Abstract
MIST1 is a transcription factor expressed in pancreatic acinar cells and other serous exocrine cells. Mice harboring a targeted deletion of the Mist1 gene (Mist1−/−) exhibit alterations in acinar regulated exocytosis and aberrant Ca2+ signaling that are normally controlled by acinar cell Ca2+-ATPases. Previous studies indicated that total sarcoendoplasmic reticulum Ca2+-ATPases (SERCA) and plasma membrane Ca2+-ATPases (PMCA) remained unaffected in Mist1−/− acinar cultures. Therefore, we have assessed the expression of Atp2c2, the gene that encodes the secretory pathway Ca2+-ATPase 2 (SPCA2). We revealed a dramatic decrease in pancreatic expression of Atp2a2 mRNA and SPCA2 protein in Mist1−/− mice. Surprisingly, this analysis indicated that the acinar-specific Atp2c2 mRNA is a novel transcript, consisting of only the 3′ end of the gene and the protein and localizes to the endoplasmic reticulum. Expression of SPCA2 was also lost in Mist1−/− secretory cells of the salivary glands and seminal vesicles, suggesting that Atp2c2 transcription is regulated by MIST1. Indeed, inducible MIST1 expression in Mist1−/− pancreatic acinar cells restored normal Atp2c2 expression, supporting a role for MIST1 in regulating the Atp2c2 gene. Based on these results, we have identified a new Atp2c2 transcript, the loss of which may be linked to the Mist1−/− phenotype.
Keywords: bHLH transcription factors, Ca2+ ATPases, Golgi apparatus, pancreatic acinar cells, regulated exocytosis, bHLHa15
Introduction
MIST1 is a transcription factor belonging to the class B family of basic helix-loop-helix (bHLH) proteins [1]. Class B bHLH proteins typically exhibit tissue specific expression and affect transcription of genes involved in cell determination, differentiation and cellular functions. Mist1 (also referred to as Basic helix loop helix a15; Bhlha15, or dimmed in Drosophila) is expressed in all serous type exocrine cells including pancreatic and salivary acini, Chief cells of the stomach, and secretory cells of the seminal vesicle, prostate and lactating mammary gland [2]. All MIST1 expressing cells rely on regulated exocytosis for their primary function. During regulated exocytosis, Ca2+ is tightly controlled with release into and uptake from the cytosol being regulated by proteins that act as ion channels and pumps, respectively [3, 4]. Upon secretagogue stimulation, Mist1−/− pancreatic acini exhibit a rapid release of Ca2+ into the cytosol but clearance of Ca2+ from the cytosol does not occur, leading to sustained high levels of cytosolic Ca2+ within Mist1−/− acinar cells [5]. The defective Ca2+ signaling in Mist1−/− acinar cells suggests that expression of proteins regulating Ca2+ movement within the cell may be compromised.
The key protein families involved in maintaining appropriate levels of cytosolic Ca2+ are the Ca2+-ATPases. Ca2+-ATPases belong to the phosphorylation-type (P-type) ATPase family and include sarcoendoplasmic reticulum Ca2+-ATPases (SERCAs), plasma membrane Ca2+-ATPases (PMCAs) and secretory pathway Ca2+-ATPases (SPCAs) [6–8]. These pumps either move activator Ca2+ out of the cell (PMCAs) or move Ca2+ from the cytosol back into Ca2+ stores such as the ER (SERCAs) and Golgi (SPCAs) [9]. Previous studies indicated no change in the total expression levels of SERCA and PMCA proteins in Mist1−/− acini cultures but the SPCA proteins were not examined [5].
In this study, we examined expression of the Atp2a, -b, -c gene family in wild type (WT) and Mist1−/− mice. In Mist1−/− pancreata, expression of Atp2c2, correlating to the SPCA2 protein, was dramatically decreased relative to WT tissue. A similar decrease was observed in all tissues examined that normally express MIST1. Loss of SPCA2 correlated with expansion of the Golgi compartment and disruption of Golgi function. Re-establishment of MIST1 expression in Mist1−/− pancreata resulted in restoration of Atp2c2 transcript levels. These results suggest that Atp2c2 is a downstream target of MIST1 transcriptional activity and may be responsible, in part, for defective Ca2+ signaling in Mist1−/− pancreata.
Experimental Procedures
All chemicals were purchased from Sigma (St. Louis, MO) unless otherwise stated.
Mice
C57 Bl/6 mice were handled according to the regulations set for The University of Western Ontario Animal Care by the Canadian Council of Animal Care (protocol #2008-116). Mice lacking Mist1 (Mist1−/−) have been previously described [10]. Transgenic mice (LSL-Mist1myc) were generated that express a myc-tagged MIST1 protein that can be selectively activated through Cre-mediated recombination. Mice harboring a targeted replacement of the Mist1 gene with CreERT2 (Mist1CreER/CreER) have been previously described [11]. Standard crosses were performed to produce a Mist1−/− mouse containing the CreERT2-inducible LSL-Mist1myc transgene (LSL-Mist1myc/Mist1CreER/CreER). Induction of Mist1CreER activity was accomplished by providing 8 week Mist1CreER/CreER/LSL-Mist1myc mice (in triplicate for each group and time point) a single dose of tamoxifen (4 mg/mouse) or control corn oil as previously described [11]. All mice were provided with a standard diet and water ad libidum.
RNA isolation and quantification
All reagents were from Promega (Ottawa, ON) unless otherwise stated. RNA was isolated from the pancreas as described [12] or from whole tissues, primary acinar cell cultures, or AR42J and ARIP cell lines using Trizol (Invitrogen, Burlington, ON) as per manufacturer’s protocol. Alternatively, pancreas RNA was isolated at 24 hr, 48 hr, and 72 hr post-tamoxifen treatment of Mist1CreER/CreER/LSL-Mist1myc mice using the RNeasy isolation system (QIAGEN, Valencia, CA) and reverse transcribed using the iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA). For RT-PCR, 1 μg of total RNA was reversed transcribed using random primers and Improm II reverse transcriptase, then amplified through 35 rounds of denaturing (94°C for 30 s), annealing (60–68°C for 40 s), and elongation (72°C for 60 s). For primer sequences and conditions see Table S1. All primers were designed using sequences from the National Center for Biotechnology Information web database (www.ncbi.nlm.nih.gov) and generated by Sigma-Genosys (Oakville, ON).
Quantitative (q) RT-PCR was performed using the Roche LightCycler® 2.0 System (Roche, Mannheim, Germany) with the LightCycler® FastStart DNA MasterPLUS SYBR Green I kit. The amplification process consisted of an initial denaturation step (95°C for 10 s), a reaction cycle (95°C for 10 s), annealing (59.9–68.1°C for 5 s), and elongation (72°C for 10 s). For primer sequences and conditions see Table S1.
Northern blot analysis was performed as described [12]. Blots were hybridized overnight at 42°C with α32P-dCTP-radiolabeled probe (1×106 cpm/ml). The template for the probe was made from DNA plasmids containing the Atp2c2 sequence.
