6a2 | − | + | + |
| col12a1 | − | + | ++ |
| col14a1 | − | + | + |
| col15a1 | − | ++ | + |
| col16a1 | − | + | + |
| col18a1 | + | ++ | +++ |
| col19a1 | ++ | + | + |
| Reference gene | |||
| β-actin | + | + | + |
Total RNA was isolated from developing zebrafish at 1 and 6 dpf, as well as from an ~ 2-year old adult. The presence of gene expression was determined by RT-PCR with specific primers using 35 cycles. The presence (+) or absence (−) of the PCR product was assessed by 2% agarose gel electrophoresis. The relative amount of each PCR product at different developmental stages was semi-quantitatively indicated by +, ++ or +++.
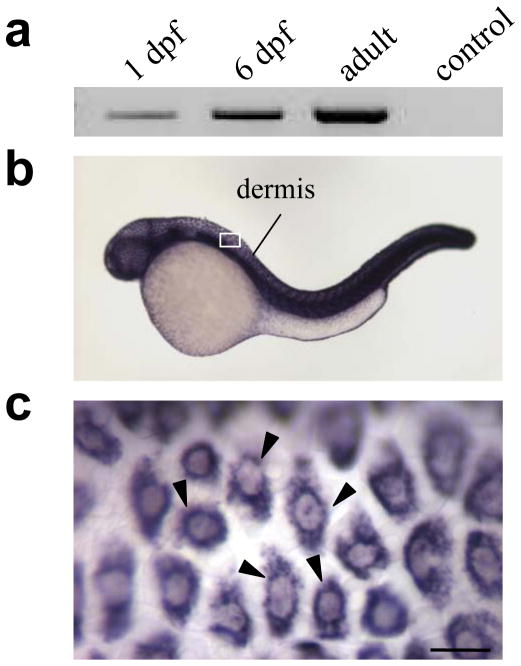
The spatial gene expression during zebrafish development can be monitored by in situ hybridizations that allow identification and location of mRNAs both during different developmental stages and also at their cellular locations (Thisse and Thisse, 2008). For example, temporal expression of the col1a1 gene can be detected as early as at ~10 hpf (Fig. 2a), and in situ hybridization localizes the corresponding mRNAs to the cellular elements in the developing skin (Fig. 2b, c). The precise identity of these cells remains to be explored, but it has been reported that the collagenous stroma of the zebrafish skin is acellular until 26 dpf and that up to this time point collagen is synthesized by keratinocytes (Le Guellec et al, 2004). Thus, the available technologies, such as RT-PCR and in situ hybridization, readily allow determination of the temporal and spatial expression of genes in developing zebrafish. Analogous techniques at the protein level, including Western blot analysis and immunohistochemistry, can also be used to study the gene expression, but these approaches are often hampered by the paucity of antibodies recognizing the zebrafish protein sequences.
Figure 2.
Demonstration of temporal and spatial expression of the col1a1 gene in the developing zebrafish. (a) Total RNA isolated from zebrafish at 1 or 6 dpf or from a ~2-year old adult fish was examined for the presence of col1a1 mRNA by RT-PCR. The control lane was amplified without RNA. (b) In situ hybridization with a col1a1 antisense RNA probe on a 42–48 hpf zebrafish embryo reveals the presence of the corresponding mRNA (purple color). (c) Higher magnification of the area within the rectangle in (b) shows the presence of mRNAs within distinct cells in the skin (arrowheads). (Scale bar = 10 μm).
