Abstract
Next-generation sequencing is rapidly transforming our ability to profile the transcriptional, genetic, and epigenetic states of a cell. In particular, sequencing DNA from the immunoprecipitation of protein-DNA complexes (ChIP-seq) and methylated DNA (MeDIP-seq) can reveal the locations of protein binding sites and epigenetic modifications. These approaches contain numerous biases which may significantly influence the interpretation of the resulting data. Rigorous computational methods for detecting and removing such biases are still lacking. Also, multi-sample normalization still remains an important open problem. This theoretical paper systematically characterizes the biases and properties of ChIP-seq data by comparing 62 separate publicly available datasets, using rigorous statistical models and signal processing techniques. Statistical methods for separating ChIP-seq signal from background noise, as well as correcting enrichment test statistics for sequence-dependent and sonication biases, are presented. Our method effectively separates reads into signal and background components prior to normalization, improving the signal-to-noise ratio. Moreover, most peak callers currently use a generic null model which suffers from low specificity at the sensitivity level requisite for detecting subtle, but true, ChIP enrichment. The proposed method of determining a cell type-specific null model, which accounts for cell type-specific biases, is shown to be capable of achieving a lower false discovery rate at a given significance threshold than current methods.
Keywords: ChIP-seq, wavelets, regression, normalization, order statistics
1 Background
Next-generation DNA Sequencing (NGS) provides enormous potential for rapidly identifying epigenetic modifications, transcription factor (TF) binding sites, and RNA transcriptional profiles. Sequencing cost continues to drop, while sequencing depth is increasing every year. In the near future, a single experiment will be able to sequence tens of giga-bases, far exceeding the size of a typical mammalian genome. Processing and interpreting such data sets present complex computational challenges at multiple levels. Although cross-hybridization problems associated with microarrays have disappeared in NGS, many of the old problems captured in terms of continuous measurements are rephrased into new discrete problems of counting short reads, and numerous biases that are intrinsic to experimental procedures still persist. It is thus critical to develop effective computational methods for processing NGS data in order to ensure the correct inference of biologically meaningful information. To facilitate the development, this paper provides rigorous theoretical studies of normalization and bias correction methods and also clarifies the suitability of various model assumptions commonly used.
Chromatin immunoprecipitated DNA sequencing (ChIP-seq) and Methylated-DNA immunoprecipitation sequencing (MeDIP-seq) are powerful tools for profiling the genome-wide binding sites of TFs and epigenetic modification sites; see Figure 1 for an illustration of the techniques. Proper data normalization is critical for comparing different biological samples. Currently, researchers typically normalize immunoprecipitation (IP) data together with corresponding control experiments (Input), or normalize multiple IP samples together, by scaling the local read density by a multiplicative ratio of total sequencing depths. This method does not account for the fact that there should be a significant enrichment of reads in regions targeted by IP compared to flanking regions. Equalizing the total number of reads can thus artificially inflate the background noise, as illustrated in Figure 2. In contrast, the method proposed here normalizes paired datasets by equalizing the expected number of reads only in background regions that contain non-specific DNA not targeted by IP.
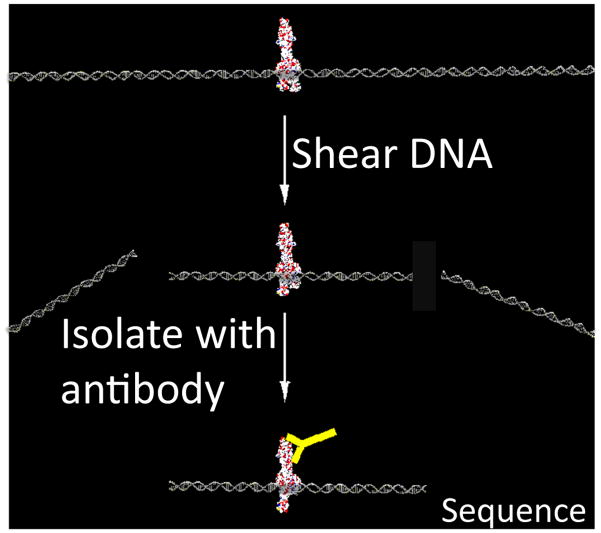
Figure 1. ChIP-seq and MeDIP-seq.
Chromatin is randomly sheared with high frequency sound waves (sonication) or digested with micrococcal nuclease (MNase). The desired Protein:DNA complex is then isolated with an antibody (yellow Y). The ends of purified DNA are then sequenced (ChIP-seq). Similarly, MeDIP-seq uses an antibody against methyl cytosine, followed by deep sequencing. Mapping the resulting short reads to the reference human genome then provides information about which genomic loci were modified or bound by a TF.
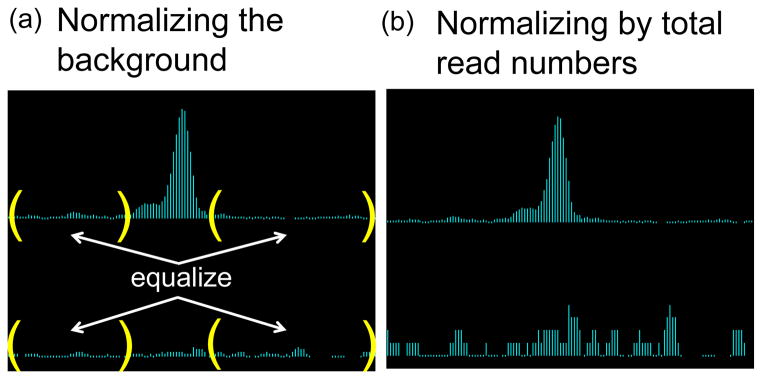
Figure 2. Comparison of scaling methods.