SPCA2 antibody generation and protein analysis
In collaboration with ProSci Inc. (Poway, CA), a peptide sequence for antibody production was determined using the sequence of the carboxy-terminal domain of the mouse SPCA2 (NCBI Accession #ABS18966). The optimal antigenic site was determined to be KLWEKFLSRARPTQMLPEAV.
Protein was isolated and western blot analysis was performed as described [13]. 10–50 μg of protein was resolved by SDS-PAGE and transferred to PVDF membrane (BioRad, Hercules, CA). The rabbit anti-SPCA2 (1:3000) and anti-β-tubulin (1:500, Santa Cruz) were recognized by HRP-conjugated goat anti-rabbit IgG (1:10000; Jackson Immunoresearch Labs, West Grove, PA). The mouse anti-adaptin γ and anti-GM130 (1:500, 1:1000, respectively, BD Biosciences, Mississauga, ON) were recognized by HRP-conjugated goat anti-mouse IgG (1:2000, Jackson Immunoresearch Labs). As a control for the specificity of the SPCA2 antibody, western blots were repeated using preimmune serum or following preabsorption with purified SPCA2 antigen.
For immunohistochemistry (IHC), pancreatic tissue was isolated from WT and Mist1−/− mice, embedded in paraffin and sectioned to 5 μm. Following deparaffinization, antigen retrieval was performed with a 20 minute incubation in sodium citrate buffer (pH=6) followed by washing in PBS, 2 × 5 minute washes with 0.2% Triton X in PBS, and more washes in PBS. Endogenous HRP activity was quenched with a 5 minute incubation in 3% H2O2. Sections were blocked for 30 minutes in 10% goat serum and incubated for one hour at room temperature in primary antibodies diluted in blocking serum. Following PBS washes, sections were incubated in biotinylated secondary antibodies diluted in blocking serum for 30 minutes at room temperature, washed and then incubated for 30 minutes at room temperature in ABC complex, prepared as described by manufacturer (Pierce, Rockford, IL). HRP was detected with a diaminobenzidine staining kit (Pierce, Rockford, IL) and then sections were washed counterstained with hematoxylin. For immunofluorescence (IF), pancreata were processed as described [10].
Primary antibodies used included rabbit anti-SPCA2 (1:250), rabbit anti-calreticulin (1:250; Chemicon), mouse anti-GM130 (1:200, BD Biosciences), mouse anti-SERCA2 (Pierce, Rockford, IL) and mouse anti-adaptin γ (1:250, BD Biosciences). Secondary antibodies included biotinylated goat-anti mouse IgG or anti rabbit IgG (1:200), fluorescein isothiocyanate (FITC) goat anti-rabbit IgG and tetramethylrhodamine isothiocyanate (TRITC) goat anti-mouse IgG (1:250; Jackson Immunoresearch Labs, West Grove, PA). In some cases, IF was combined with FITC-conjugated wheat germ agglutinin (WGA; incubated 1:1000). IF was visualized with a Leica DMIOS upright microscope or an Olympus Fluoview FV1000 confocal microscope. Images were captured using the Open Lab 3.1.5 imaging program (Quorom Techniologies, Guelph, ON) or FV 10-ASW 1.6 (FluoView 1000 software, Olympus corporation) and Image-Pro Analyzer (version 6.2, Media Cybernetics) software, respectively.
In situ hybridization
In situ hybridization was performed on 7 μm cryosections as described [13]. Digoxigenin-labelled RNA probes were synthesized by in vitro transcription of region-spanning parts of the insulin and amylase coding regions from pancreatic cDNA and the Atp2c2 clone (#4984604) (for primer sequences, see Table S1) using T3 RNA polymerase. Staining was visualized using OpenLab 4.0.3 Modular Software for Scientific Imaging.
Construct design and cloning
To generate FLAG-tagged, C-terminal SPCA2 expression vector, primers were designed to amplify exons 24 to 27 of the Atp2c2 gene (Table S1). FLAG-Spca2760-944 was amplified from pCMV-SPORT6-3′Spca2 plasmid (clone # 4984604) and ligated into the pSC-A vector using the Strataclone™ PCR Cloning kit (Stratagene, La Jolla, CA). pSC-A-FLAG-Spca2760-944 was digested with XbaI and BamHI and ligated into pcDNA3.0(+) (cut with the same enzymes) using the Roche Rapid DNA ligation kit and transformed into DH5α E. coli competent cells (Invitrogen).
Cell culture and transfection
All cell culture reagents were purchased from Gibco (Burlington, ON) unless otherwise stated. Primary cell cultures were established as described [14]. ARIP and AR42J were maintained in F12 Kaighn’s Modification (F12-K) media containing 4 mM L-glutamine, 1% Pencillin/Streptomycin (Penstrep) and 10% (ARIP) or 20% (AR42J) fetal bovine serum (FBS). HEK 293T (ATCC, Manassas, VA) were maintained in 1X Dulbecco’s Modified Eagle Medium (DMEM) high glucose media containing 4.5 g/L D-glucose, 4 mM L-glutamine, 10% FBS and 1% Penstrep. Transfections of HEK 293T cells were performed at 50–60% confluency in 6-well plates with pcDNA3.0-FLAG-SPCA2760-944, pcDNA3-Mist1HA and pcDNA3-GFP constructs. A total of 1 μg of DNA was added per well in 1X HEPES Buffered Saline Solution (HBSS) and 125 mM CaCl2. The solution was gently agitated and added to each well in a dropwise manner. Media was changed the following day and protein isolated or cells were fixed for IF 48 hours after transfection.
