Abstract
Cutaneous T-cell lymphomas (CTCLs) are the most frequent primary skin lymphomas. Nevertheless, diagnosis of early disease has proven difficult because of a clinical and histologic resemblance to benign inflammatory skin diseases. To address whether microRNA (miRNA) profiling can discriminate CTCL from benign inflammation, we studied miRNA expression levels in 198 patients with CTCL, peripheral T-cell lymphoma (PTL), and benign skin diseases (psoriasis and dermatitis). Using microarrays, we show that the most induced (miR-326, miR-663b, and miR-711) and repressed (miR-203 and miR-205) miRNAs distinguish CTCL from benign skin diseases with > 90% accuracy in a training set of 90 samples and a test set of 58 blinded samples. These miRNAs also distinguish malignant and benign lesions in an independent set of 50 patients with PTL and skin inflammation and in experimental human xenograft mouse models of psoriasis and CTCL. Quantitative (q)RT-PCR analysis of 103 patients with CTCL and benign skin disorders validates differential expression of 4 of the 5 miRNAs and confirms previous reports on miR-155 in CTCL. A qRT-PCR–based classifier consisting of miR-155, miR-203, and miR-205 distinguishes CTCL from benign disorders with high specificity and sensitivity, and with a classification accuracy of 95%, indicating that miRNAs have a high diagnostic potential in CTCL.
Introduction
Cutaneous T-cell lymphomas (CTCLs) are the most frequent primary lymphomas of the skin,1 with mycosis fungoides (MF) being the most prevalent clinical form, accounting for ∼ 60% of new cases.2 In early disease stages, which can last several years, MF presents as flat erythematous skin patches resembling inflammatory diseases such as dermatitis or psoriasis. In later stages, MF lesions gradually form plaques and overt tumors and may disseminate to lymph nodes and internal organs. The early skin lesions contain numerous inflammatory cells, including a large quantity of T cells with a normal phenotype as well as a small population of T cells with abnormal morphology and a malignant phenotype. The infiltrate primarily consists of nonmalignant T helper 1 cells, regulatory T cells, and cytotoxic CD8+ T cells that to some degree seem to control the malignant T cells.3,4 The malignant T cells typically exhibit the phenotype of mature CD4+ memory T cells and are normally of clonal origin.5 T cells with a malignant phenotype are characterized by epidermotropism and are preferentially present in the upper parts of the skin, whereas T cells with a normal phenotype primarily are detected in the lower portions of the dermis. The epidermal T cells are sometimes found in patterns of Pautrier microabscesses that are collections of T cells adherent to dendritic processes of Langerhans cells. During disease development, the epidermotropism is gradually lost concomitant with an increase in malignant, and a decrease in nonmalignant, infiltrating T cells.
The etiology of CTCL remains poorly understood, and occupational exposures, infectious agents, and genetic mutations have been proposed as etiologic factors, but no evidence of causation has been provided.6 Instead, an aberrant expression and function of transcription factors and regulators of signal transduction are characteristic features of CTCL. Accordingly, it has been hypothesized that a dysfunctional regulation of signal molecules and cytokines plays a key role in the malignant transformation and epigenetic modifications such as aberrant gene methylation and histone deacetylation are clearly involved in the pathogenesis of CTCL.7–10 Recent data indicate that several small, noncoding RNA molecules, microRNAs (miRNAs), are differentially expressed and possibly also involved in the pathogenesis of this disease.11,12 Thus, miR-21 expression is up-regulated and seems to play a role in the regulation of apoptosis in malignant T cells obtained from patients with Sézary Syndrome (SS), a leukemic variant of CTCL. These findings are in keeping with studies in other cancers where miRNAs have been ascribed a key role in cancer development and metastasis. Indeed, specific miRNAs are directly involved in the malignant transformation because they can function as oncogenes and tumor suppressors.13
Early diagnosis of CTCL has important consequences concerning therapeutic options and determination of prognosis. Currently, the diagnosis is primarily based on clinical observations and histologic examinations of skin biopsies as well as additional laboratory tests such as analysis of TCR clonality by PCR. Unfortunately, early diagnosis of CTCL has proven difficult because of the great clinical, pathologic, and histologic resemblance to benign inflammatory skin diseases and because inflammatory skin disorders can be associated with clonal TCR rearrangements.14–17 From the initial appearance of skin lesions to the time a certain MF diagnosis is established takes a median of almost 6 years. Because miRNA expression profiling can distinguish other cancers according to diagnosis and developmental stage of the tumor to a greater degree of accuracy than traditional mRNA analysis,18 the present investigation was undertaken to address whether miRNA expression profiling has a diagnostic potential in relation to CTCL.
Methods
Selection of patients
The study includes formalin-fixed and paraffin-embedded biopsies from 63 patients with CTCL; 39 patients with nodal PTL (not otherwise specified [NOS]); and 96 patients with benign skin diseases, including psoriasis, atopic dermatitis, contact dermatitis, and unspecified dermatitis as well as skin from 2 healthy volunteers. Biopsies from the lymphoma patients were sampled from 1979 to 2004 and were collected from the archives at the Departments of Pathology at Rigshospitalet, Bispebjerg Hospital, Aalborg Sygehus, and Herlev Hospital. From all lymphoma cases, tissue samples were reviewed by histology and immunohistochemistry, using as a minimum CD3, CD4, CD8, CD30, CD56, TIA-1, and Granzyme B stains. The samples were then classified in accordance with the World Health Organization-European Organization for Research and Treatment of Cancer16,19–21 guidelines, and the clinical characteristics of the cohort were reviewed to establish the final diagnoses. Biopsies from the patients with benign skin diseases and healthy controls were collected after informed consent in accordance with the Declaration of Helsinki at the Departments of Dermato-Allergology, Gentofte Hospital; Dermatology, Bispebjerg Hospital; and Pathology, Rigshospitalet and as part of clinical trials at LEO Pharma A/S, and they were approved by the local ethical committees (H-B-2009-045 and H-1-2009-111) and the Data Protection Agency (Datatilsynet J.NR. 2010-41-4303).
Human psoriasis xenograft mouse models
Three patients with chronic plaque-psoriasis were used as donors for the psoriatic keratome biopsies (thickness, 0.5 mm). Keratome biopsies were taken from infiltrated red plaques located on the anterior or lateral aspect of the femoral region.22 Each biopsy was divided into 4 pieces of 1.5 × 1.5 cm. As recipients, female CB.17 SCID mice (M&B Taconic) aged 6 weeks were used. The split human keratome biopsies were then grafted onto the back of the anesthetized animals, and the grafts were protected by a bandage during the next 2 weeks in a specific-pathogen-free environment as described previously.23 After 2 weeks, the animals were randomized into 2 groups. The groups were treated with betamethasone (0.5 mg/g [as diproprionate]) in ointment (n = 6) or ointment vehicle (n = 6) twice daily. After 4 weeks of treatment, the animals were bled and killed, and a 4-mm punch biopsy was taken from each xenograft. Biopsies were fixed in 10% neutral buffered formalin for a maximum of 48 hours and were processed according to standard histologic procedures and embedded in paraffin.