Morpholino Knockdown of Zebrafish Gene Expression
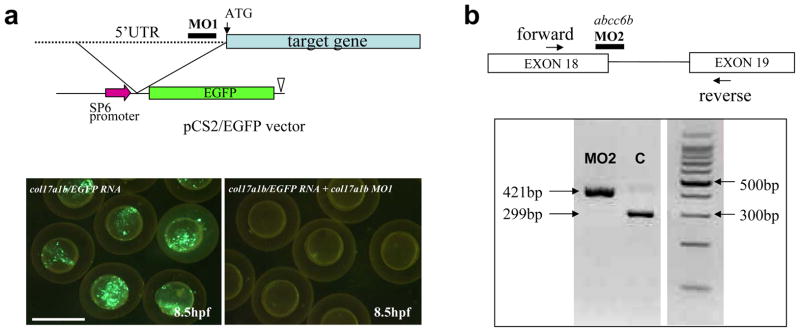
One of the convenient features of zebrafish as a model system to study heritable skin diseases is that the expression of specific genes can be manipulated by injection of 1–4 cell embryos with morpholino-based antisense oligonucleotides (Eisen and Smith, 2008; Shan, 2010). These oligonucleotides can be targeted either to correspond to the sequences around or slightly upstream from the translation initiation codon (AUG) to prevent translation of the mRNA, or to the splice junction sequences to prevent processing of the pre-mRNA to the corresponding mature mRNA (Fig. 3). In case of morpholinos targeting the upstream regulatory sequences, the efficacy of the down-regulation of the corresponding gene expression can be monitored by co-injection of the morpholino with an expression construct containing the 5′ regulatory elements linked to a green fluorescent protein reporter (Fig. 3a). In the case of splice junction morpholinos, the efficiency of the morpholino-mediated knockdown of the gene expression can be monitored by RT-PCR of the RNA sequences to determine whether the morpholino has been effective in preventing the splicing of the targeted intron (Fig. 3b). For example, we have been successful in efforts to knockdown the col17a1b gene expression in zebrafish by a specific morpholino targeting the 5′ regulatory sequences, with essentially 100 percent efficiency, as shown in Fig. 3 (Kim et al, 2010). Similarly, the efficacy of 100 percent of a morpholino placed on the exon 18/intron 19 junction of the abcc6b gene is illustrated in Fig. 3 (Li et al, 2010).
Figure 3.
Morpholino-mediated knockdown of zebrafish genes. (a) A morpholino (MO1) corresponding to the col17a1b gene was used to target the 5′ untranslated region of the corresponding mRNA to prevent translation. To determine the efficacy of morpholino in down-regulating the translation, an expression construct consisting of SP6 promoter, 5′ UTR of the col17a1b gene, and downstream enhanced green fluorescent protein (EGFP) reporter gene was generated. Microinjection of mRNA transcribed in vitro from the pCS2/EGFP vector to 1–4 cell stage embryos, shows green fluorescence at 8.5 hpf (lower left panel). Co-injection of this mRNA together with the MO1 morpholino completely abolished the fluorescence, indicating inhibition of the translation (Scale bar = 1 mm). (b) A morpholino (MO2) corresponding to the zebrafish abcc6b gene was placed on the exon 18/intron 18 splice junction. Efficiency of the morpholino in preventing splicing of the abcc6b pre-mRNA into mature mRNA was monitored by RT-PCR using primers placed on exon 18 (forward) and exon 19 (reverse). PCR of the genomic sequence resulted in a 421 bp fragment while fully spliced cDNA yields a 299 bp fragment devoid of intron 18 (122 bp). RT-PCR of morpholino (MO2)-treated zebrafish embryo reveals the presence of the 421 bp mRNA sequence only, indicating complete inhibition of the removal of intron 18 by splicing. Since intron 18 sequence is out-of-frame, this results in complete absence of the abcc6b protein product. (Figures 3a and b are reproduced from Kim et al, 2010, and Li et al, 2010, respectively, with permission).
The specificity of the morpholino knockdown can be confirmed by co-injection of the corresponding mRNA or protein from another species, such as mouse or human, which counteracts the development of the phenotype (Eisen and Smith, 2008). Furthermore, use of a standard control morpholino, which is biologically inactive since there is no target sequence for it in the zebrafish genome, can confirm the specificity of a morpholino (Robu et al, 2007). It should be noted that microinjection of purified mRNA (without morpholinos) can also be used to monitor the effects of overexpression of the corresponding gene during zebrafish development.