Results
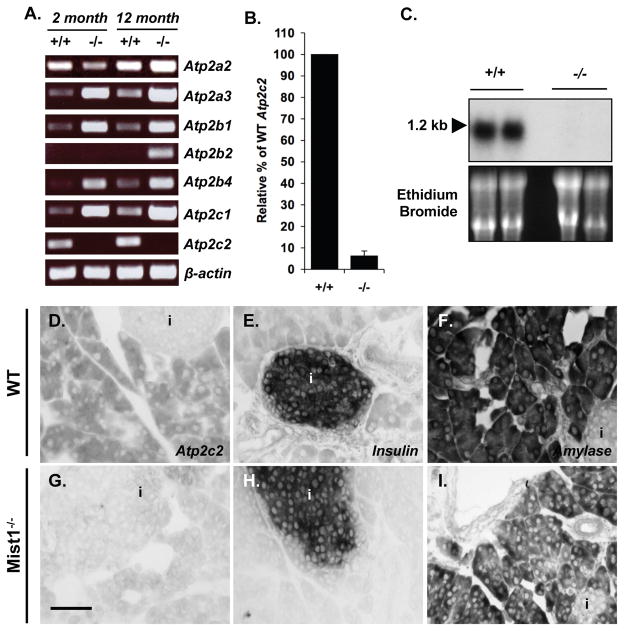
Loss of MIST1 results in altered Ca2+ movement and decreased regulated exocytosis in pancreatic acinar cells [5] (Supplemental Figure S1). Analysis of individual proteins involved in Ca2+ movement within the cell revealed a decrease in IP3R3 accumulation but no change in the protein levels of the SERCA or PMCA pumps in isolated acini [5, 15]. The maintained expression of the SERCA and PMCA pumps was surprising since Mist1−/− acinar cells exhibit severe defects in moving Ca2+ back out of the cytosol following secretagogue stimulation. To address this issue, we examined expression of the Atp2 gene family in WT and Mist1−/− pancreatic tissue from 2- and 12-month old mice. As shown in Figure 1A, a marked increase in expression of many Atp2 genes was observed in Mist1−/− tissues, suggesting that Mist1−/− acinar cells attempt to compensate the increased cytosolic Ca2+ pools by increasing Atp2 levels within the cell. Indeed, increased cytosolic Ca2+ has been shown to increase expression of several Ca2+-ATPases in other models [16, 17], so a compensatory increase in the SERCA and PMCA pumps would not be surprising. The only exception to the compensation mechanism was the Atp2c2 gene where transcripts were readily detected in all WT pancreatic samples but were absent in Mist1−/− pancreata (Figure 1A). The decrease in Atp2c2 expression was confirmed by qRT-PCR (Figure 1B), Northern blot (Figure 1C) and in situ hybridization analyses (Figure 1D–I) where Atp2c2 transcripts were exclusively restricted to acinar cells (Figure 1D).
Figure 1. Atp2c2 mRNA is reduced in Mist1−/− pancreatic tissue.
(A) RT-PCR for Atp2a2 and 3, Atp2b1, 2 and 4, or Atp2c1 and 2 on two and twelve month-old WT (+/+) and Mist1−/− (−/−) pancreatic RNA. β-actin was used as a positive control for RT reactions. (B) Quantitative RT-PCR for Atp2c2 expression in WT and Mist1−/− pancreatic RNA. The WT amount is set to 100%. (C) Northern blot analysis on WT and Mist1−/− pancreatic RNA using a probe specific to the 3′ end of the Atp2c2 sequence. (D–I) In situ hybridization on WT (D–F) and Mist1−/− (G–I) pancreatic tissue sections using probes for the 3′ end of Atp2c2 (D, G), insulin (E, H) and amylase (F, I). Scale bar = 95 μm; i - islet.
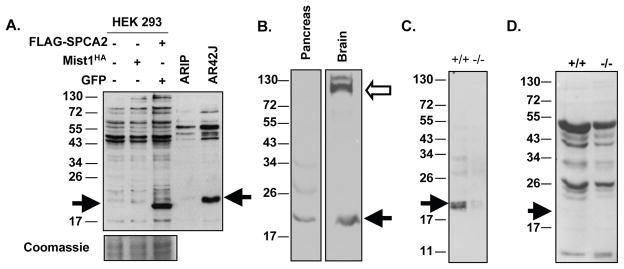
To further examine SPCA2, which is the protein encoded by the Atp2c2 gene, we generated an antibody specific for its C-terminal region. Western blots of protein extracts from HEK 293T cells transfected with expression vectors for a truncated FLAG-tagged SPCA2 (FLAG-SPCA2760-944), HA-tagged MIST1 or control GFP protein identified the expected 20 kDa band corresponding to the truncated FLAG-SPCA2760-944 protein (Figure 2A). Similarly, the SPCA2 antibody recognized a protein in the acinar cell line AR42J, but not in ARIP cells which are MIST1 negative [2]. Surprisingly, the protein size in AR42J cells was only slightly larger than the truncated FLAG-SPCA2760-944 protein. This protein was not observed following Western blot analysis after preabsorption with a SPCA2 blocking peptide or with preimmune serum (Supplemental Figure S2). The lower molecular weight isoform of SPCA2 was detected in whole pancreatic and brain protein samples (Figure 2B) but not in Mist1−/− protein extracts (Figure 2C), suggesting that SPAC2 in pancreatic cells is much smaller than its NCBI-predicted molecular mass. Again, preabsorption with the SPCA2 blocking peptide completely abolished detection of the 20 kDa protein even with extended exposures (Figure 2C). Interestingly, we did not detect the published full-length (103 kDa) SPCA2 protein in pancreatic extracts, although this larger isoform was detected in brain samples (Figure 2B).
Figure 2. A truncated isoform of SPCA2/Atp2c2 is expressed in pancreatic acinar cells.
(A) Western blot for SPCA2 on protein extracts from HEK 293T cells transfected with pcDNA3.0-FLAG-SPCA2760-944, pcDNA3-Mist1HA or pcDNA3-GFP or from AR42J and ARIP cells. Coomassie staining was used to verify equal loading. A protein between 17–26 kDa (arrow) was identified in AR42J cells and not in ARIP cells. FLAG-SPCA2760-944 produces a protein of approximately 20 kDa. (B) Western blot analysis on pancreatic and brain protein extracts identified the lower molecular weight isoform in both tissues (arrow) and the larger SPCA2 isoform only in the brain (open arrow). (C) Western blot analysis for SPCA2 reveals this smaller isoform only in WT (arrow) but not Mist1−/− pancreata. (D) Preabsorption with a SPCA2 blocking peptide completely abolished detection of the protein. Higher molecular weight bands begin to appear at longer exposures of the blots.
In an effort to establish if the smaller pancreas SPCA2 protein was the product of degradation, we assessed the size of Atp2c2 transcripts. Northern blot analysis revealed that the pancreas produced a much smaller Atp2c2 transcript than the expected full length transcript (Figure 1C). Quantitative RT-PCR confirmed that transcripts encoding exons 24–27 were approximately 100-fold more abundant in pancreas and 5-fold more abundant in salivary gland samples when compared to transcripts encoding exons 15–17 (Supplemental Figure S3). Importantly, this difference in amplification was not observed in RNA samples from testes, where both sets of primers amplified equivalent amounts of Atp2c2 transcripts. Together, these data suggest that an alternative size transcript of Atp2c2 exists in the pancreas whose expression is regulated by MIST1.