CTCL xenograft model
The murine xenograft model of CTCL is based on the immunodeficient NOD.Cg-Prkdcscid B2mtm1Unc/J strain (NOD/SCID-B2m−/−; The Jackson Laboratory) as described in detail previously.24 In brief, NOD/SCID-B2m−/− mice were inoculated subcutaneously into each flank with 1 × 106 MyLa2059 cells, and when a mouse had established palpable tumors it was allocated alternately to a group receiving vehicle or a group receiving celecoxib (Selleck Chemicals). The mice received 10 μg/g celecoxib or vehicle intraperitoneally daily, and the tumor growth was monitored continuously by slide caliper measurements. All animal experiments were carried out in accordance with the local ethics committee and with animal welfare guidelines provided by the Animal Experiments Inspectorate, Ministry of Justice, Denmark.
RNA extraction
Total RNA was isolated from six 10-μm tissue sections using the RecoverAll total nucleic acid isolation kit (Applied Biosystems/Ambion) according to the manufacturer's guidelines. Total RNA quantity and quality were checked by an ND-1000 spectrophotometer (NanoDrop).
miRNA microarray
From each sample, 100 ng of total RNA was labeled with Hy3 fluorescent dye using the miRCURY LNA array power labeling kit (Exiqon A/S). All samples were labeled the same day with the same master mix, to minimize technical variation. The Hy3-labeled samples were hybridized to miRCURY LNA arrays (Version 11.0; Exiqon A/S), containing capture probes targeting all human miRNAs registered in the miRBASE Version 15.0 at the Sanger Institute. The hybridization was performed overnight at 56°C according to the manufacturer's specifications using an HS4800 hybridization station (Tecan). Because it was not possible to hybridize all arrays at the same time, samples were randomly split into 5 batches to minimize day-to-day variation in the hybridization process. After hybridization, the microarray slides were scanned using a G2565BA Microarray Scanner system (Agilent Technologies) at 5-μm resolution, and the resulting TIFF images were analyzed using the ImaGene 8.0 software on standard settings (BioDiscovery). All microarray data are deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.gov/geo/query/acc.cgi2acc-GSE31408).
miRNA real-time qRT-PCR
Ten nanograms of RNA was reverse transcribed in triplicate 10-μL reactions using miRCURY LNA Universal RT miRNA PCR, polyadenylation, and cDNA synthesis kit (Exiqon A/S). cDNA was diluted 50 × and assayed in 10-μL PCR reactions according to the protocol for miRCURY LNA Universal RT miRNA PCR; each miRNA was assayed once by qPCR. Negative controls excluding template from the reverse transcription reaction were performed and profiled in parallel. The amplification was performed in a LightCycler Version 1.5.0.SP3 software 480 real-time PCR system (Roche) in 384-well plates. The amplification curves were analyzed using Roche LC software, both for determination of crossing point (Cp, by the second derivative method) and for melting curve analysis. All assays were inspected for distinct melting curves, and the melting point was checked to be within known specifications for the assay. Furthermore, assays must be detected with 3 Cps less than the negative control, and with Cp < 39 to be included in the data analysis. Data that did not pass these criteria were omitted from any further analysis. miR-103 and miR-423-5p were stably expressed and the average of their Cps in each sample was used as normalization factor.
Microarray data preprocessing
Probe signals were background corrected by fitting a convolution of normal and exponential distributions to the foreground intensities using the background intensities as a covariate.25 Four technical replicate spots for each probe were combined to produce 1 signal by taking the logarithmic base-2 mean of reliable spots. If all 4 replicates for a given probe were judged unreliable, that probe was removed from further analysis.A reference data vector R was calculated as the median signal of each probe across all samples. For all probe signals in a given sample, represented by the sample data vector S, a curve F was determined by locally weighted polynomial regression so as to provide the best fit between S and R.26 A normalized sample vector M was calculated from this by transforming it with the function F, so that M = F(S). In this manner, all samples were normalized to the reference R. This normalization procedure largely follows that outlined in Rosenfeld et al.27 Remote data points (probes in sparsely sampled intensity regions with <15 probes per signal unit) were considered unreliably adjusted by this method and removed before further analysis.
Classifier statistics
Significance of differences in expression levels was assessed by a 2-sided unpaired t test. Class prediction was done using nearest shrunken centroid classification.28 In brief, a standardized centroid is computed for each class as the average expression of each miRNA in each class divided by the within-class standard deviation for that miRNA. The miRNA expression profile of a new sample is then compared with each of these class centroids, and the class whose centroid is closest in Euclidean distance is the predicted class for that new sample. The algorithm is trained by shrinking class centroids toward the overall centroid for all classes by a threshold amount that minimizes the misclassification error as determined through 10-fold cross validation on the training set.
Results
miRNA expression profiling using microarrays
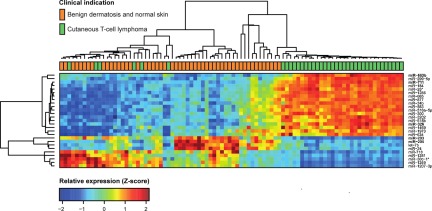
To address whether miRNA expression profiling can differentiate CTCL from benign chronic skin disorders such as psoriasis and dermatitis, as well as from normal skin, we initially used microarrays to perform miRNA profiling of 148 formalin-fixed paraffin-embedded samples. Of these samples, 63 were from patients with various forms of CTCL and 85 were from patients with benign inflammatory skin diseases or healthy individuals (BDN; Table 1). The samples were divided into 3/5 for training (n = 90) and 2/5 for testing (n = 58), with approximately equal proportion of CTCL to BDN samples in both sets. This division follows the 5 microarray production batches used in the study (Table 1). Of the 688 miRNAs that passed preprocessing filtering criteria, initial statistical analysis of the training set identified 27 miRNAs showing strong (at least 50% change) and highly significant (Bonferroni corrected P values < .001 from t test) differences between CTCL and benign skin diseases and normal skin (Figure 1). Thus, the expression levels of a large number of miRNAs differ considerably between patients with CTCL and patients with BDN. Essentially similar results were obtained using unsupervised hierarchical clustering based on the 209 most variable miRNAs (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Table 1.