(a) Scaling IP (top row) and Input (bottom row) samples to equalize the read counts only in the background (enclosed by parentheses) preserves the statistical significance of the IP peak shown. (b) On the other hand, forcing the total number of reads to be equal between IP and Input would artificially redistribute the counts that accumulated within the IP peak to background regions, thus inflating the noise level in Input. Some true peaks can be lost in this process.
Several studies have recently focused on removing sequence-dependent biases and inferring transcriptional levels from RNA-seq data (Hansen et al., 2010, Anders and Huber, 2010, Robinson and Smyth, 2007). However, rigorous analysis methods are still limited for ChIP-seq and MeDIP-seq. ChIP-seq has numerous sources of bias stemming from differential protection against sonication across the genome, variable antibody quality, sequence-dependent PCR amplification, and differential mappability of short reads to repeat-rich genomic regions (Teytelman et al., 2009, Aird et al., 2011). The general pattern of these complex biases has not yet been characterized, and appropriate correction methods are still lacking. This paper develops signal processing and statistical methods for detecting and adjusting for numerous biases found in ChIP-seq data.
The paper is organized as follows: in the next section we propose a method for ChIP-seq and MeDIP-seq data normalization. After describing the algorithm and associated significance test, we then demonstrate the power of this procedure to identify enriched loci which would otherwise be drowned out by background noise under scaling by sequencing depth. In the third section, we motivate the need for bias correction by analyzing an ensemble of 62 publicly available ChIP-seq data sets using test statistics designed to rigorously qualify the common assumption that read count is Poisson distributed. A technique from signal processing known as spectral analysis is used to decompose an alignment density into peaks of various amplitudes and frequencies. This information can then be correlated across control experiments from the same cell type to generate a cell type-specific model of sonication bias. We demonstrate this technique in the fourth section, where we advocate the use of nonlinear regression to model the local propensity of chromatin to sonicate as a response to sequence-dependent and cell type-dependent predictors. We compare the performance of this approach in detecting histone methylation in wild type mouse neural stem cells with that of a commonly used Poisson model.
2 Normalization
Proper normalization methods are crucial for comparing two independent ChIP-seq data sets. For example, researchers are often interested in combining the whole-genome profiles of two different transcription factors in order to study their interactions. Different antibodies have different affinities, and experimental conditions are intrinsically variable. Consequently, independent ChIP-seq experiments can produce quite different distributions of reads, and arbitrarily choosing a common p-value cutoff for two experiments may unfairly bias the peak-calling algorithm towards one experiment. Standardizing ChIP-seq data thus remains an important unsolved problem. Let N be the total number of reads in the IP channel and M that in the Input channel. A simple algorithm is to multiply the Input read density by N/M. We refer to this method as sequencing depth scaling (SDS). The distribution of alignments across the genome is never uniform, and increasing sequencing depth does not uniformly increase alignment density genome wide; however, this is precisely the underlying assumption in SDS. Scaling the Input density by the ratio N/M often incorrectly estimates the background, inducing false negatives and false positives in peak detection, as illustrated in Figure 2. The IP channel is actually composed of two pools of DNA fragments in unknown ratio: the genomic background and those DNA fragments pulled down by the antibody. The optimal scaling factor should thus normalize Input only to the background component of the IP, and not the entire IP data. We here propose a method of signal extraction scaling (SES) which estimates this background component. Other currently used approaches employ chromosome-wise regression of IP and Input data (Rozowsky et al., 2009) or normalization of low count regions that are below a pre-defined significance level (Kharchenko et al., 2008). However, our analysis shows that regression-based scaling factors can be sensitive to outliers and may over-estimate the scaling factor. Furthermore, it is not clear why the same significance level predefined by an arbitrarily chosen p-value cutoff should be applied to all ChIP-seq data, since the quality of each antibody may influence the minimum enrichment level of targeted loci. We thus propose a robust, data-driven normalization scheme based on the order statistics of binned count data.
Given a reference genome, we partition it into n non-overlapping windows of fixed width. We then count the number of alignments which fall within a given window. Let [Yi]:= [Y1, …, Yn] and [Xi]:= [X1, …, Xn] be the resulting lists of alignment counts for the IP and Input, respectively. We then sort [Yi] in increasing order to obtain the list [Y(i)] of order statistics, where (i) denotes the i-th element of the sorted vector. Let [X(i)] be the list of Input channel counts reordered to match [Y(i)]; i.e., X(i) and Y(i) count alignments in the same window. Denote the partial sums of [Y(i)] and [X(i)] by and , respectively, and consider the percentage pj = Ȳj/Ȳn of reads from the IP channel and qj = X̄j/X̄n from the Input channel, allocated to the first j ordered bins (1),…,(j). Our working assumption is that IP bins of relatively low tag count correspond to background regions that are not directly targeted by the antibody. Consequently, the percentage pj of tags allocated to the first j ordered bins from the IP channel should not exceed the percentage allocation qj of tags from the Input channel, since the Input channel is entirely comprised of background, while the IP channel is a mixture of background and immunoprecipitated DNA. Moreover, the difference |qj − pj|, which gives the differential cumulative percentage tag count between IP and Input, will initially increase from 0 as j increases. However, once we begin to include bins of sufficiently large tag count corresponding to ChIP-enriched loci, the difference will rapidly drop to zero. Therefore, an estimate of the background component in IP data can be obtained by identifying the bin cutoff k at which the percentage allocation of tags in the Input channel maximally exceeds that of the IP channel. That is, we propose to choose the cutoff k = argmaxj|qj − pj| and normalize the Input density with a multiplicative scaling factor α = Ȳk/X̄k.