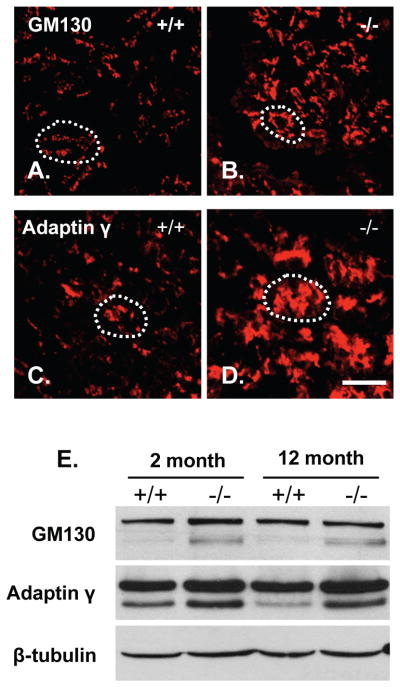
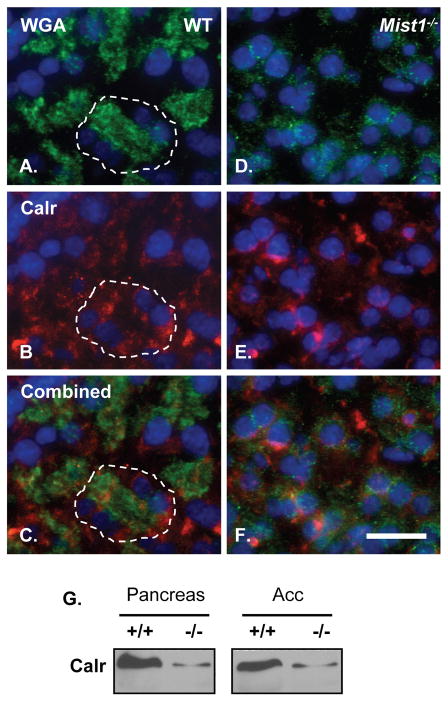
To date, no targeted knockout of Atp2c2 has been reported. However, ablation of the closely related Atp2c1 gene resulted in expansion of the Golgi complex and an altered ER stress response [18]. Previous studies from our group showed that Mist1−/− acini also exhibit an altered ER stress response [12], suggesting that pancreatic SPCA2 may play a similar role as SPCA1. To address this, we next examined the overall organization of acinar Golgi in WT and Mist1−/− pancreata using cis (GM130) and trans (adaptin γ) Golgi-specific markers. Confocal immunofluorescence (IF) analysis revealed an expansion of the Golgi complexes in Mist1−/− acini (Figure 3A–D). Western blots confirmed an increase in adaptin γ and GM130 proteins in Mist1−/− tissue (Figure 3E). To determine if Golgi function was disrupted in acinar cells lacking MIST1, WT and Mist1−/− pancreas sections were incubated with a FITC-conjugated wheat germ agglutinin (WGA), which recognizes sialic acid and N-acetylglucosaminyl sugar residues on glycosylated proteins that are processed through the Golgi. Fluorescent imaging revealed a marked decrease in lectin accumulation in Mist1−/− acinar cells compared to WT pancreata (Figures 4 and S4). FITC-WGA primarily detects acinar cell zymogen granules and does not co-localize with the endoplasmic reticulum (Figure 4). The decrease in FITC-WGA binding in Mist1−/− samples supports the idea of improper Golgi function in Mist1−/− acinar cells and this alteration correlates with the absence of SPCA2. Interestingly, Mist1−/− pancreata also have decreased calreticulin accumulation (Figure 4A, D, G), suggesting widespread defects in the Golgi and ER compartments of these cells.
Figure 3. Mist1−/− acinar cells have an expanded Golgi apparatus.
(A–D) IF on pancreatic sections for Golgi-specific markers GM130 (A, B) or adaptin γ (C, D). White dotted lines indicate an acinus. Scale bars = 40 μm. (E) Western blot analysis for GM130 and adaptin γ on 2 and 12 month WT (+/+) and Mist1−/− (−/−) pancreatic protein. β-tubulin was used as a loading control.
Figure 4. Altered cellular function in Mist1−/− acini.
Localization of FITC-WGA (A, D) combined with IF analysis for calreticulin (Calr; B, E) in WT (A–C) and Mist1−/− (D–F) pancreatic acini. Images were merged in panels C and F. DAPI was used to stain the nuclei (blue). An acinus is indicated in WT sections by the white dashed lines. Scale bars = 20 μm. (G) Western blot analysis for Calreticulin (Calr) in WT (+/+) and Mist1−/− (−/−) pancreata or purified acinar cell (Acc) extracts.
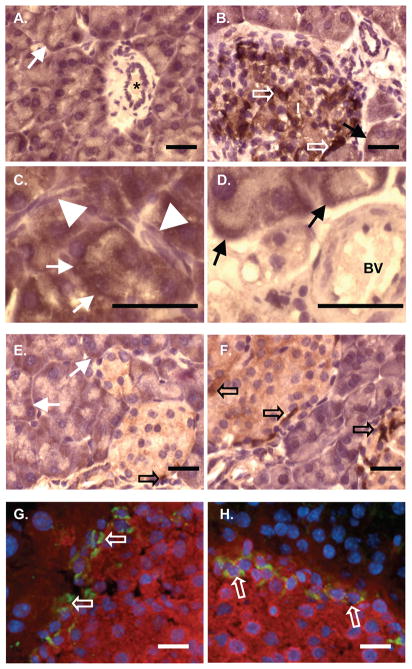
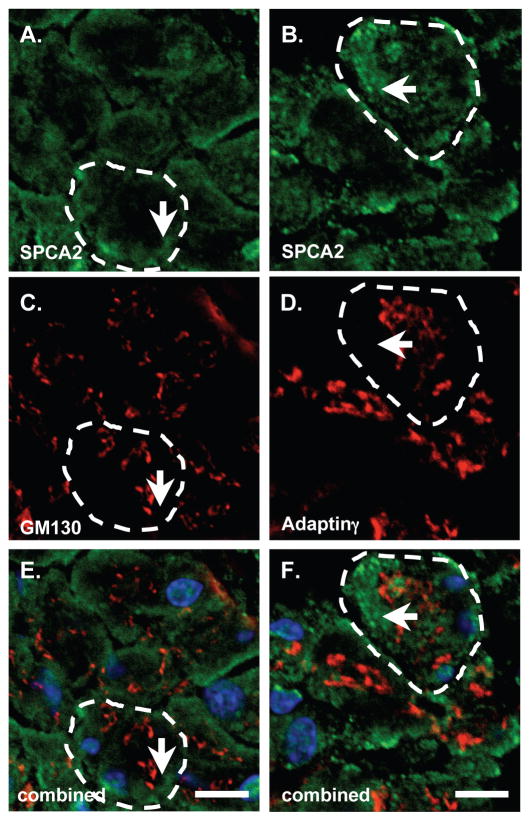
To assess the location of SPCA2 accumulation in the pancreas, IHC analysis was performed on paraffin sections (Figure 5). Expression was observed throughout acinar tissue (Figure 5A) and in a subset of islet cells (Figure 5B) typically restricted to cells at the periphery of the islets. Pancreatic ducts (Figure 5A), intercalated ducts (Figure 5C) and blood vessels (Figure 5D) were all negative for SPCA2 accumulation. Interestingly, the loss of SPCA2 in Mist1−/− tissue is specific to acinar cells as SPCA2+ islet cells are still observed in the islets of Mist1−/− pancreata (Figure 5F). These cells are not β cells as they did not stain for insulin expression based on co-IF analysis (Figure 5G, H). Surprisingly, the majority of SPCA2 appeared to accumulate in the basolateral region of pancreatic acinar cells (Figure 5), consistent with localization to the ER. To assess intra-acinar localization (Figure 6), SPCA2 accumulation was compared to markers for the Golgi apparatus (adaptin γ) and ER (calreticulin and SERCA2). Pancreatic acinar cells have a highly organized intracellular structure with the Golgi apparatus positioned between the basally-localized nuclei and the apical-located zymogen granules [19]. SPCA2 accumulation (Figure 6A) was consistent with calreticulin (Figure 6D) and SERCA2 (Figure 6E) localization, limited to the basal portion of the acinus. Adaptin γ (Figure 6C) accumulated just apical to the ER. To verify that pancreas SPCA2 does not localize to the Golgi, we next performed co-IF using antibodies against SPCA2 and either GM130 or adaptin γ (Figure 7). In confirmation of the IHC results, SPCA2 did not co-localize with GM130 (Figure 7A,C,E) or adaptin γ (Figure 7B,D,F) in the Golgi, but instead remained associated with the basal aspect of the cell.
