Abstract
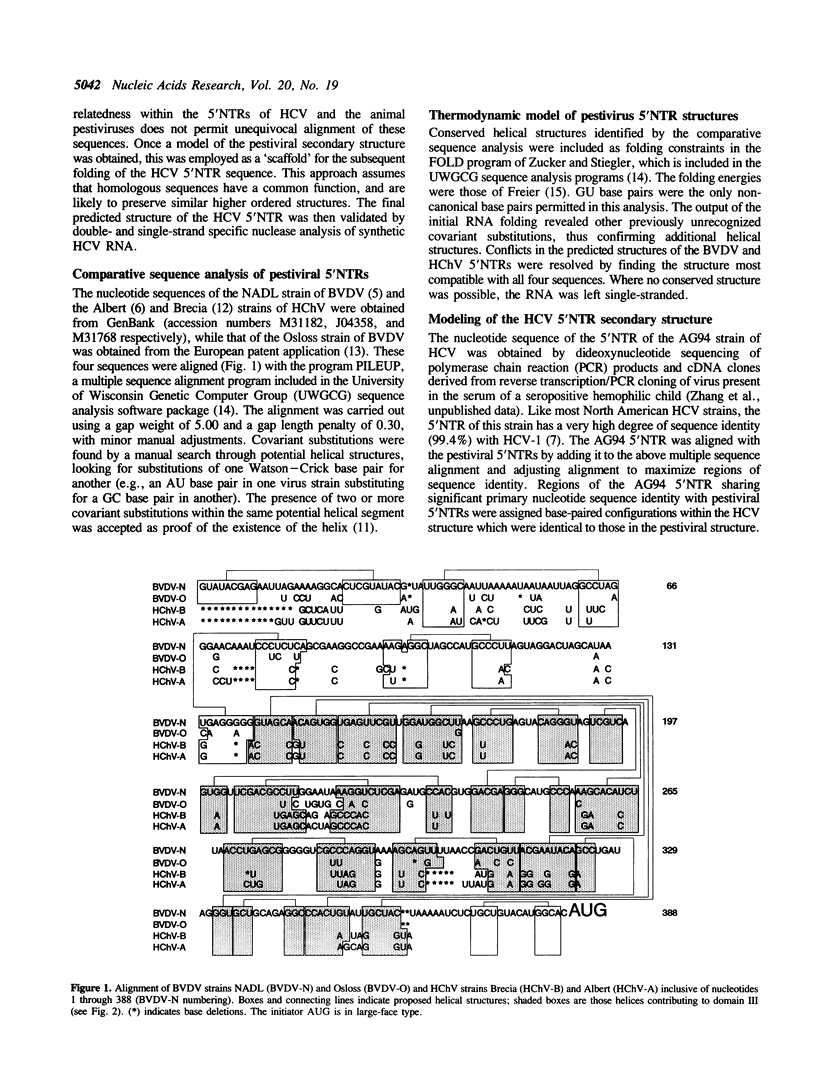
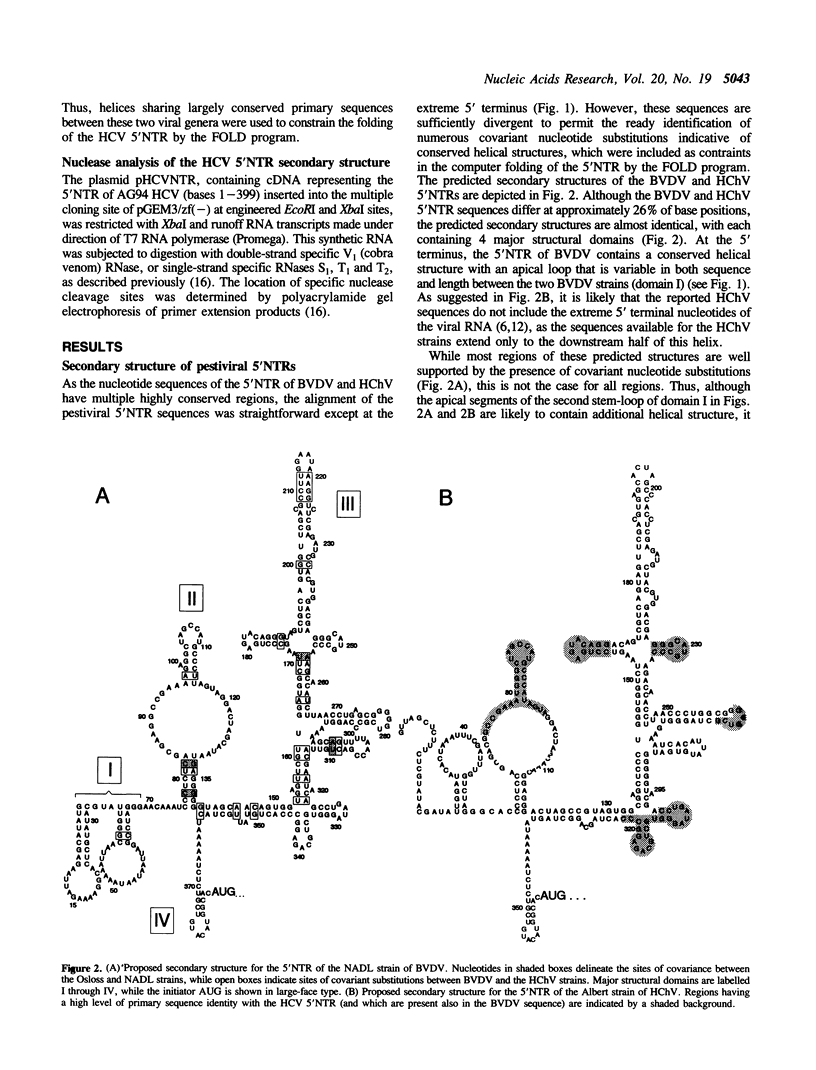
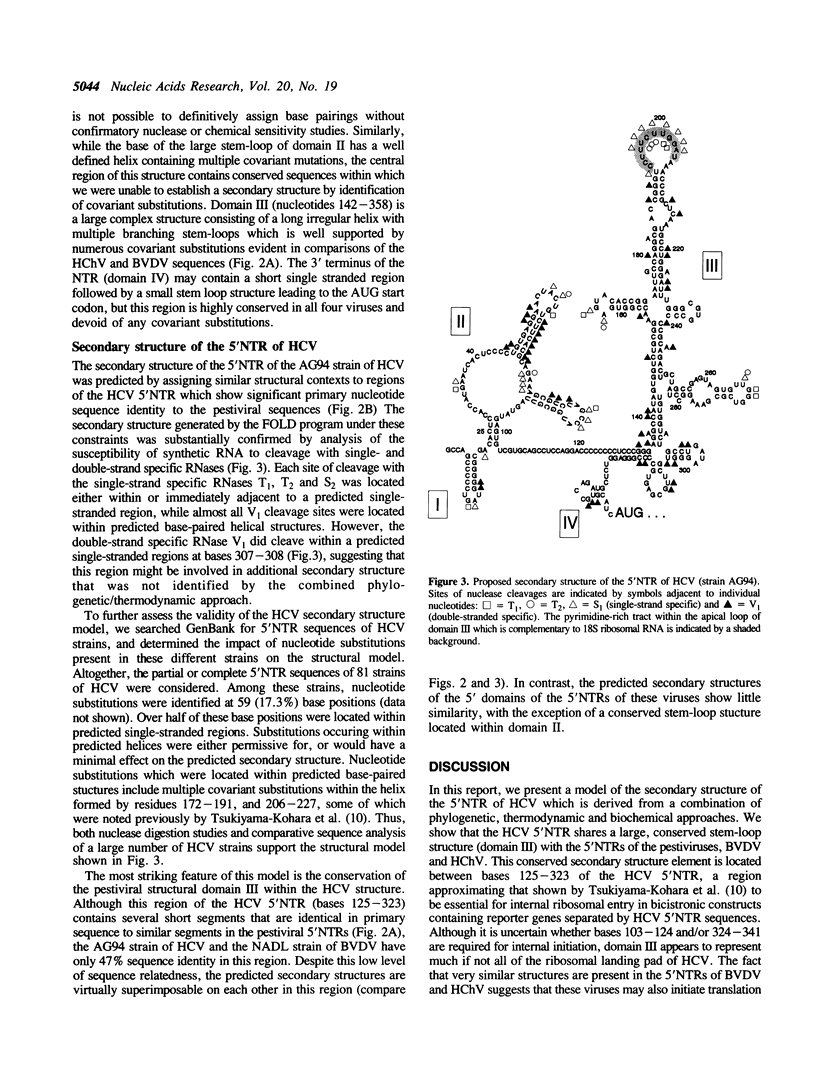
The RNA genomes of human hepatitis C virus (HCV) and the animal pestiviruses responsible for bovine viral diarrhea (BVDV) and hog cholera (HChV) have relatively lengthy 5' nontranslated regions (5'NTRs) sharing short segments of conserved primary nucleotide sequence. The functions of these 5'NTRs are poorly understood. By comparative sequence analysis and thermodynamic modeling of the 5'NTRs of multiple BVDV and HChV strains, we developed models of the secondary structures of these RNAs. These pestiviral 5'NTRs are highly conserved structurally, despite substantial differences in their primary nucleotide sequences. The assignment of similar structures to conserved segments of primary nucleotide sequence present in the 5'NTR of HCV resulted in a model of the secondary structure of the HCV 5'NTR which was refined by determining sites at which synthetic HCV RNA was cleaved by double- and single-strand specific RNases. These studies indicate the existence of a large conserved stem-loop structure within the 3' 200 bases of the 5'NTRs of both HCV and pestiviruses which corresponds to the ribosomal landing pad (internal ribosomal entry site) of HCV. This structure shows little relatedness to the ribosomal landing pad of hepatitis A virus, suggesting that these functionally similar structures may have evolved independently.