This approach is similar to the approaches of CCAT (Xu et al., 2010) and SPP (Kharchenko et al., 2008), both of which attempt to identify a subset of relatively low tag count background bins from the IP channel. This background region is then normalized to the Input channel, as in our approach. SPP arbitrarily defines background regions as those whose tag count has a Poisson p-value greater than 10−5. It is not clear why the same p-value cutoff should be applied to every ChIP-seq dataset or why this particular choice is guaranteed to identify regions devoid of signal. Also, our analysis of the Poisson model described in the next section suggests that this model is sensitive to scaling; and, thus, which bins are included as background under SPP is sensitive to sequencing depth. The CCAT method determines a background set as the complement of a signal set on which what they call “signal-to-noise-ratio” (SNR) between the IP and Input is maximized. Their definition of SNR is similar to the ratio pj/qj, with the key distinction that we sum over indices defined by the order statistic of the IP, while they allow sums over arbitrary sets of bin indices. We identify a background partition by maximizing qj − pj over the order statistic. This choice maximizes qj/pj, the “noise-to-signal-ratio,” over low order bins identified by the lower component of the order statistic and consequently maximizes SNR on the complement set of signal bins. The CCAT algorithm attempts to identify a SNR maximizing subset by heuristically generating a sequence of genomic partitions. There is no guarantee that the CCAT algorithm will converge as they acknowledge by including a trip count threshold in their termination criterion. Moreover, upon termination, there is no guarantee that the resulting partition will contain all background bins. Ideally, in order to determine the set of background bins, one would enumerate all partitions of the genome and choose the partition which maximized SNR. Unfortunately, complete subset enumeration requires an operation count exponential in the number of bins and is computationally not feasible. For this reason, we limit our search to partitions induced by the order statistic of the IP. Under the assumption that background bins have relatively lower tag counts than signal bins, our method identifies the smallest subset of the order statistic which contains the background partition. Our method is thus motivated by a well-developed theory of order statistics which we describe below, is guaranteed by construction to maximize SNR over all partitions of the genome induced by the order statistic, and has a significance test associated with it, which can be used for quality control also described below.
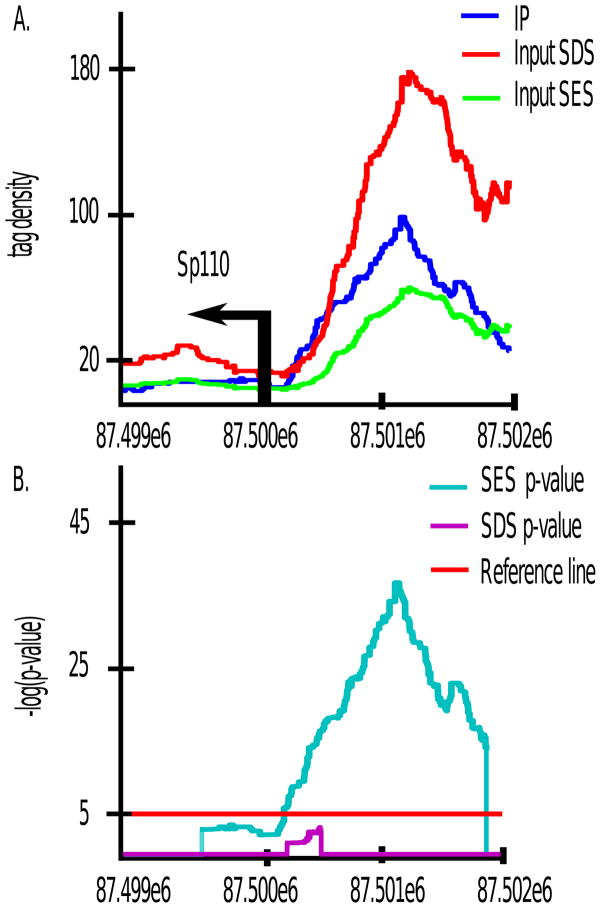
To demonstrate this technique, we applied our method to histone 3 lysine 4 tri-methylation (H3K4me3) ChIP-seq data in mouse neural stem cells (NSC). We partitioned the MM9 mouse reference genome into 1 kb non-overlapping bins and generated the IP alignment density order statistics as above. In Figure 3, pj and qj are ploted as a function of j/n, where n is the total number of bins. The scaling factor computed via the SES method is Ȳk/X̄k ≈ 0.3394. CCAT and SPP produced similar estimates of 0.36 and 0.38 respectively. In contrast, normalizing by the total sequencing depths would scale Input by a factor of 1.05, severely inflating the Input channel. A regression-based method (Rozowsky et al., 2009) also yielded an inflated scaling factor of 1.8. This dramatic difference stems from the fact that the H3K4me3 antibody is very strong and that up to 80% of reads in the IP channel localize within H3K4me3 peaks (data not shown). Our method will thus be able to recover many false negatives that result when improperly scaling by the total sequencing depth. For example, gene expression profiles determined via Affymetrix microarrays show that the expression level of Sp110 is 2.4 fold higher than the median expression level of all genes, suggesting that Sp110 is being transcribed. H3K4me3 is an epigenetic mark of active transcription, yet Figure 4 shows that under the sequencing depth scaling of 1.05, there is almost no enrichment of IP over Input in the promoter region (Figure 4(A)), and the associated p-value is not statistically significant (Figure 4(B)). The commonly used peak caller MACS (Zhang et al., 2008) for example does not call a peak here under the default settings. Under our method of SES, however, the promoter region shown in Figure 4 exhibits statistically significant enrichment of H3K4me3 over Input.