Figure 5. SPCA2 is expressed in acinar cells and a subset of islet cells in the pancreas.
(A–F) IHC for SPCA2 on pancreatic tissue shows expression in acinar cells (arrows) and a subset of islet (I) cells (open arrows, B), but not in ducts (*, A), intercalated duct (arrowheads, C) or blood vessels (BV, D). IHC for SPCA2 or co-IF (G, H) for SPCA2 (green) and insulin (red) in wild type (E, G) and Mist1−/− (F, H) tissue reveals maintenance of SPCA2 in the islets even after deletion of Mist1. Magnification bars = 20 μm.
Figure 6. SPCA2 is localized to the basal aspect of acinar cells typical of ER localization.
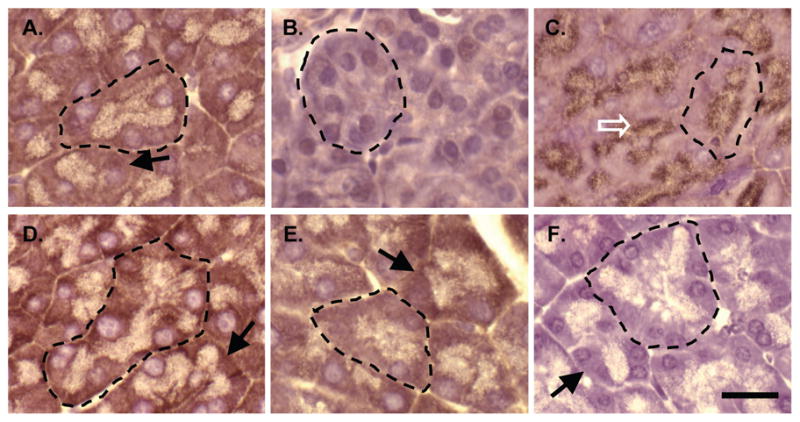
IHC for SPCA2 on wild type (A) or Mist1−/− (B) pancreatic tissue, or for adaptin γ (C), calreticulin (D), SERCA2 (E) or with no primary antibody (F). Individual acini have been outlined with a dotted line and staining consistent with the ER (arrows) or Golgi apparatus (open arrow) indicated in an adjacent acinus. Magnification bar = 20 μm.
Figure 7. SPCA2 is not localized to the Golgi apparatus in pancreatic acinar cells.
Confocal co-IF on pancreatic sections for Golgi-specific markers, GM130 (C) or adaptin γ (D) and SPCA2 (A, B). White dotted lines indicate a single acinus. Scale bars = 10 μm. DAPI was used to stain the nuclei (blue). (E, F) Represent the merged images of SPCA2 and the Golgi markers.
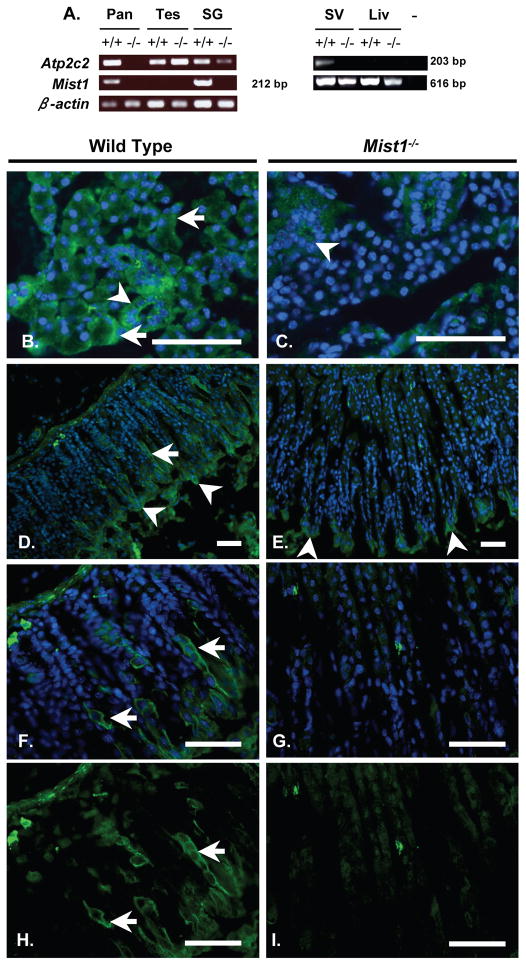
The absence of SPCA2 in Mist1−/− pancreatic tissue suggests that the Atp2c2 gene is a downstream transcriptional target of MIST1. Consistent with a role for MIST1 in the regulation of Atp2c2 expression, other tissues that normally express MIST1, including the salivary glands and seminal vesicle, also showed a decreased level of Atp2c2 transcripts in Mist1−/− tissues (Figure 8A). In contrast, Atp2c2 gene expression was similar in WT and Mist1−/− testes, a tissue that does not express MIST1. IF localization confirmed that Mist1−/− salivary glands (Figure 8B–C) and gastric glands of the stomach (Figure 8D–I), exhibited a dramatic reduction in SPCA2 levels. Importantly, SPCA2 levels were not altered in the surface epithelial cells of the stomach (arrowheads in Figure 8D, E), which never express MIST1.
Figure 8. Atp2c2/SPCA2 accumulation is reduced in other MIST1-expressing tissue in Mist1−/− mice.