Clinical characteristics of patients in the study
| Cutaneous lymphoma (n = 63) |
Benign skin disease or normal skin (n = 85) |
P | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MF (n = 39) | SS (n = 7) | CALCL (n = 8) | NOS (n = 9) | Σ | AD (n = 20) | ND (n = 4) | PP (n = 42) | PN (n = 17) | NN (n = 2) | Σ | ||
| Age, y* | < .001 | |||||||||||
| Younger than 30 | 0 | 0 | 1 | 0 | 1 | 19 | 0 | 4 | 4 | 2 | 29 | |
| 30-44 | 5 | 0 | 0 | 0 | 5 | 1 | 0 | 6 | 3 | 0 | 10 | |
| 45-59 | 9 | 1 | 1 | 2 | 13 | 0 | 1 | 19 | 7 | 0 | 27 | |
| 60-74 | 14 | 6 | 2 | 2 | 24 | 0 | 3 | 12 | 2 | 0 | 17 | |
| Older than 75 | 10 | 0 | 2 | 5 | 17 | 0 | 0 | 1 | 1 | 0 | 2 | |
| Sex* | .999 | |||||||||||
| Male | 23 | 7 | 5 | 5 | 40 | 8 | 1 | 30 | 16 | 2 | 57 | |
| Female | 15 | 0 | 1 | 4 | 20 | 12 | 3 | 12 | 1 | 0 | 28 | |
| Microarray batch | .999 | |||||||||||
| 1 | 8 | 2 | 1 | 2 | 13 | 4 | 1 | 8 | 3 | 1 | 17 | |
| 2 | 7 | 2 | 2 | 1 | 12 | 4 | 1 | 8 | 3 | 1 | 17 | |
| 3 | 7 | 1 | 2 | 2 | 12 | 4 | 1 | 8 | 3 | 0 | 16 | |
| 4 | 9 | 1 | 2 | 2 | 14 | 4 | 0 | 9 | 4 | 0 | 17 | |
| 5 | 8 | 1 | 1 | 2 | 12 | 4 | 1 | 9 | 4 | 0 | 18 | |
Indications are stratified according to age, sex, and the microarray production batch. P values were calculated using Fisher's exact test on sums across subindications (the Σ columns).
CALCL indicates cutaneous anaplastic large cell lymphoma; AD, atopic dermatitis; ND, unspecified dermatosis; PP, lesional skin from psoriasis patients; PN, nonlesional skin from psoriasis patients; and NN, normal skin from healthy controls.
Sex and age of 2 CALC and 1 MF samples are unknown.
Figure 1.
Expression profiles in training set for highly significant miRNAs. We analyzed microarray measurements of 688 miRNAs in the training set of 90 samples with t tests to discover differences in expression between samples of subjects with CTCL and those of BDN subjects. The 27 miRNAs that displayed highly significant (Bonferroni corrected P < .001) and strong differences (at least 50% change) are presented in the heat map. Samples are arranged in columns, miRNAs in rows, and both are hierarchically clustered using Euclidean distance with average linkage of nodes. Red-to-yellow shades indicate increased relative expression; blue shades indicate reduced expression; green indicates median expression. The top 5 most significantly induced or repressed miRNAs are shown in bold.
Identification of a CTCL-specific miRNA signature
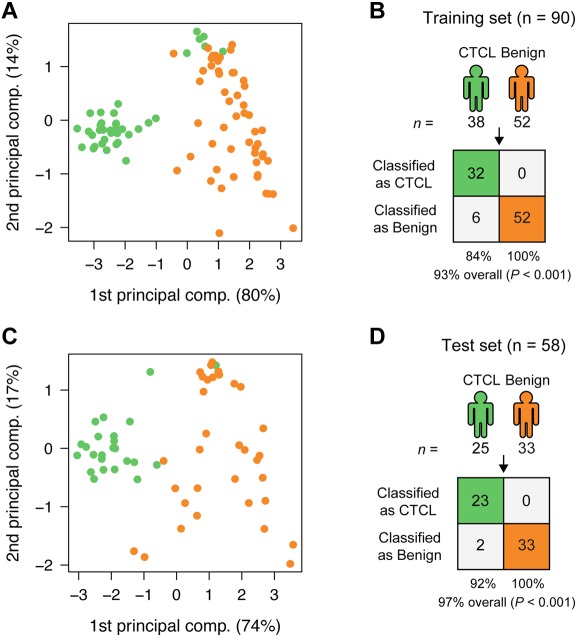
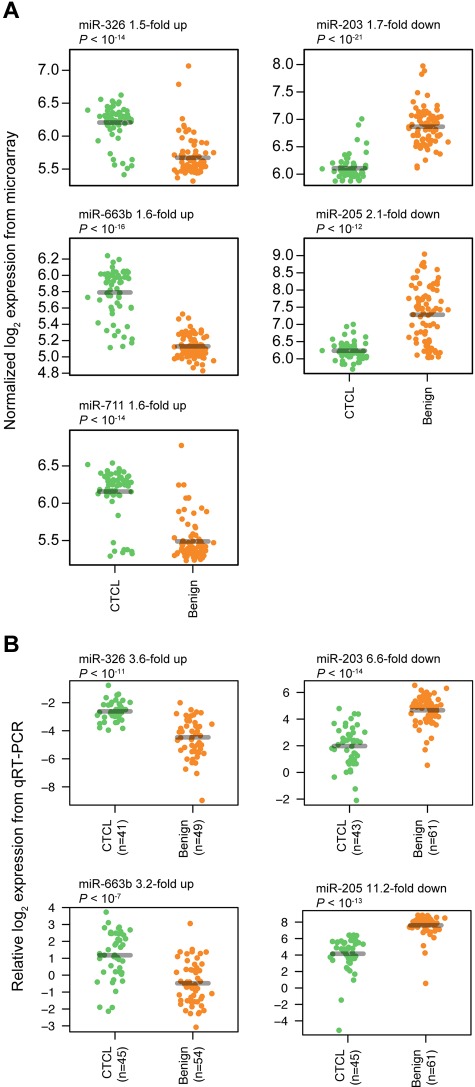
To find a CTCL-specific signature, we analyzed the training set with a nearest shrunken centroid algorithm,28 using only the top 3 most induced (miR-326, miR-663b, and miR-711) and repressed (miR-203, miR-205, and miR-718) miRNAs among the 27 highly significant miRNAs as identified in Figure 1. All 6 miRNAs had Bonferroni corrected P values < 10−8 by t test. After shrinking of centroids (supplemental Figure 2), samples in the training set could be classified with 93% accuracy (84% sensitivity and 100% specificity; P < .001 by Fisher exact test; Figure 2A-B). Shrinkage of centroids identified an optimal threshold value at 5.135 (dashed vertical line in supplemental Figure 2A). At this threshold value, the relative contribution of individual miRNAs in the classifier is shown in supplemental Figure 2B. As shown in supplemental Figure 2B, miR-718 was shrunk to 0, indicating that it did not contribute to the classification, and, accordingly, it was removed from the classifier. The final classifier consequently consisted of 5 miRNAs. To assess the performance of the 5 miRNAs in the classification of unknown samples, we used the already trained classifier on the 58 test set samples, and here achieved 97% classification accuracy (92% sensitivity and 100% specificity; P < .001 by Fisher exact test; Figure 2C-D). Figure 3A shows the expression of the individual miRNAs in the classifier. For each of the 5 miRNAs, the normalized log2 expression values are grouped according to patient type (CTCL and BDN, respectively; Figure 3A).