Figure 3. Signal extraction scaling algorithm.
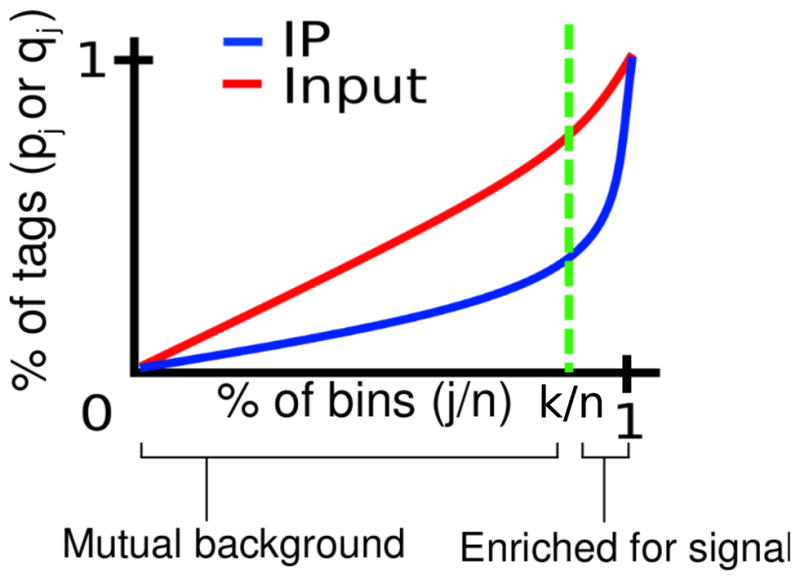
We maximize the difference between the cumulative percentage tag allocation in Input (red) and IP (blue), over all partitions of the genome ordered by the IP read density. The maximizing index divides the genome into two sets of loci: high tag count bins for which, on average, percentage IP tag density will exceed percentage Input tag density, and low tag count background bins for which the opposite is true.
Figure 4. SES recovers SDS false negatives.
sp100 is expressed at 2.4 fold above the median expression level of all genes in NSC. Yet, under SDS the promoter region shows no statistically significant enrichment of H3K4me3 IP over Input, despite it being an epigenetic mark of active transcription. Under SES, the sp110 promoter region is detected as methylated. Note that p-values were not computed at positions where Input density exceeds IP density.
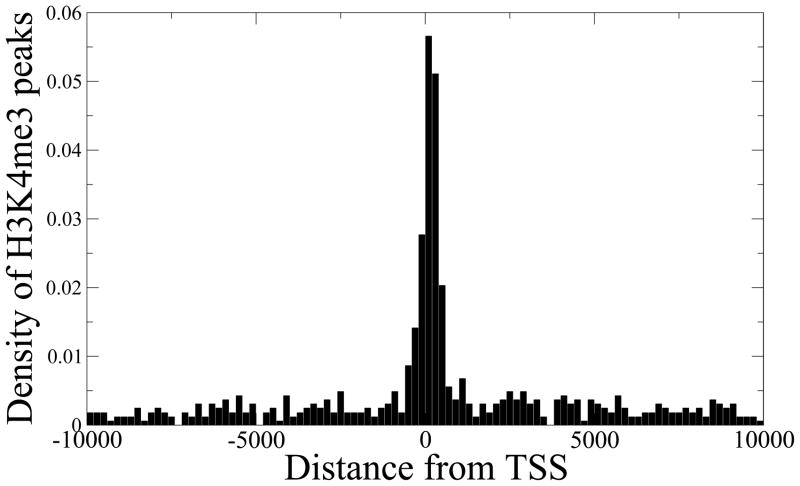
To further demonstrate the advantage of our method, we ran MACS (Zhang et al., 2008) and PeakSeq (Rozowsky et al., 2009) on our H3K4me3 data. Both MACS and PeakSeq overestimate the scaling factor for Input by 2–5 fold compared to our method, thus potentially missing many peaks with subtle H3K4me3 enrichment. Figure 5 shows the genomic location distribution of H3K4me3 peaks detected by our method, but missed by PeakSeq. A similar distribution is obtained when comparing with MACS. As expected, the missed peaks strongly localize at the transcription start sites (TSS) of known genes, and those genes have ~3 fold higher expression index than the genes that do not have any H3K4me3 peak (Wilcoxon test p-value = 2.0 × 10−81). Similarly, PeakSeq’s scaling factor for H3K27me3 ChIP-seq in neural stem cells was more than twice our estimated scaling factor. Our method identified 1,177 H3K27me3 peaks that were not found in the list of 281,720 peaks detected by PeakSeq at a very loose p-value cutoff of 0.03, corresponding to 5% FDR, and the peaks again tended to localize near TSS, as shown in Figure 6. The corresponding genes also have ~4 fold lower expression than the genes that have H3K4me3 peaks (Wilcoxon test p-value = 1.3 × 10−25). This analysis demonstrates the global effect of over-scaling the Input channel on suppressing potentially true signals.