(A) RT-PCR for Atp2c2, Mist1 or β-actin in RNA extracts from pancreas (Pan), testes (Tes), salivary glands (SG), seminal vesicle (SV) and liver (Liv). No RT (−) was added to the negative control. (B–I) IF for SPCA2 on salivary glands (B, C) and stomach (D–I) sections from WT (B, D, F, H) and Mist1−/− (C, E, G, I) mice. Arrows in B indicates basal localization of SPCA2 in salivary gland acini. Arrowheads in B and C indicate SPCA2 expression in duct cells of the salivary gland, which are not positive for MIST1. Arrowheads in D and E indicate SPCA2+ cells at the surface of epithelial crypts in both strains of mice while arrows indicate SPCA2 positive cells with the epithelial crypts only in WT stomach sections. Images F–I are higher magnifications of D and E to better reveal SPCA2+ cells. All panels identify nuclei through DAPI staining expect H and I. Scale bar = 50 μM.
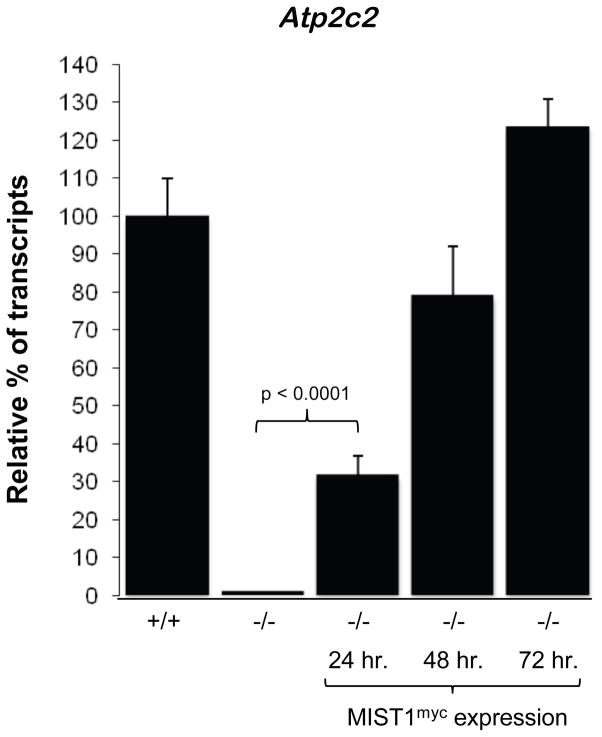
To support Atp2c2 as a downstream target of MIST1, we employed a genetic mouse model in which expression of a MIST1 transgene (LSL-Mist1myc) can be selectively activated through Cre-mediated recombination (unpublished data). LSL-Mist1myc/Mist1CreER/CreER (Mist1−/−) mice were then generated through standard crosses producing a Mist1−/− mouse containing a CreERT2-inducible transgene. As shown in Figure 7, Atp2c2 transcripts were virtually undetectable in Mist1−/− pancreata. However, 24 hr following a single dose of tamoxifen, MIST1myc expression was induced in ~85% of acinar cells (data not shown) and this led to a 37% increase in endogenous Atp2c2 transcript levels, which continued to increase to WT levels over the next two days (Figure 9). Thus, Atp2c2 gene expression was completely restored in Mist1−/− acinar cells upon Mist1myc transgene expression, strongly suggesting that Atp2c2 is a direct target of the MIST1 protein.
Figure 9. MIST1 re-expression rescues Atp2c2 expression.
Quantitative RT-PCR for Atp2c2 following a single dose of tamoxifen in Mist1CreER/CreER/LSL-Mist1myc mice. WT levels are set at 100% and compared to wild type (+/+), Mist1−/− (−/−) or 24, 48 or 72 hours after induction of MIST1myc expression. Error bars represent mean ± SE; n=3.
Discussion
MIST1 is a transcription factor required for complete acinar cell maturation and function. Mist1−/− acini display disrupted Ca2+ movement upon secretagogue stimulation, an altered exocytosis ability and increased sensitivity to caerulein-induced pancreatitis [5, 12] suggesting that Mist1−/− acinar cells have defects in the expression of Ca2+ handling proteins. However, to date no alterations in Ca2+ATPase proteins have been documented in this model. In the current study, we identified a specific loss of the Ca2+ATPase Atp2c2/SPCA2 in Mist1−/− pancreatic acinar cells as well as in other secretory cells that normally express MIST1, suggesting a link. While the loss of SPCA2 correlated with expansion of the Golgi compartment and disruption in protein glycosylation, the pancreatic isoform of SPCA2 does not localize to the Golgi, nor does it contain many of the domains required for Ca2+ pump action. Therefore, we have identified a novel isoform of the Atp2c2 gene that may be, in part, responsible for the altered acinar cell phenotype observed in Mist1−/− mice.
Unlike past studies on Ca2+ ATPase expression, we observed increased accumulation of Atp2a (SERCA) and Atp2b (PMCA) pumps. There are two possibilities for the differences in expression. First, this study examines mRNA levels in vivo, while Luo et al (2005) examined protein levels in cultured acini. There may be differences in transcript or protein accumulation or the culturing of the cells could account for differences in expression. Second, the antibodies used by Luo et al (2005) were not specific to individual PMCA or SERCA isoforms. Since we have not performed an exhaustive study for all SERCA and PMCA isoforms, it is possible that the total levels are equal. In any event, the only Atp2 gene that is reduced in Mist1−/− pancreatic tissue is Atp2c2. Mouse genetic studies showed that reactivation of MIST1 was closely followed by increased Atp2c2 transcript levels suggesting that Atp2c2 may be a downstream target of MIST1 transcriptional activity. Indeed, the overall expression pattern for Atp2c2 correlated predominantly with MIST1 expression. Previous studies have revealed that Atp2c2 is expressed in a number of tissues, most notably in the testes, brain and lactating mammary gland [20, 21]. While MIST1 is not found in testes or brain, it is expressed transiently in epithelial alveolar cells during lactation [20]. Similar to Atp2c2, expression of Mist1 decreases to negligible levels upon mammary gland involution [22, 23]. Our studies have now extended the expression pattern of SPCA2 to include the secretory cells of pancreatic acini, salivary gland acini, seminal vesicles and stomach epithelium. In all cases, Atp2c2/SPCA2 expression correlates with MIST1 expression. Importantly, loss of MIST1 in these tissues produced dramatically decreased levels of Atp2c2 and this expression pattern could be rescued in the pancreas within hours by inducing Mist1myc transgene expression. Thus, although it remains possible that the absence of Atp2c2/SPCA2 reflects incomplete maturation of the Mist1−/− secretory cells and thus not a direct target of MIST1, given the rapid rescue response to MIST1myc expression, this seems highly unlikely. Unfortunately, specific regulatory regions within the Atp2c2 gene have not been identified so future studies employing MIST1 chromatin immunoprecipitation to show direct MIST1-promoter interactions will have to await further characterization the Atp2c2 gene.
The full length SPCA2 is a Ca2+ATPase that is thought to pump Ca2+ and Mn2+ from the cytosol into the Golgi where proper Ca2+ concentrations are required for accurate post-translational modifications [7]. The SPCA proteins are the mammalian equivalent to the PMR1 protein found in yeast and defective glycosylation and an altered secretory pathway are observed in Pmr1-deficient yeast, a phenotype similar to that found in Mist1−/− acini [24–26]. In addition, targeted ablation of the closely related mouse Atp2c1 gene results in expansion of the Golgi compartment consistent with that observed in Mist1−/− acini [27]. It is interesting that a simlar phenotype is observed in Mist1−/− tissue without a decrease in Atp2c1 expression levels.