It should be noted that the morpholinos have a relatively short half life (up to 5 days), and therefore this approach is most suitable for evaluation of the effects of a morpholino on the early zebrafish development. Thus, the morpholino knockdown phenotype is most likely to reproduce the clinical manifestations in human diseases which develop during prenatal development or shortly after birth. An example of such situation is the development of epidermal perturbations in zebrafish as a result of knockdown of abca12, the gene harboring mutations in the harlequin ichthyosis (Frank et al, 2010). Specifically, the morphant fish demonstrated absence of microridges and development of scale-like spicules on the surface of the skin, somewhat resembling the scales in human ichthyosis (Frank et al, 2010). Another example of zebrafish phenotype mimicking a human skin disease is the morpholino knockdown of the col17a1a gene expressed in the skin in hemidesmosomal complexes. These knockdown fish manifest with blistering of the epidermis similar to a form of junctional EB due to mutations in the COL17A1 gene (Kim et al, 2010). In contrast, diseases that are of late onset or slowly progressive may not be evident in the zebrafish model system. An example of such conditions is PXE, a slowly progressive, ectopic mineralization disorder with late onset. Specifically, the clinical diagnosis of PXE in humans is not frequently made until an individual is in the teenage years or early twenties. Similarly, KO mice, in which the Abcc6 gene has been inactivated by targeted ablation, characteristic mineralization is not evident until at 5–6 weeks of life (Jiang et al, 2007, 2009). Injection of the abcc6a morpholino in the zebrafish resulted in a early phenotype of pericardial edema and curled tail, associated with death by 8 dpf, but there was no evidence of ectopic mineralization at this stage (Li et al, 2010). The mineralization phenotype, if developmentally corresponding to human or mouse pathogenesis of PXE, might occur later in life. Therefore, the discordance between the zebrafish phenotypes with that in human and mouse, raises the possibility that the ABCC6 gene products might have different biological functions depending on the species, in which case the abcc6 knockdown in zebrafish is not an appropriate model for PXE (Li et al, 2010).
An unexplored facet of the zebrafish as a model system for heritable skin disorders is the potential influence of the genetic background. The genetic background of the mouse strains may have a major influence on the development of the disease phenotype, as has been shown, for example, in the case of PXE-like mineralization of connective tissues in the Abcc6 KO mice (Li and Uitto, 2010; Hovnanian, 2010). In case of zebrafish, the genetic background of strains (and substrains) has been reported to be highly variable, thus possibly leading to modulation of the phenotype (Guryev et al, 2006).
Forward Genetics for Identification of Disease-causing Genes
An approach complementary to morpholino knockdown in using zebrafish as a model system to study heritable skin diseases involves forward genetics, in which the fish can be screened for cutaneous phenotypes after ethylnitrosourea (ENU) induced mutagenesis to induce point mutations. Similar random mutagenesis can also be carried out in zebrafish using retroviral methods, which have the advantage of tagging each insertion site, thus facilitating the identification of mutated genes (Jao et al, 2008). Following mutagenesis, a large number of embryos and larvae can be screened for phenotypic manifestations in the skin, allowing a large scale screen without elaborate infrastructure or equipment. Such a screen, which is facilitated by transparency of the developing larvae, has an advantage over other vertebrate systems including the mouse (Table 2). These large-scale forward-genetic screens have allowed identification of a number of mutated genes orthologous to those causing human heritable diseases, with phenotypic similarities (Amsterdam and Hopkins, 2006; Lieschke and Currie, 2007). In addition, this approach can be used to identify novel candidate genes that can then be tested in patients with similar phenotypes but with no known mutations. Once the zebrafish disease model has been established, the embryos are readily amenable to further investigation for the molecular and cellular bases of the disease. It should be noted that these approaches have also been used to examine more complex pathological pathways, such as those involved in carcinogenesis and melanoma formation, and the zebrafish models of human diseases can also be used for small-molecular-weight compound screening and for drug discovery programs.
Acknowledgments
Our original studies were supported by the United States Department of Health and Human Services, NIH/NIAMS (JU), the Pilot Research Award Program of Thomas Jefferson University (QL), the Dermatology Foundation Research Career Development Award (QL), and the University of Virginia (CT, BT). Carol Kelly assisted in preparation of this publication.