Figure 5. SES preserves subtle H3K4me3 peaks.
The majority of the H3K4me3 peaks detected by our method, but missed by PeakSeq, are found near transcription start sites (TSS) of known genes. The corresponding genes also show significantly higher expression levels compared to the genes that do not have any H3K4me3 peak (Wilcoxon test p-value = 2.0 × 10−81).
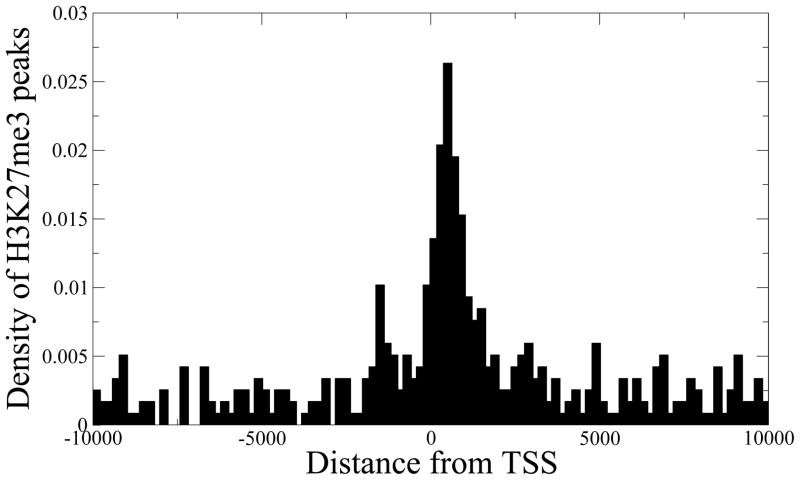
Figure 6. SES preserves subtle H3K27me3 peaks.
The majority of the H3K27me3 peaks detected by our method, but missed by PeakSeq, are found near transcription start sites (TSS) of known genes. The corresponding genes also show significantly lower expression levels compared to the genes that have H3K4me3 peaks (Wilcoxon test p-value = 1.3 × 10−25).
Our method of signal extraction scaling is motivated by the theory of order statistics. Consider the following observation for normally distributed random variables: let Y1, Y2, …,Yn be independent identically distributed normal random variables with mean μ and variance σ2, and let Y(1), Y(2), …, Y(n) be their order statistics, i.e. the rearrangements of Yi such that Y(1) ≤ Y(2) ≤ ··· ≤ Y(n). If we partition the reference genome into n equal-sized bins, then for sufficiently large μ, Poisson count data are approximately normal, and we can think of Yi as counting reads in the i-th bin. By binning in ~1–2 kb windows, we found that a typical ChIP-seq nowadays yields 10–20 reads per bin on average, sufficient for a normal approximation. For example, our H3K4me3 Input data has 46 million uniquely mapped reads, and ~70% of 1kb bins have more than 5 reads, with a mean count of 17.5 reads per 1kb bin. In reality, adjacent bins are correlated, and different regions in the genome may have different Poisson mean μ. However, our analysis below shows that ChIP-seq data can be reliably modeled as continuous mixtures of Poisson distributions, with extra zero counts in unmappable regions. In our experience with numerous ChIP-seq data, the discussion below also holds for such distributions. In particular, low-count regions will affect only the left tail of the ratio of partial sums of order statistics. Our scaling factor, however, is based on the asymptotic behavior of the ratio as ordered genomic bins are added and is obtained by computing the rank of order statistics where the peaks in IP channel begin to accumulate significantly greater number of reads than the Input channel. Our estimation is thus robust against extra zero counts. Although we cannot prove it analytically, we empirically observed that continuous mixtures of Poisson, such as ChIP-seq data, also have a linear regime for the ratio of partial sums of order statistics, to be described below.
Define the partial mean Sj/n as
Because the order statistics are ranked in an increasing order, it can be seen that the partial mean is an increasing function of j. In fact, in the limit of large sample size n, the partial mean is almost a linear function of j with a positive slope. More precisely, for large n,
where f is the probability density of Yi, β = j/n, and γβ is the β-th quantile of the standard normal distribution (Burrows, 1971). Expanding this asymptotic form around β = 1/2, one can show that the partial sum satisfies
which is almost linear in β and can be fitted with linear regression with R2 > 0.99.
Similarly, consider a set of bivariate normal random variables Zi = (Yi, Xi), where Yi are defined as above, and X1, X2, …, Xn are independent identically distributed normal random variables that are uncorrelated with Yi. We can think of Yi as binned immunoprecipitated DNA counts and Xi as binned Input DNA counts. Then, we define the i-th order statistic Z(i) to be the pair (Yt, Xt), such that Yt corresponds to the i-th order statistic Y(i); i.e., the order statistics are obtained by sorting Zt with respect to the first entry. Because we have assumed that X and Y are uncorrelated, is an unbiased estimate of the expectation E[X], and for sufficiently large j, the ratio of partial sums thus approaches , which is proportional to the partial mean . Consequently, for large n, the above analysis shows that Rj/n can be approximated by a linear function of β = j/n. Importantly, our simulation study shows that this linearity also holds for weakly correlated X and Y. The correlation between nonzero IP and Input bins is typically ~0.3, and our simulation of bivariate normal (X, Y) with 0.4 correlation showed an excellent linear fit that was virtually identical to the case of independent X and Y.