While a model with MIST1 affecting exocrine cell function through the regulation of Atp2c2 is attractive, we have uncovered novel findings regarding the structure and localization of the SPCA2 protein that adds considerable complexity. IF analysis indicated that SPCA2 does not co-localize with GM130 or adaptin γ at the Golgi apparatus. Instead, we show for the first time that SPCA2 localizes in the same location as the ER in both pancreas and salivary gland acinar cells. In agreement with our current observations, studies examining SPCA2 in neuronal cells also found that SPCA2 localizes to cellular regions outside of the Golgi apparatus [21]. Interestingly, localization of SPCA2 to the ER is more consistent with a role for this protein in regulated exocytosis. The Ca2+ required for exocytosis is contained within the ER of acinar cells, and during physiological stimulation, Ca2+ oscillations are observed within the apical portion of the acinar cells [28], suggesting that rapid release and uptake from the ER is essential for efficient transport of digestive enzymes out of the cell.
Even more intriguing is that our studies have uncovered a previously unknown pancreatic SPCA2 isoform that is significantly smaller (17–20 kDa) than the NCBI predicted SPCA2 protein (103 kDa). While it is possible that this protein represents a degradation product, our data support the existence of an alternative SPCA2 protein. First, immunoabsorption with the peptide used to generate the antibody eliminates the 20 kDa band observed in pancreatic protein extracts. Second, the 17–20 kDa SPCA2 band is detected in protein extracts from WT pancreata and in the acinar cell line AR42J. In contrast, no SPCA2 is detected in Mist1−/− samples or in the duct-like cell line ARIP. Finally, analysis of Atp2c2 transcripts revealed a similar truncated mRNA only in WT RNA samples and not in Mist1−/− samples. The existence of different Ca2+-ATPase isoforms is not unusual, as multiple isoforms exist for the SERCA, PMCA and SPCA1 pumps [17]. However, this particular isoform of Atp2c2 is significantly different as it represents only the carboxy terminus of the protein. Based on RT-PCR and EST data, the pancreatic Atp2c2 consists of at least the last six exons of the gene. This would include the 6th transmembrane domain and the carboxy domain for ion binding but exclude the Ca2+ transport site, phsophorylation sites, most transmembrane domains and the ATP binding site. Based on its structure, it is unlikely to act as an ion pump, sequester Ca2+ in the ER or even bind Ca2+ on its own.
We have attempted to identify the complete sequence of this novel Atp2c2 transcript through 5′ RACE, degenerate primers, or mass spectrometry of protein extracts resolved by two-dimensional gel elctrophoresis. However, to date, most of our success stems from bioinformatic analysis of EST databases. ACEview suggests that 4 transcripts for Atp2c2 exist, but none of these transcripts correlate to the size of mRNA observed in the pancreas. Based on ACEview, 75 ESTs exist with sequences identical to the Atp2c2 gene. Eight of these ESTs were derived from pancreatic tissue, all aligned to the 3′ end of the gene. Not one of these ESTs would produce an alternative transcript that corresponds to the predicted molecular weight of the protein, which is in the range of 17 to 22 kDa. The largest pancreatic EST is AK007419, which was derived from a tiling array of cDNA from 10 day-old mouse pancreatic tissue. The EST is 704 bps and, when translated, produces a 104 amino acid protein that aligns with the carboxy terminus of the protein. All other pancreatic ESTs are less than 400 bp in length. There is a single EST identified that would produce a protein the size of the predicted pancreatic SPCA2 isoform. Accession #BI414588 was cloned from pooled lung tissue and is 935 bp long. It encodes exons 20–26 and translation from the first identified Kozak sequence would produce, in frame, a 185 amino acid protein. Further studies are necessary to determine if this represents the full length protein found in the pancreas.
In conclusion, we have identified a novel protein that is identical to the carboxy terminus of SPCA2. While lacking the functional domains required for Ca2+-ATPase function, this protein appears to accumulate in the ER, and is correlated to the expression of MIST1 in pancreatic acinar cells. Future work should focus on understanding how this alternative SPCA2 functions in the secretory pathway of MIST1-expressing cells and how MIST1 transcriptionally regulates Atp2c2 gene expression.
Supplementary Material
Acknowledgments
Grant Support: This work was funded by operating grants from the Canadian Institutes of Health Research (MOP 53083), Children’s Health Research Institute, and Lawson Health Research Institute to C.P. as well as by NIH DK55489 and NIH CA124586 to S.F.K. D.D. was supported by a Purdue Research Foundation Fellowship through the Purdue Center for Cancer Research. C.P. is supported by a salary award from the CIHR.
We would like to thank Drs. Gabriel DiMattia and Tom Drysdale and members of the Pin laboratory for the comments on suggestions on these studies, Dr. John Di Guglielmo for the kind gift of the GM130 antibody and to the Drysdale lab for their help and expertise with in situ hybridization.
Abbreviations
- SPCA
secretory pathway Ca2+ ATPase
- PMCA
plasma membrane Ca2+ ATPase
- SERCA
sarcoendoplasmic reticulum Ca2+ ATPase
- P-type
phosphorylation type
- bHLH
basic helix-loop-helix
- CCK
cholecystokinin
- WT
wild type
- ACC
acinar cell cultures
- ER
endoplasmic reticulum
Contributor Information
Stephen F. Konieczny, Email: sfk@purdue.edu.
Christopher L. Pin, Email: cpin@uwo.ca.