Figure 2.
Classification of CTCL and BDN. (A) PCA plot of samples from subjects with CTCL (green) and those of BDN subjects (orange) in the training set based on the 5-miRNA profile identified by the nearest shrunken centroids (NSC) algorithm. Percentages indicate percentage of variance explained by that component. (B) Classification performance in the training set using the NSC algorithm. P values were calculated using Fisher exact test. (C) PCA plot of samples in the test set based on the 5-miRNA profile identified from the training set. (D) Classification performance in the test set using the trained NSC algorithm.
Figure 3.
Classifier miRNA expressions measured by microarray and qRT-PCR. For each miRNA, expressions are grouped according to patient type (CTCL and BDN, respectively), with a small scatter on the x-axis within each group to allow better visualization of all measurements. P values were calculated using a t test. (A) Expressions measured by microarray. (B) Expressions measured by qRT-PCR.
Next, we evaluated the robustness of the classifier by 10-fold cross-validation, each time selecting different batches as training and test set (but keeping the ratio 3/5 to 2/5). This approach identified miR-203, miR-663b, miR-205, and miR-711 in almost all cases (supplemental Table 1). One miRNA, miR-326, was only selected in 2 of 10 divisions. Importantly, no matter which division was chosen, the classification accuracy in the test set was consistently > 90% (93.1% on average, with a 99% confidence interval between 90.5% and 95.6%; supplemental Table 1).
As shown in Figure 1, several other miRNAs (besides those included in the classifier) discriminate to some degree between CTCL and BDN. To evaluate the potential of these miRNAs as classifiers, we repeated the selection and training procedure as outlined in the preceding paragraph, but we excluded the 5 miRNAs from the input set. This identified a new set of miRNAs for which we recorded the classification accuracies in the training and test set. We repeated this 4 more times, each time excluding the miRNAs selected for the classifier in the last round. As shown in supplemental Figure 3, the accuracy in the test set decreased rapidly: When the best 23 miRNAs were excluded, the 4 miRNAs selected only achieve 67% accuracy in the test set. Even the second-best set of classifier miRNAs (those identified in round 2) clearly performed worse in the test set than the 5-miRNA classifier actually used (supplemental Figure 3). In addition, combining the miRNAs identified in rounds 1 and 2 into a 9-miRNA classifier did not improve the performance either (data not shown). Taken together, these findings show that the 5-miRNA classifier consisting of miR-326, miR-663b, miR-711, miR-203, and miR-205 performed best compared with the other classifiers examined.
Extension and validation of the miRNA classifier
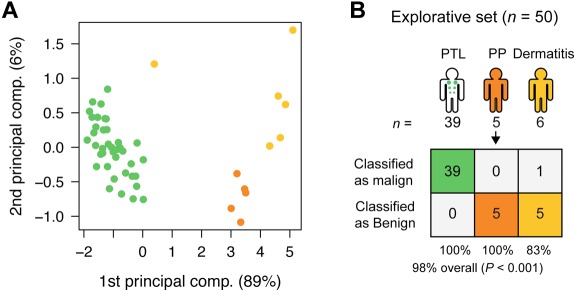
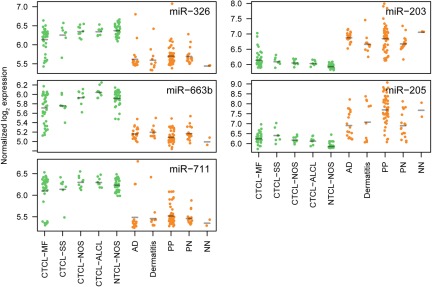
CTCL is characterized by primary involvement of the skin, but other PTLs may secondarily disseminate to the skin and give rise to skin lesions with resemblance to CTCL. To address whether the miRNA classifier also distinguishes between PTLs and benign inflammatory skin disorders, microarray analysis was performed on lesions from patients with PTL (NOS; 39 patients), and on 11 patients with benign disorders (dermatitis [6 patients] and psoriasis [5 patients]). As shown in Figure 4, using the 5-miRNA classifier, 10 of 11 patients with benign skin disorders were classified as benign, whereas 39 of 39 PTL patients were classified as malignant, resulting in an overall classification accuracy of 98% (P < .001; Figure 4). Figure 5 shows the expression levels of each individual classifier miRNA in all examined patients divided into clinical subgroups. The overall dichotomy between malignant and benign disorders also applied to each clinical subgroup of patients. However, the expression of individual miRNAs clearly varied within each subgroup, and there was also a considerable overlap in expression levels of each individual miRNA in malignant and benign disorders (Figure 5). Recently, miR-21 was identified as differentially expressed and involved in antiapoptosis in malignant T cells from SS patients compared with peripheral CD4+ T cells in healthy individuals.29 We also observed an increased expression of miR-21 in CTCL compared with most benign disorders (supplemental Figure 4). However, miR-21 was also highly increased in psoriatic lesions and in some dermatitis lesions (supplemental Figure 4), confirming other studies30 and explaining why miR-21 is not identified as a classifier miRNA.
Figure 4.
Classification of PTL and dermatitis. (A) PCA plot of samples from subjects with PTL (green), dermatitis (light orange), and psoriasis (yellow) for comparison, in the explorative set, based on the 5-miRNA profile. (B) Classification performance in the explorative set using the trained NSC algorithm. See Table 1 for explanation of PP.
Figure 5.
Overview of miRNA expressions measured by microarray for the miRNAs in the classifier for all sub indications included in the study. For each miRNA, expressions are grouped according to sub indication (same abbreviations as in Table 1).
Classification of treated and untreated CTCL and psoriasis from xenograft models and patients
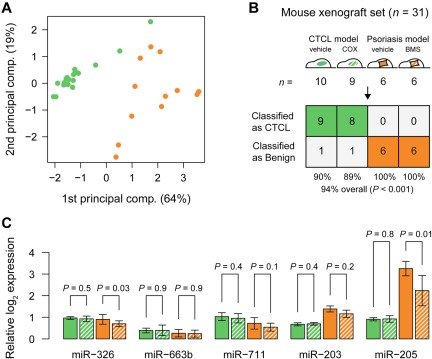
Because xenograft mouse models are momentous for the development of novel therapies, we examined the expression of the classifier miRNAs in tumors and skin lesions from human xenograft mouse models of CTCL and psoriasis, respectively.25 A principal component analysis (PCA) plot of samples from 19 mice with human xenograft CTCL lesions and 12 mice with human psoriatic skin xenograft lesions based on the 5-miRNA profile (Figure 6A) shows similar organization and separation of samples as with the clinical samples (Figures 2A,C and 4A). Accordingly, when the 5-miRNA classifier is applied, 17 of 19 samples from the xenograft CTCL model and 12 of 12 samples from the xenograft psoriasis model were classified as malignant and benign, respectively, that is, with a 94% overall accuracy (P < .001; Figure 6B). Interestingly, the most differentially expressed of the 5 miRNAs, miR-203 and miR-205, in the xenografts were also the most differentially expressed miRNAs is the clinical samples (Figure 6C vs Figure 5).