In a more realistic situation, the distribution of the IP channel data Yi can be modeled as a mixture of two Poisson distributions, e.g. one component representing the basal level of background noise and the second component representing the enrichment of actual immunoprecipitated DNA. For sufficiently large mean, Poisson distributions approach normal distributions, and the above analysis still holds; but, the ratio Rβ in this case begins to diverge from linearity at β roughly equal to the mixing probability. By computing this critical value of β = k/n as described above, we can thus approximate the proportion of background noise in the IP channel data Y1, Y2, …, Yn.
The statistical significance of a particular choice of cutoff k can be assessed by interpreting the corresponding percentages pk and qk as the probabilities of Bernoulli trials. For each k, we have a partition of the genome into two disjoint subsets: high count signal bins and low count background bins. We can test the null hypothesis that the IP and Input channels identically allocate their reads across these two subsets against the alternative hypothesis that the allocation of tags in the IP differs from that of Input. Under this interpretation, our method maximizes the difference between the IP and Input binary probability distributions and is therefore an instance of statistical inference in the sense of Shannon (Shannon, 1948, Jaynes, 1957). Distances between distributions in this context are typically measured in terms of a divergence metric such as the Jenson-Shannon divergence. In fact, the Jenson-Shannon divergence or any other distance metric can be used as an alternative to the above absolute value in determining the optimal cutoff k. The statistical significance of nonzero Jenson-Shannon divergence can be computed via a divergence test which tests the hypothesis that the divergence is zero to linear order (Trapnell et al., 2010). This methodology has been also applied in the popular software Cufflinks to detect statistically significant changes in the distributions of reads amongst isoforms in RNA-seq data (Trapnell et al., 2010).
3 Testing the Poisson Assumption
Many ChIP-seq peak callers are model based, assessing the statistical significance of enrichment in the IP channel at a given genomic locus by assuming a null distribution with mean estimated from Input. Since sequencing yields count data, it is natural to approximate the null distribution as Binomial, Poisson or a generalization of Poisson. In this section, we systematically analyze a large ensemble of ChIP-seq data to rigorously determine when and to what extent the Poisson distribution and its generalizations are appropriate. We begin by recapitulating the distributional assumptions used by a few of the current peak callers: PeakSeq, BayesPeak, MACS, and MOSAiCS. PeakSeq (Rozowsky et al., 2009) scores target sites relative to control under the null hypothesis of a binomial distribution of tags with a mean estimated from the number of tags in the Input sample at the same site. The binomial distribution can be approximated by the Poisson distribution in the usual asymptotic limit. As we will demonstrate below, the Poisson model is subject to false discoveries induced by overdispersion and zero-inflation.
MACS also uses a variable rate Poisson model, where the model mean is determined from Input by taking the maximum of average read counts computed on 1kb, 5kb, 10kb, and genome-wide intervals (Zhang et al., 2008). Although MACS does perform a library swap procedure, reversing the roles of IP and Input, in order to estimate false discovery rate (FDR), this is still problematic for two reasons. Firstly, while controlling for false discovery by thresholding peaks based on an estimated FDR does eliminate some false positives, it does not eliminate false negatives and the Poisson null hypothesis’ sensitivity to scaling induces false negatives, as evidenced by the SP110 false negative example in section 2 as well as the mathematical analysis presented below. Secondly, during the library swap procedure, IP and Input are reversed and control peaks are called under the Poisson null hypothesis; as a consequence, the same scaling issues which induce false discoveries will persist in the library swap in an unknown way. BayesPeak (Spyrou et al., 2009) uses a negative binomial regression model, formulated as a Poisson-Gamma mixture, with parameters estimated from the Input channel via Monte Carlo Markov chain methods. Although a negative binomial model will in principle be more flexible with regards to overdispersion, it may not accommodate the zero-inflation we demonstrate to be present in the data. Moreover, the BayesPeak model parameters are estimated as a response to the raw Input and IP tag counts alone and do not compensate for sequence dependent biases.
Another software which uses a negative binomial regression model and which does attempt to compensate for sequence properties is MOSAiCS (Kuan et al., 2009). This approach is perhaps the closest to our approach, with some key differences. Here a negative binomial mixture model of the IP is regressed on GC content, mapability, and a monomial in Input tag count. The model mean is defined piecewise. For bins with tag count less than a tuning parameter s, the mean μ is modeled as a function of mapability, a univariate spline of GC content, and Xd where X is the Input tag count and d is a heuristically chosen parameter. For bins of size larger than s, the Input tag count monomial Xd alone is used. This formulation is problematic for several reasons. Firstly, the parameters s and d are tuning parameters that have to be arbitrarily chosen by the user, and no method is given for choosing them in general. They were chosen by trial and error in their test case, and it is unclear how different choices will affect enrichment estimates for arbitrary datasets. Secondly, replacing GC content values with a univariate spline results in loss of information, since the graph of μ as a function of the three covariates of GC, mapability, and Xd is a subset of ℝ4, and the univariate spline construction amounts to projecting this graph into the μ-GC plane. An alternative formulation of this approach, which was perhaps undertaken to reduce computational complexity, would be predicated on a multivariate spline of all predictors (Chui, 1987). We have indeed found regression modeling on typical ChIP-seq datasets to be computationally cumbersome on single to quad-core processors and have opted for implementations utilizing GPU computing (Sarkar et al., 2010). Lastly, like BayesPeak, it is unclear whether MOSAiCS will accommodate zero-inflation in two-sample analysis. However, as we now show there is roughly a 1 in 4 chance that a randomly chosen bin in a typical ChIP-seq data set contains a zero count. We now examine how and why real ChIP-seq data exhibit several features which violate the Poisson assumption.