Reference List
- 1.Lemercier C, To RQ, Swanson BJ, Lyons GE, Konieczny SF. Mist1: a novel basic helix-loop-helix transcription factor exhibits a developmentally regulated expression pattern. Developmental biology. 1997;182:101–113. doi: 10.1006/dbio.1996.8454. [DOI] [PubMed] [Google Scholar]
- 2.Pin CL, Bonvissuto AC, Konieczny SF. Mist1 expression is a common link among serous exocrine cells exhibiting regulated exocytosis. The Anatomical record. 2000;259:157–167. doi: 10.1002/(SICI)1097-0185(20000601)259:2<157::AID-AR6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Cancela JM, Van Coppenolle F, Galione A, Tepikin AV, Petersen OH. Transformation of local Ca2+ spikes to global Ca2+ transients: the combinatorial roles of multiple Ca2+ releasing messengers. The EMBO journal. 2002;21:909–919. doi: 10.1093/emboj/21.5.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee MG, Xu X, Zeng W, Diaz J, Kuo TH, Wuytack F, Racymaekers L, Muallem S. Polarized expression of Ca2+ pumps in pancreatic and salivary gland cells. Role in initiation and propagation of [Ca2+]i waves. The Journal of biological chemistry. 1997;272:15771–15776. doi: 10.1074/jbc.272.25.15771. [DOI] [PubMed] [Google Scholar]
- 5.Luo X, Shin DM, Wang X, Konieczny SF, Muallem S. Aberrant localization of intracellular organelles, Ca2+ signaling, and exocytosis in Mist1 null mice. The Journal of biological chemistry. 2005;280:12668–12675. doi: 10.1074/jbc.M411973200. [DOI] [PubMed] [Google Scholar]
- 6.MacLennan DH, Green NM. Structural biology. Pumping ions. Nature. 2000;405:633–634. doi: 10.1038/35015206. [DOI] [PubMed] [Google Scholar]
- 7.Wuytack F, Raeymaekers L, Missiaen L. PMR1/SPCA Ca2+ pumps and the role of the Golgi apparatus as a Ca2+ store. Pflugers Arch. 2003;446:148–153. doi: 10.1007/s00424-003-1011-5. [DOI] [PubMed] [Google Scholar]
- 8.Cartwright EJ, Oceandy D, Neyses L. Plasma membrane calcium ATPase and its relationship to nitric oxide signaling in the heart. Annals of the New York Academy of Sciences. 2007;1099:247–253. doi: 10.1196/annals.1387.007. [DOI] [PubMed] [Google Scholar]
- 9.Missiaen L, Dode L, Vanoevelen J, Raeymaekers L, Wuytack F. Calcium in the Golgi apparatus. Cell calcium. 2007;41:405–416. doi: 10.1016/j.ceca.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Pin CL, Rukstalis JM, Johnson C, Konieczny SF. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. The Journal of cell biology. 2001;155:519–530. doi: 10.1083/jcb.200105060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi G, Zhu L, Sun Y, Bettencourt R, Damsz B, Hruban RH, Konieczny SF. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology. 2009;136:1368–1378. doi: 10.1053/j.gastro.2008.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalik AS, Johnson CL, Chadi SA, Weston JY, Fazio EN, Pin CL. Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein-induced pancreatitis. American journal of physiology. 2007;292:G1123–1132. doi: 10.1152/ajpgi.00512.2006. [DOI] [PubMed] [Google Scholar]
- 13.Johnson CL, Weston JY, Chadi SA, Fazio EN, Huff MW, Kharitonenkov A, Koester A, Pin CL. Fibroblast growth factor 21 reduces the severity of cerulein-induced pancreatitis in mice. Gastroenterology. 2009;137:1795–1804. doi: 10.1053/j.gastro.2009.07.064. [DOI] [PubMed] [Google Scholar]
- 14.Yule DI, Williams JA. U73122 inhibits Ca2+ oscillations in response to cholecystokinin and carbachol but not to JMV-180 in rat pancreatic acinar cells. The Journal of biological chemistry. 1992;267:13830–13835. [PubMed] [Google Scholar]
- 15.Johnson CL, Kowalik AS, Rajakumar N, Pin CL. Mist1 is necessary for the establishment of granule organization in serous exocrine cells of the gastrointestinal tract. Mechanisms of development. 2004;121:261–272. doi: 10.1016/j.mod.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Carafoli E, Genazzani A, Guerini D. Calcium controls the transcription of its own transporters and channels in developing neurons. Biochemical and biophysical research communications. 1999;266:624–632. doi: 10.1006/bbrc.1999.1879. [DOI] [PubMed] [Google Scholar]
- 17.Zacharias DA, Strehler EE. Change in plasma membrane Ca2(+)-ATPase splice-variant expression in response to a rise in intracellular Ca2+ Curr Biol. 1996;6:1642–1652. doi: 10.1016/s0960-9822(02)70788-4. [DOI] [PubMed] [Google Scholar]
- 18.Vanoevelen J, Dode L, Van Baelen K, Fairclough RJ, Missiaen L, Raeymaekers L, Wuytack F. The secretory pathway Ca2+/Mn2+-ATPase 2 is a Golgi-localized pump with high affinity for Ca2+ ions. The Journal of biological chemistry. 2005;280:22800–22808. doi: 10.1074/jbc.M501026200. [DOI] [PubMed] [Google Scholar]
- 19.Petersen OH, Burdakov D, Tepikin AV. Polarity in intracellular calcium signaling. Bioessays. 1999;21:851–860. doi: 10.1002/(SICI)1521-1878(199910)21:10<851::AID-BIES7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 20.Anantamongkol U, Takemura H, Suthiphongchai T, Krishnamra N, Horio Y. Regulation of Ca2+ mobilization by prolactin in mammary gland cells: possible role of secretory pathway Ca2+-ATPase type 2. Biochemical and biophysical research communications. 2007;352:537–542. doi: 10.1016/j.bbrc.2006.11.055. [DOI] [PubMed] [Google Scholar]
- 21.Xiang M, Mohamalawari D, Rao R. A novel isoform of the secretory pathway Ca2+,Mn(2+)-ATPase, hSPCA2, has unusual properties and is expressed in the brain. The Journal of biological chemistry. 2005;280:11608–11614. doi: 10.1074/jbc.M413116200. [DOI] [PubMed] [Google Scholar]
- 22.Reinhardt TA, Lippolis JD. Mammary gland involution is associated with rapid down regulation of major mammary Ca2+-ATPases. Biochemical and biophysical research communications. 2009;378:99–102. doi: 10.1016/j.bbrc.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Johansson C, Tran T, Bettencourt R, Itahana Y, Desprez PY, Konieczny SF. Identification of a basic helix-loop-helix transcription factor expressed in mammary gland alveolar cells and required for maintenance of the differentiated state. Molecular endocrinology (Baltimore, Md. 2006;20:2187–2198. doi: 10.1210/me.2005-0214. [DOI] [PubMed] [Google Scholar]
- 24.Durr G, Strayle J, Plemper R, Elbs S, Klee SK, Catty P, Wolf DH, Rudolph HK. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Molecular biology of the cell. 1998;9:1149–1162. doi: 10.1091/mbc.9.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudolph HK, Antebi A, Fink GR, Buckley CM, Dorman TE, LeVitre J, Davidow LS, Mao JI, Moir DT. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- 26.Antebi A, Fink GR. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Molecular biology of the cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okunade GW, Miller ML, Azhar M, Andringa A, Sanford LP, Doetschman T, Prasad V, Shull GE. Loss of the Atp2c1 secretory pathway Ca(2+)-ATPase (SPCA1) in mice causes Golgi stress, apoptosis, and midgestational death in homozygous embryos and squamous cell tumors in adult heterozygotes. The Journal of biological chemistry. 2007;282:26517–26527. doi: 10.1074/jbc.M703029200. [DOI] [PubMed] [Google Scholar]
- 28.Wasle B, Edwardson JM. The regulation of exocytosis in the pancreatic acinar cell. Cellular signalling. 2002;14:191–197. doi: 10.1016/s0898-6568(01)00257-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.