Figure 6.
Classification of treated and untreated CTCL and psoriasis from mouse xenograft models. (A) PCA plot of all samples from mouse xenograft CTCL model (green), and psoriasis model (orange), based on the 5-miRNA profile. Solid bars indicate vehicle controls, whereas hatched green bars indicate systemic treatment with a COX-2 inhibitor (celecoxib) and hatched orange bars indicate topical treatment with steroid (betamethasone). (B) Classification performance in the mouse xenograft set using the trained 5-miRNA classifier. (C) Bar plots showing normalized log2 expression levels relative to average of lowest expressions measured across all samples. Error bars denote ± 1 SD. For each miRNA, P values were calculated using a t test between treated and untreated samples from each model, respectively.
Nine of the 19 mice in the CTCL xenograft group had been treated with daily intraperitoneal injections of 10 μg/g celecoxib, an inhibitor of the cyclooxygenase-2 (COX-2) that significantly inhibited tumor growth.31 Likewise, 6 of the 12 mice in the psoriasis xenograft group were treated daily with topical glucocorticoid (betamethasone dipropionate). As shown in Figure 6C, the expression levels of 3 of the miRNAs (miR-203, miR-663b, and miR-711) were not influenced by the celecoxib or betamethasone, whereas the expression of miR-205 and miR-326 was inhibited by betamethasone but unaffected by celecoxib. However, the changes were relatively small and did not interfere with the malignant or benign classification (Figure 6B), indicating that the 5-miRNA classifier was relatively insensitive to treatment-induced changes in miRNA expression.
To address whether betamethasone treatment influenced the expression of the 5 miRNAs in psoriasis patients, we compared the expression in 5 patients treated with vehicle and 3 patients treated daily with topical betamethasone. As shown in supplemental Figure 5, 4 weeks of topical steroid treatment improved the clinical score but had little influence on the miRNA expression levels and miRNA-based classification in psoriasis patients, confirming the observations in xenograft human psoriatic skin (Figure 6).
Identification of a qRT-PCR–based miRNA classifier
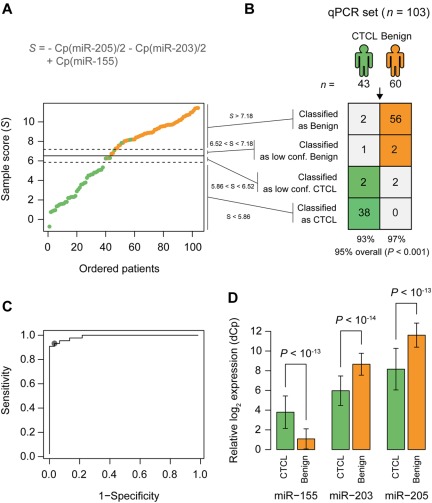
For diagnostic purposes, qRT-PCR is often considered more sensitive, specific, and applicable than microarrays. To confirm our microarray results, expression levels were measured for the 5-miRNA signature on a subset of 103 samples by qRT-PCR. Samples were selected based on high RNA content and covered both training and test set samples. miR-103 and miR-423-5p were identified as the most stably expressed references across samples, and their average Cp value was used as normalization factor when calculating ΔCp.32 The differential expression was clearly confirmed for miR-203, miR-205, and miR-326, with P values below 10−13, and for miR-663b, with a P value below 10−7 (Figure 3B), whereas the last miRNA, miR-711, could not be measured reliably above background fluorescence (supplemental Figure 6). Essentially similar results were obtained in an independent series of qRT-PCR experiments on 44 patient samples using a different qRT-PCR platform (TaqMan; data not shown). Recent studies on skin lesions from tumor stage MF and blood samples from SS patients reported on a differential expression of miR-155, miR-21, miR-24, miR-34b, miR-191, miR-486, miR-214, Let-7b, and other miRNAs.33–36 Accordingly, we performed qRT-PCR for these miRNAs and confirmed a differential expression of miR-155, miR-24, miR-191, and Let-7b (Figure 7; data not shown). In contrast, miR-34b did not achieve significance in the qRT-PCR measurements, whereas miR-21 was increased in CTCL but also in a fraction of psoriasis patients (supplemental Figure 4; data not shown). The nearest shrunken centroid algorithm identified miR-155, miR-203, and miR-205 as the most discriminative set of miRNAs. By rewriting the equations for nearest centroid classification as an equivalent linear combination, and simplifying, we write the discriminant function, or sample score, as S = Cp(miR-155) − Cp(miR-203)/2 − Cp(miR-205)/2.
Figure 7.
qRT-PCR–based classification of samples from patients with CTCL and benign skin disease. (A) A Cp based sample score (S) were calculated for each sample. Patients are ordered by increasing values of this score. The solid line shows the cutoff between patients with CTCL (green) and patients with benign skin disease (orange). The dotted lines show the cutoffs for the low confidence region. (B) Classification performance using the cutoffs defined in panel A. P values were calculated using Fisher exact test. (C) ROC curve ROC showing the sensitivity and specificity for various cutoff values on the sample score of the samples. (D) Relative expression of the 3 miRNAs used in the classification in samples from patients with CTCL and benign skin disease. Error bars indicate ± 1 SD.
We chose thresholds by inspecting the distribution of samples scores (Figure 7A-B) and the receiver operator characteristic (ROC) curve (Figure 7C), and we introduced a low-confidence region around the threshold (Figure 7A-B). Because miR-203 and miR-205 expression was decreased in CTCL (Figure 7D) and miR-155 expression increased in CTCL (Figure 7D), the score S was smaller in CTCL compared with benign skin disorders (Figure 7A-B). In 103 samples, this qRT-PCR-based “minimal” miRNA classifier (miR-155, miR-203, and miR-205) distinguished patients with CTCL from benign skin diseases with 95% classification accuracy (P < .001; Figure 7B) and high sensitivity and specificity as illustrated by the ROC graph in Figure 7C (dot indicates 91% sensitivity at 97% specificity). Importantly, MF patients were classified as malignant independently of the disease stage (supplemental Figure 7).