Consider the Poisson null model , with mean μ estimated from the Input channel. This model’s variance σ2 = μ is often unable to accommodate the drastic oscillations in read density observed in ChIP-seq data. The p-value associated with observing y or more tags in the IP channel with respect to the Poisson model is . If we scale the IP and Input channel simultaneously by a factor of t, then the associated p-value is . This formula is dominated by the behavior of the exponential term, and as a result, the p-value is a decreasing function of t. This type of scaling occurs implicitly when normalizing by SDS and also in biased loci that artificially accumulate a large number of reads. To illustrate this phenomenon, we called peaks on the H3K4me3 ChIP-seq data in NSC using MACS (Zhang et al., 2008). MACS uses a variable rate Poisson model where the model mean is determined from Input by taking the maximum of average read counts computed on 1kb, 5kb, 10kb, and genome-wide intervals (Zhang et al., 2008). We first called H3K4me3 peaks on chromosome 1 by using default parameters. We then scaled the IP and Input alignment densities by factors of 10 and 100, and generated alignments by re-sampling from the scaled distribution. The number of called peaks at a fixed p-value cutoff of 10−5 increased approximately 500% after scaling by a factor of 10 and almost 4450% after a scaling of 100. These scaling factors are obviously higher than would be used in practice, but nonetheless demonstrate how scaling can artificially induce false positives. Our simulation study shows that the FDR computed by MACS 1.4.1 is also affected by the IP sequencing depth, as would be expected for most algorithms. For example, upon randomly removing 25% of reads from H3K4me3 IP data, 2.8% of the peaks that were originally called at a 5% FDR cutoff in the full dataset had FDR greater than 5% in the trimmed dataset. When 50% of reads were removed from IP, 4.2% of the peaks that were originally called at a 5% FDR cutoff in the full dataset had FDR greater than 5% in the trimmed dataset. Similarly, scaling up both IP and Input by a factor of 5 to 10-fold increased the number of peaks called by MACS 1.4.1 at 1% FDR by roughly 20%.
Furthermore, ChIP-seq data are often zero-inflated and over-dispersed. That is, the number of zero count bins is typically in excess of what is expected under a Poisson model, and the variance σ2 in count far exceeds the mean count μ. These phenomena can introduce a significant bias in Poisson models, generating both false positives and false negatives in peak detection. To study these artifacts, we examined 62 ChIP-seq Input data sets from the UCSC ENCODE Yale transcription factor binding sites repository (Kent et al., 2002, Birney and al., 2007) consisting of two replicates from each of 31 cell lines. We binned uniquely mapped alignments into non-overlaping 1kb windows and examined the distribution of counts. The ENCODE data demonstrate typical characteristics that deviate from Poisson modeling assumptions. Table 1 summarizes the distribution of counts in the ENCODE data. Approximately 23% of the windows have a zero count, suggesting potential zero inflation. Regression models have been previously used to study over-dispersion and bias in RNA-seq analysis (Hansen et al., 2010, Anders and Huber, 2010, Robinson and Smyth, 2007, Li et al., 2010). We apply a similar methodology here. In order to rigorously study zero inflation and over-dispersion in ChIP-seq data, we fitted each of the ENCODE data sets with regression models representing a progressive relaxation of the Poisson assumptions. Since ChIP-seq read density has been shown to be biased with respect to GC content and mappability (Aird et al., 2011, Li et al., 2010), we chose GC content and mappability as predictors in four regression models: 1. a variable rate Poisson model (P), 2. a variable rate negative binomial model (NB), 3. a zero-inflated Poisson model (ZIP), and 4. a zero-inflated negative binomial model (ZINB). While the Poisson models assume that the model mean μ is equal to the variance σ2, the negative binomial models relax the condition with a quadratic model of variance σ2 = μ + φμ2, where φ is a constant dispersion parameter estimated from the data (Hilbe, 2011). Poisson models are special cases of NB models with φ = 0. We will use this property to assess the validity of the Poisson variance assumption in ENCODE data by testing the significance of nonzero φ. The model definitions for P and NB are:
| (1) |
| (2) |
Table 1.
The distribution of the alignment count per 1kb window for a pool of 62 ChIP-seq Input channel datasets.
| Distribution of read counts c.
| |
|---|---|
| Range | Percentage of 1 kb bins |
| c = 0 | 22.905% |
| 1 ≤ c < 5 | 55.225% |
| 5 ≤ c < 10 | 17.922% |
| 10 ≤ c < 25 | 3.8136% |
| 25 ≤ c < 50 | 0.1126% |
| 50 ≤ c < 100 | 0.0142% |
| 100 ≤ c < 1000 | 0.0067% |
| c ≥ 1000 | 0.0006% |
The NB model can be viewed as a Poisson-Gamma mixture (Hilbe, 2011), and P can be thus viewed as a restricted NB model. ZIP and ZINB generalize P and NB respectively to mixture models with two components. The first component is one of the above probability masses and the second component, in both cases, is a point mass concentrated at zero with probability δ. δ is modeled as a logistic function of a given set of regressors. In all four models the mean μ is taken to be log linear in the regressors. Model parameters were estimated via maximum likelihood estimation. The details of the model, how the regressors were aggregated, and the methods used to estimate the model parameters are described in detail in the methods section. Tables 2 and 3 describe the results of this analysis.