Abbreviations
- BMZ
basement membrane zone
- EB
epidermolysis bullosa
- PXE
pseudoxanthoma elasticum
- KO
knock-out
- hpf and dpf
hours and days post-fertilization, respectively
Footnotes
Conflict of Interest
The authors state no conflict of interest.
References
- Amsterdam A, Hopkins N. Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet. 2006;22:473–478. doi: 10.1016/j.tig.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Brittijn S, Duivesteijn SJ, Belmamoune M, et al. Zebrafish development and regeneration: new tools for biomedical research. Int J Dev Biol. 2009;53:835–850. doi: 10.1387/ijdb.082615sb. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: Don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Feramisco JD, Sadreyev RI, Murray ML, Grishin NV, Tsao H. Phenotypic and genotypic analyses of genetic skin disease through the Online Mendelian Inheritance in Man (OMIM) database. J Invest Dermatol. 2009;129:2628–2638. doi: 10.1038/jid.2009.108. [DOI] [PubMed] [Google Scholar]
- Froehlicher M, Liedtke A, Groh KJ, Neuhauss SC, Segner H, Eggen RI. Zebrafish (Danio rerio) neuromast: Promising biological endpoint linking development and toxicological studies. Aquat Toxicol. 2009;95:307–319. doi: 10.1016/j.aquatox.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Frank M, Li Q, Akiyama M, et al. The abca12 Gene is Required for Normal Zebrafish Skin Development – a Model System for Harlequin Ichthyosis. J Invest Dermatol. 2010;130(Suppl 1):Abstract 516, S86. [Google Scholar]
- Guryev V, Koudijs MJ, Berezikov E, et al. Genetic variation in the zebrafish. J Invest Dermatol. 2006;16:491–497. doi: 10.1101/gr.4791006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Has C, Kern JS. Collagen XVII. Dermatol Clin. 2010;28:61–66. doi: 10.1016/j.det.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Hovnanian A. Modifier genes in pseudoxanthoma elasticum: Novel insights from the Ggcx mouse model. J Mol Med. 2010;88:149–153. doi: 10.1007/s00109-009-0576-7. [DOI] [PubMed] [Google Scholar]
- Jao LE, Maddison L, Chen W, Burgess SM. Using retroviruses as a mutagenesis tool to explore the zebrafish genome. Brief Funct Genomic Proteomic. 2008;7:417–443. doi: 10.1093/bfgp/eln038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Li Q, Uitto J. Aberrant mineralization of connective tissues in a mouse model of pseudoxanthoma elasticum. Systemic and local regulatory factors. J Invest Dermatol. 2007;127:1392–1402. doi: 10.1038/sj.jid.5700729. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Endo M, Dibra F, Wang K, Uitto J. Pseudoxanthoma elasticum is a metabolic disease. J Invest Dermatol. 2009;129:348–354. doi: 10.1038/jid.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-H, Choi HY, So J-H, et al. Zebrafish type XVII collagen: Gene structures, expression profiles, and morpholino “knock-down” phenotypes. Matrix Biol. 2010;29:629–637. doi: 10.1016/j.matbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Le Guellec D, Morvan-Dubois G, Sire J-Y. Skin development in bony fish with particular emphasis on collagen deposition in the dermis of the zebrafish (Danio rerio) Int J Dev Biol. 2004;48:217–231. doi: 10.1387/ijdb.15272388. [DOI] [PubMed] [Google Scholar]
- Lee Y, Nachtrab G, Klinsawat PW, Hami D, Poss KD. Ras controls melanocyte expansion during zebrafish fin stripe regeneration. Dis Model Mech. 2010;3:496–503. doi: 10.1242/dmm.004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, MacDonald JR, Wei RY, et al. Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse. BMC Genomics. 2007;8:92. doi: 10.1186/1471-2164-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Jiang Q, Pfendner E, et al. Pseudoxanthoma elasticum: clinical phenotypes, molecular genetics and putative pathomechanisms. Exp Dermatol. 2009;18:1–11. doi: 10.1111/j.1600-0625.2008.