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agol V. I. The 5'-untranslated region of picornaviral genomes. Adv Virus Res. 1991;40:103–180. doi: 10.1016/S0065-3527(08)60278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. A., Day S. P., Jansen R. W., Lemon S. M. The 5' nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J Virol. 1991 Nov;65(11):5828–5838. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q. L., Richman K. H., Han J. H., Berger K., Lee C., Dong C., Gallegos C., Coit D., Medina-Selby R., Barr P. J. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colett M. S., Larson R., Gold C., Strick D., Anderson D. K., Purchio A. F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988 Jul;165(1):191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. H., Shyamala V., Richman K. H., Brauer M. J., Irvine B., Urdea M. S., Tekamp-Olson P., Kuo G., Choo Q. L., Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5' untranslated region and poly(A) tails at the 3' end. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Pace N. R. Phylogenetic comparative analysis of RNA secondary structure. Methods Enzymol. 1989;180:227–239. doi: 10.1016/0076-6879(89)80104-1. [DOI] [PubMed] [Google Scholar]
- Jang S. K., Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990 Sep;4(9):1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- Meyers G., Rümenapf T., Thiel H. J. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology. 1989 Aug;171(2):555–567. doi: 10.1016/0042-6822(89)90625-9. [DOI] [PubMed] [Google Scholar]
- Miller R. H., Purcell R. H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moenning V. Pestiviruses: a review. Vet Microbiol. 1990 Jun;23(1-4):35–54. doi: 10.1016/0378-1135(90)90135-i. [DOI] [PubMed] [Google Scholar]
- Moormann R. J., Warmerdam P. A., van der Meer B., Schaaper W. M., Wensvoort G., Hulst M. M. Molecular cloning and nucleotide sequence of hog cholera virus strain Brescia and mapping of the genomic region encoding envelope protein E1. Virology. 1990 Jul;177(1):184–198. doi: 10.1016/0042-6822(90)90472-4. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Kurai K., Okada S., Yamamoto K., Lizuka H., Tanaka T., Fukuda S., Tsuda F., Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992 May;188(1):331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988 Jul 28;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Gmyl A. P., Maslova S. V., Svitkin Y. V., Sinyakov A. N., Agol V. I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992 Jan 10;68(1):119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992 Mar;66(3):1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]