Discussion
Here, we used microarrays for an initial screening of miRNAs with a potential ability to distinguish between malignant (CTCL) and benign inflammatory disorders such as psoriasis, atopic dermatitis, and contact dermatitis. Five miRNAs (miR-203, miR-205, miR-326, miR-663b, and miR-711) were identified that discriminated with high accuracy (> 90%) between malignant and benign conditions in a total of 198 patients, including an initial training set of 90 patients, a test set of 58 patients, and an independent cohort of 50 patients. Importantly, the expression pattern of 4 of 5 miRNAs was verified using qRT-PCR on 103 patients. Likewise, 31 samples from human xenograft mouse models of CTCL and psoriasis also were classified with > 90% accuracy. Moreover, the classification was relatively resistant to topical treatment of psoriatic skin with steroids. Taken together, these findings indicate that miRNAs can distinguish between CTCL and benign skin disorder with a high accuracy and a high level of robustness.
Recent studies on subpopulations of CTCL patients identified miR-21 and miR-155 as differentially expressed in SS patients and advanced (tumor-stage) MF, respectively.29,37 The aberrant expression of miR-21 expression was driven by signal transducer and activator of transcription-3, which is aberrantly activated in CTCL and involved in antiapoptosis.38 Accordingly, it was a surprise that we did not identify miR-21 as differentially expressed in CTCL. However, miR-21 expression was indeed significantly increased in CTCL compared with dermatitis and nonlesional skin from psoriasis patients. In contrast, miR-21 was not increased in CTCL compared with lesional skin from psoriasis patients. On the contrary, miR-21 expression was equal to and often higher in psoriatic lesions than in CTCL. The increased miR-21 expression in lesional psoriasis might easily explain why we did not pick-up miR-21 as one of the highly differentially expressed miRNAs in CTCL. Our observation of a high miR-21 expression in psoriatic lesions is also in agreement with recent reports on an increased miR-21 expression in psoriatic skin lesions.30 Our findings do not, however, argue against an important role of miR-21 in the pathogenesis in CTCL, but they suggest that miR-21 might not be suited for this diagnostic purpose.
In advanced MF, van Kester et al37 identified several miRNAs as differentially expressed in tumor stage MF from 19 patients compared with 12 patients with benign skin diseases. The majority of miRNAs also were identified as differentially expressed in our array analysis. Because they did not report on miR-203, miR-205, miR-326, miR-663b, and miR-711 (and raw data were not presented), we have not been able to verify whether these miRNAs also were able to distinguish between CTCL and benign disorders in their patient cohort. Interestingly, miR-155 was the most significantly up-regulated in the microarray data by van Kester et al,37 and a series of studies have previously identified miR-155 as differentially expressed and linked to the pathogenesis of other lymphomas. We were therefore puzzled that we did not identify miR-155 as differentially expressed in CTCL in our microarray analysis. A possible explanation could relate to differences in patient subgroups and control populations as observed for miR-21. This was, however, not the case. Accordingly, we performed qRT-PCR for miR-155, and we were able to confirm and extend the findings of van Kester et al37 that miR-155 was aberrantly expressed in all subgroups of CTCL. Importantly, our qRT-PCR results also showed that miR-155 was one of the most significantly differentially expressed miRNAs in CTCL. At present, it is unclear why miR-155 was not identified as differentially expressed in our microarray analysis. Because the LNA-based microarray platform used here has been used previously to identify miR-155 as differentially expressed in the NCI-60 cell line panel,39 it is unlikely that the difference is because of platform related issues. We therefore speculate that cross-hybridization or presence of pre-miR-155 might have masked the signal of the mature miR-155 because the hybridization probes do not discriminate between mature and pre-miRNAs in the microarray platform. Our data strongly underline the importance of qRT-PCR validation of microarray data and support the notion that in case of miR-155, qRT-PCR assays are more sensitive than microarrays.
For diagnostic purposes, qRT-PCR is not only more sensitive and specific but also more applicable than microarrays. Using the nearest shrunken centroid algorithm, miR-155, miR-203, and miR-205 were identified as the most discriminative set of miRNAs, and a score formulated as the difference between the Cp of miR-155 and the average Cp of miR-203 and miR-205 found to distinguish patients with CTCL from benign skin diseases with very high sensitivity, specificity, and classification accuracy (95%). Our observation that the classifier miRNAs were differentially expressed in almost all CTCL patients suggests that they might play a key role in the pathogenesis of CTCL. As mentioned, miR-155 is differentially expressed in other lymphomas and hematologic malignancies and is linked to the malignant transformation.40 Accordingly, it was not unexpected that miR-155 was differentially expressed in CTCL. Several miR-155 targets have been identified and miR-155 modifies oncogenesis at multiple levels, including down-regulation of negative regulators such as Src homology-2 domain-containing inositol 5-phosphatase 1 and increased expression of autocrine growth factors.41 Studies are in progress to unravel the function of miR-155 in the malignant transformation in CTCL.
Our finding of an increased expression of miR-203 in benign disorders is in line with previous reports on miR-203 expression in psoriasis and atopic dermatitis.42 MiR-203 is of particular interest to skin diseases because it plays a key role in keratinocyte biology.43 As mentioned, miR-203 is highly up-regulated in keratinocytes in psoriatic lesions and involved in the repression of suppressor of cytokine signaling-3 (SOCS3).44 Among other cytokine signaling pathways, SOCS3 inhibits signaling from the IFNα receptor. Because psoriasis patients exhibit hypersensitivity to IFNα that is partly caused by a deficient SOCS3 expression, and IFNα triggers or exuberates psoriasis in cancer patients,45 studies are in progress to address whether the aberrant miR-203 expression plays a role in IFNα hypersensitivity in psoriatic cells. A decreased expression of miR-203 has not previously been reported in CTCL, but Bueno et al46 provided evidence that miR-203 is a tumor suppressor and miR-203 silencing promotes lymphoma development via increased expression of oncogenes such as ABL.46 Although these oncogenes are not expressed in CTCL it is possible that miR-203 silencing also plays a direct role in malignant transformation in CTCL. Malignant T cells often display an aberrant expression of SOCS3 that protects against IFNα-mediated growth inhibition.47 Therefore, it is possible that miR-203 silencing (via an increased SOCS3 expression) plays a role in resistance to IFNα treatment in CTCL patients. A few verified targets of miR-205 have recently been reported the most interesting of which (in a CTCL context) is VEGF48 that is aberrantly expressed in all stages of CTCL.49 Accordingly, it is possible that silencing of miR-205 could play a role in the spontaneous VEGF production in CTCL lesions. Thus, it seems likely that all of the 3 miRNAs in the “minimal” qRT-PCR classifier play important roles as oncogenic (miR-155) and tumor suppressor (miR-203 and miR-205) miRNAs. We speculate that such features explain why the miRNA classifier is so robust and conserved across all stages of CTCL.
In conclusion, we provide the first evidence that a miRNA classifier can distinguish CTCL from benign inflammatory skin disorders with very high accuracy, suggesting a diagnostic potential in CTCL.