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Uitto J. The mineralization phenotype in Abcc6−/− mice is affected by Ggcx gene deficiency and genetic background – a model for pseudoxanthoma elasticum. J Mol Med. 2010;88:173–181. doi: 10.1007/s00109-009-0522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Sadowski S, Frank M, et al. The abcc6a gene expression is required for normal zebrafish development. J Invest Dermatol. 2010;130:2561–2568. doi: 10.1038/jid.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke G, Currie PD. Animal models of human disease: Zebrafish swim into view. Nature Rev. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- O’Reilly-Pol T, Johnson SL. Melanocyte regeneration reveals mechanisms of adult stem cell regulation. Semin Cell Dev Biol. 2009;20:117–124. doi: 10.1016/j.semcdb.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH. The zebrafish genome in context: Ohnologs gone missing. J Exp Zool. 2007;308B:563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Ringpfeil F, Uitto J. Progress in heritable skin diseases: Molecular bases and clinical implications. J Am Acad Dermatol. 2002;47:91–104. doi: 10.1067/mjd.2002.120601. [DOI] [PubMed] [Google Scholar]
- Rakers S, Gebert M, Uppalapati S, et al. “Fish matters”: The relevance of fish skin biology to investigative dermatology. Exp Dermatol. 2010;19:313–324. doi: 10.1111/j.1600-0625.2009.01059.x. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, et al. p53 activation by knock down technologies. PLos Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch GJ, Wu S, Sherwood NM. Hormones and receptors in fish: Do duplicates matter? Gen Comp Endocrinol. 2009;161:3–12. doi: 10.1016/j.ygcen.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Seppänen A, Autio-Harmainen H, Alafuzoff I, et al. Collagen XVII is expressed in human CNS neurons. Matrix Biol. 2006;25:185–188. doi: 10.1016/j.matbio.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Shan G. RNA interference as gene knockdown technique. Int J Biochem Cell Biol. 2010;42:1243–1251. doi: 10.1016/j.biocel.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Sire J-Y, Akimendo MA. Scale development in fish: A review, with description of sonic hedgehog (shh) expression in the zebrafish (Danio rerio) Int J Dev Biol. 2004;48:233–247. [PubMed] [Google Scholar]
- Sonawane M, Carpio Y, Geisler R, Schwarz H, Maischein H-M, Nüsslein-Volhard C. Zebrafish penner/lethal giant larvae 2 functions in hemidesmosome formation, maintenance of cellular morphology and growth regulation in the developing basal epidermis. Development. 2005;32:3255–3265. doi: 10.1242/dev.01904. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Sprecher E. Familial tumoral calcinosis: From characterization of a rare phenotype to the pathogenesis of ectopic calcification. J Invest Dermatol. 2010;130:652–660. doi: 10.1038/jid.2009.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J. Progress in heritable skin diseases: Translational implications of mutation analysis and prospect of molecular therapies. Acta Derm Venereol. 2009;89:228–235. doi: 10.2340/00015555-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J, Christiano AM. Molecular genetics of the cutaneous basement membrane zone. Perspectives on epidermolysis bullosa and other blistering skin diseases. J Clin Invest. 1992;90:687–692. doi: 10.1172/JCI115938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J, McGrath JA, Rodeck U, Bruckner-Tuderman L, Robinson EC. Progress in epidermolysis bullosa research: Toward treatment and cure. J Invest Dermatol. 2010a;130:1778–1784. doi: 10.1038/jid.2010.90. [DOI] [PubMed] [Google Scholar]
- Uitto J, Li Q, Jiang Q. Pseudoxanthoma elasticum: molecular genetics and putative pathomechanisms. J Invest Dermatol. 2010b;130:661–670. doi: 10.1038/jid.2009.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanchieri C. Move over, mouse: Make way for the woodchucks, ferrets, and zebrafish. J Natl Cancer Inst. 2001;93:418–419. doi: 10.1093/jnci/93.6.418. [DOI] [PubMed] [Google Scholar]