Supplementary Material
Acknowledgments
This work was supported in part by research funding from The Danish Foundation for Advanced Technology (Højteknologifonden), The Carlsberg Foundation (Carlsbergfondet), The Danish Research Councils, The Lundbeck Foundation, The Danish Cancer Society (Kræftens Bekæmpelse), Dansk Kræftforsknings Fond, The Novo Nordic Foundation, Fabrikant Vilhelm Pedersen og Hustrus Mindelegat, The A. P. Moeller Foundation, The University of Copenhagen, the National Cancer Institute (grant CA89194, M.A.W.), LEO Pharma A/S, and Exiqon A/S.
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: U.R. designed and performed research (qRT-PCR), performed sample handling (malignant biopsies), analyzed data, and wrote the paper; P.H.H. designed research (statistical models), handled and analyzed data, and wrote the paper; N.B. performed research (qRT-PCR); M.B.L. sample handling (benign skin disease biopsies) and performed research; C.B.A. performed research (qRT-PCR); L. Svensson performed patient and sample handling and research (animal model); K.L.K. designed and performed research (animal models); M.T.V. performed research (qRT-PCR; animal model CTCL); B.L. designed and performed research (animal models); J.R.Z. designed research (RNA extraction) and analyzed data; T.K. designed and performed research (animal models) and analyzed data; C.M.B. performed research and analyzed data and wrote paper; L.-M.G. supplied and analyzed clinical data for the malignant biopsies; T. Labuda contributed to the research design (qRT-PCR); A.-M.M. designed and performed research (animal models); K.G. contributed to overall research design; M.A.W. contributed to the overall research design, analyzed data, and wrote the paper; T. Litman contributed to the overall research design, array analysis, and research on miR-targets; M.S.-W. supplied the malignant biopsies; C.Q.-R. supplied and analyzed clinical data for the biopsies from benign skin disorders; R.G. supplied and analyzed clinical data for the malignant biopsies; E.R. contributed to the overall research design, supplied malignant biopsies, analyzed data, and determined histologic classification of malignant biopsies; R.S. performed data analysis of qRT-PCR, array analysis, annotation, and submission to Gene Expression Omnibus; C. Geisler contributed to the overall research design (arrays and qRT-PCR) and analyzed data; A.W. contributed to the overall research design, analyzed data (animal models), and wrote the paper; C. Glue contributed to the research design (arrays and qRT-PCR) and analyzed data; M.A.R. contributed to the overall research design, supplied benign skin disease biopsies, analyzed data, and wrote the paper; L. Skov contributed to the overall research design, supplied benign skin disease, performed biopsies histologic classification of benign biopsies, analyzed data, and the wrote paper; and N.O. contributed to overall research design, analyzed data, primus motor and chairman, and wrote the paper.
Conflict-of-interest disclosure: C. Glue, R.S., and T. Litman are or have been employed by Exiqon A/S, and M.A.R., P.H.H., M.B.L., T. Labuda, L. Svensson, and J.R.Z. are or have been employed by LEO Pharma A/S. Some of the information presented is part of a patent application filed by the institutions and companies engaged in the project. The remaining authors declare no competing financial interests.
Correspondence: Niels Odum, Departments of Biology and International Health, Immunology and Microbiology, Immunology, ISIM, 22.5.34 Panum, University of Copenhagen, Blegdamsvej 3c, DK-2200 Copenhagen N, Denmark; e-mail: ndum@sund.ku.dk.
References
- 1.Bradford PT, Devesa SS, Anderson WF, Toro JR. Cutaneous lymphoma incidence patterns in the United States a population-based study of 3884 cases. Blood. 2009;113(21):5064–5073. doi: 10.1182/blood-2008-10-184168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trautinger F, Knobler R, Willemze R, et al. EORTC consensus recommendations for the treatment of mycosis fungoides/Sézary syndrome. Eur J Cancer. 2006;42(8):1014–1030. doi: 10.1016/j.ejca.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 3.Lee BN, Duvic M, Tang CK, et al. Dysregulated synthesis of intracellular type 1 and type 2 cytokines by T cells of patients with cutaneous T-cell lymphoma. Clin Diagn Lab Immunol. 1999;6(1):79–84. doi: 10.1128/cdli.6.1.79-84.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gjerdrum LM, Woetmann A, Odum N, et al. FOXP3+ regulatory T cells in cutaneous T-cell lymphomas association with disease stage and survival. Leukemia. 2007;21(12):2512–2518. doi: 10.1038/sj.leu.2404913. [DOI] [PubMed] [Google Scholar]
- 5.Rosen ST, Querfeld C. Primary cutaneous T-cell lymphomas. Hematol Am Soc Hematol Educ Program. 2006;323:330. doi: 10.1182/asheducation-2006.1.323. [DOI] [PubMed] [Google Scholar]
- 6.Dereure O, Levi E, Vonderheid EC, Kadin ME. Infrequent Fas mutations but no Bax or p53 mutations in early mycosis fungoides a possible mechanism for the accumulation of malignant T lymphocytes in the skin. J Invest Dermatol. 2002;118(6):949–956. doi: 10.1046/j.1523-1747.2002.01794.x. [DOI] [PubMed] [Google Scholar]
- 7.van Doorn R, van Kester MS, Dijkman R, et al. Oncogenomic analysis of mycosis fungoides reveals major differences with Sézary syndrome. Blood. 2009;113(1):127–136. doi: 10.1182/blood-2008-04-153031. [DOI] [PubMed] [Google Scholar]
- 8.Girardi M, Heald PW, Wilson LD. The pathogenesis of mycosis fungoides. N Engl J Med. 2004;350(19):1978–1988. doi: 10.1056/NEJMra032810. [DOI] [PubMed] [Google Scholar]
- 9.Kadin ME, Vonderheid EC. Targeted therapies: denileukin diftitox–a step towards a “magic bullet” for CTCL. Nat Rev Clin Oncol. 2010;7(8):430–432. doi: 10.1038/nrclinonc.2010.105. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Q, Wang HY, Woetmann A, et al. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108(3):1058–1064. doi: 10.1182/blood-2005-08-007377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garzon R, Calin G a, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 12.Tessel MA, Krett NL, Rosen ST. Steroid receptor and microRNA regulation in cancer. Curr Opin Oncol. 2010;22(6):592–597. doi: 10.1097/CCO.0b013e32833ea80c. [DOI] [PubMed] [Google Scholar]
- 13.Krejsgaard T, Vetter-Kauczok CS, Woetmann A, et al. Ectopic expression of B-lymphoid kinase in cutaneous T-cell lymphoma. Blood. 2009;113(23):5896–5904. doi: 10.1182/blood-2008-09-181024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pimpinelli N, Olsen EA, Santucci M, et al. Defining early mycosis fungoides. J Am Acad Dermatol. 2005;53(6):1053–1063. doi: 10.1016/j.jaad.2005.08.057. [DOI] [PubMed] [Google Scholar]
- 15.Glusac EJ. Criterion by criterion, mycosis fungoides. Am J Dermatopathol. 2003;25(3):264–269. doi: 10.1097/00000372-200306000-00014. [DOI] [PubMed] [Google Scholar]
- 16.Olsen E, Vonderheid E, Pimpinelli N, et al. Revisions to the staging and classification of mycosis fungoides and Seìzary syndrome a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood. 2007;110(6):1713–1722. doi: 10.1182/blood-2007-03-055749. [DOI] [PubMed] [Google Scholar]
- 17.Keehn CA, Belongie IP, Shistik G, Fenske NA, Glass LF. The diagnosis, staging, and treatment options for mycosis fungoides. Cancer Control. 2007;14(2):102–111. doi: 10.1177/107327480701400203. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 19.Willemze R, Jaffe ES, Burg G, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. 2005;105(10):3768–3785. doi: 10.1182/blood-2004-09-3502. [DOI] [PubMed] [Google Scholar]
- 20.Foss FM, Zinzani PL, Vose JM, et al. Peripheral T-cell lymphoma. Blood. 2011;117(25):6756–6767. doi: 10.1182/blood-2010-05-231548. [DOI] [PubMed] [Google Scholar]
- 21.Burg G, Kempf W, Cozzio A, et al. WHO/EORTC classification of cutaneous lymphomas 2005 histological and molecular aspects. J Cutan Pathol. 2005;32(10):647–674. doi: 10.1111/j.0303-6987.2005.00495.x. [DOI] [PubMed] [Google Scholar]
- 22.Stenderup K, Rosada C, Worsaae A, et al. Interleukin-20 plays a critical role in maintenance and development of psoriasis in the human xenograft transplantation model. Br J Dermatol. 2009;160(2):284–296. doi: 10.1111/j.1365-2133.2008.08890.x. [DOI] [PubMed] [Google Scholar]
- 23.Kvist PH, Svensson L, Hagberg O, et al. Comparison of the effects of vitamin D products in a psoriasis plaque test and a murine psoriasis xenograft model. J Trans Med. 2009;7:107. doi: 10.1186/1479-5876-7-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krejsgaard T, Kopp K, Ralfkiaer E, et al. A novel xenograft model of cutaneous T-cell lymphoma. Exp Dermatol. 2010;19(12):1096–1102. doi: 10.1111/j.1600-0625.2010.01138.x. [DOI] [PubMed] [Google Scholar]
- 25.Ritchie ME, Silver J, Oshlack A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23(20):2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 26.Cleveland WS, Grosse E, Shyu WM. Local regression models. In: Chambers JM, Hastie TJ, editors. Statistical Models in S. Pacific Grove, CA: Wadsworth & Brooks/Cole; 1992. pp. 309–376. [Google Scholar]
- 27.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 28.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Fits L, van Kester MS, Qin Y, et al. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sézary syndrome. J Invest Dermatol. 2011;131(3):762–768. doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- 30.Zibert JR, Løvendorf MB, Litman T, et al. MicroRNAs and potential target interactions in psoriasis. J Dermatol Sci. 2010;58(3):177–185. doi: 10.1016/j.jdermsci.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Kopp KLM, Dabelsteen S, Krejsgaard T, et al. COX-2 is a novel target in therapy of mycosis fungoides. Leukemia. 2010;24(12):2127–2129. doi: 10.1038/leu.2010.221. [DOI] [PubMed] [Google Scholar]
- 32.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;(3):research0034–research0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Odenike O, Rowley JD. Leukaemogenesis more than mutant genes. Nat Rev Cancer. 2010;10(1):23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballabio E, Mitchell T, van Kester MS, et al. MicroRNA expression in Sézary syndrome identification, function, and diagnostic potential. Blood. 2010;116(7):1105–1113. doi: 10.1182/blood-2009-12-256719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holst LM, Kaczkowski B, Gniadecki R. Reproducible pattern of microRNA in normal human skin. Exp Dermatol. 2010;19(8):e201–e205. doi: 10.1111/j.1600-0625.2009.01049.x. [DOI] [PubMed] [Google Scholar]
- 36.Narducci MG, Arcelli D, Picchio MC, et al. MicroRNA profiling reveals that miR-21, miR-486 and miR-214 are upregulated and involved in cell survival in Sézary syndrome. Cell Death Dis. 2011;2:e151. doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Kester MS, Ballabio E, Benner MF, et al. miRNA expression profiling of mycosis fungoides. Mol Oncol. 2011;5(3):273–280. doi: 10.1016/j.molonc.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sommer VH, Clemmensen OJ, Nielsen O, et al. In vivo activation of STAT3 in cutaneous T-cell lymphoma. evidence for an antiapoptotic function of STAT3. Leukemia. 2004;18(7):1288–1295. doi: 10.1038/sj.leu.2403385. [DOI] [PubMed] [Google Scholar]
- 39.Søkilde R, Kaczkowski B, Podolska A, et al. Global microRNA analysis of the NCI-60 cancer cell panel. Mol Cancer Ther. 2011;10(3):375–384. doi: 10.1158/1535-7163.MCT-10-0605. [DOI] [PubMed] [Google Scholar]
- 40.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15(4):352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen IM, Otero D, Kao E, et al. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol Med. 2009;1(5):288–295. doi: 10.1002/emmm.200900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs novel regulators in skin inflammation. Clin Exp Dermatol. 2008;33(3):312–315. doi: 10.1111/j.1365-2230.2008.02804.x. [DOI] [PubMed] [Google Scholar]
- 43.Wei T, Orfanidis K, Xu N, et al. The expression of microRNA-203 during human skin morphogenesis. Exp Dermatol. 2010;19(9):854–856. doi: 10.1111/j.1600-0625.2010.01118.x. [DOI] [PubMed] [Google Scholar]
- 44.Sonkoly E, Wei T, Janson PCJ, et al. MicroRNAs novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2(7):e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eriksen KW, Woetmann A, Skov L, et al. Deficient SOCS3 and SHP-1 expression in psoriatic T cells. J Invest Dermatol. 2010;130(6):1590–1597. doi: 10.1038/jid.2010.6. [DOI] [PubMed] [Google Scholar]
- 46.Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13(6):496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 47.Brender C, Lovato P, Sommer VH, et al. Constitutive SOCS-3 expression protects T-cell lymphoma against growth inhibition by IFNalpha. Leukemia. 2005;19(2):209–213. doi: 10.1038/sj.leu.2403610. [DOI] [PubMed] [Google Scholar]
- 48.Baranwal S, Alahari SK. MiRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126(6):1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krejsgaard T, Vetter-Kauczok CS, Woetmann A, et al. Jak3- and JNK-dependent vascular endothelial growth factor expression in cutaneous T-cell lymphoma. Leukemia. 2006;20(10):1759–1766. doi: 10.1038/sj.leu.2404350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.