Table 2.
Two estimates of dispersion and two statistical tests were used to assess dispersion in chip-seq data. All datasets examined demonstrate considerable over-dispersion.
| Dispersion test statistics.
| ||||
|---|---|---|---|---|
| Cell line | NB φ | ZINB φ | Dean’s score (p-value) | Likelihood ratio (p-value) |
| Gm10847InputIggrab | 2.1126 | 2.7791 | 39491 (≤ 2.2e-16) | 2890196 (≤2.2e-16) |
| Gm10847MusiggMusigg | 2.14083 | 2.8056 | 33662 (≤2.2e-16) | 2806455 (≤2.2e-16) |
| Gm12878InputIggrab | 1.89733 | 3.3883 | 63821 (≤2.2e-16) | 7362938 (≤2.2e-16) |
| Gm12878MusiggMusigg | 2.26196 | 2.8738 | 34888 (≤2.2e-16) | 1987654 (≤2.2e-16) |
| Gm12891InputIggrab | 1.061948 | 1.1758 | 537964 (≤2.2e-16) | 9907326 (≤2.2e-16) |
| Gm12891MusiggMusigg | 2.0107 | 2.5456 | 34253 (≤2.2e-16) | 2809971 (≤2.2e-16) |
| Gm12892InputIggrab | 1.48523 | 1.6718 | 65456 (≤2.2e-16) | 3952893 (≤2.2e-16) |
| Gm12892MusiggMusigg | 1.19205 | 1.2857 | 37652 (≤2.2e-16) | 4650547 (≤2.2e-16) |
| Gm15510InputIggrab | 2.11635 | 2.7549 | 36472 (≤2.2e-16) | 2722262 (≤2.2e-16) |
| Gm15510MusiggMusigg | 1.46499 | 1.817 | 52124 (≤2.2e-16) | 6385709 (≤2.2e-16) |
| Gm18505InputIggrab | 2.11635 | 2.7549 | 36472 (≤2.2e-16) | 27222261 (≤2.2e-16) |
| Gm18505MusiggMusigg | 1.75816 | 1.9236 | 942055 (≤2.2e-16) | 3943887 (≤2.2e-16) |
| Gm18526InputIggrab | 2.37477 | 3.1339 | 29622 (≤2.2e-16) | 2167641 (≤2.2e-16) |
| Gm18526MusiggMusigg | 1.56948 | 1.7764 | 38400 (≤2.2e-16) | 3821897 (≤2.2e-16) |
| Gm19099InputIggrab | 2.50625 | 3.683 | 21514 (≤2.2e-16) | 2163575 (≤2.2e-16) |
| Gm19099MusiggMusigg | 1.63685 | 2.2079 | 48046 (≤2.2e-16) | 5631237 (≤2.2e-16) |
| Gm19193InputIggrab | 1.91547 | 2.4246 | 39662 (≤2.2e-16) | 2962330 (≤2.2e-16) |
| Gm19193MusiggMusigg | 1.93517 | 2.6687 | 20847 (≤2.2e-16) | 3184818 (≤2.2e-16) |
| Helas3LargeFragment | 1.70227 | 2.6181 | 33941 (≤2.2e-16) | 5147206 (≤2.2e-16) |
| Helas3MnaseV2 | 1.33332 | 1.5637 | 59031 (≤2.2e-16) | 8096157 (≤2.2e-16) |
| Helas3MouseiggV2 | 1.72362 | 1.5637 | 32414 (≤2.2e-16) | 8342183 (≤2.2e-16) |
| Helas3Nakeddna | 1.91147 | 3.0919 | 31069 (≤2.2e-16) | 5932107 (≤2.2e-16) |
| Hepg2ControlForskln | 1.15222 | 1.2729 | 120398 (≤2.2e-16) | 5122610 (≤2.2e-16) |
| Hepg2ControlInskln | .632829 | .6485 | 38852 (≤2.2e-16) | 6410233 (≤2.2e-16) |
| Hepg2InputV2 | 1.05644 | 1.1843 | 12938 (≤2.2e-16) | 1727295 (≤2.2e-16) |
| HuvecInputStd | 1.75729 | 2.9294 | 161597 (≤2.2e-16) | 15952170 (≤2.2e-16) |
| K562InputV3 | 1.35451 | 1.8738 | 516901 (≤2.2e-16) | 19144353 (≤2.2e-16) |
| K562MusiggMusigg | 1.48218 | 2.2728 | 54932 (≤2.2e-16) | 12902485 (≤2.2e-16) |
| Mcf7Input | 1.32286 | 1.5971 | 63674 (≤2.2e-16) | 12890295 (≤2.2e-16) |
| Nt2d1InputV2 | 2.2687 | 3.7535 | 55442 (≤2.2e-16) | 4433772 (≤2.2e-16) |
Table 3.
A comparison of the expected and observed percentage of zeros in several regression models of the ENCODE dataset.
| Observed vs. expected percentage of zero counts | |||||
|---|---|---|---|---|---|
| Cell line | Observed | P | NB | ZIP | ZINB |
| Gm10847InputIggrab | 22.07% | 9.85% | 20.89% | 23.513% | 23.239% |
| Gm10847MusiggMusigg | 22.013% | 9.94% | 20.868% | 23.361% | 23.22% |
| Gm12878InputIggrab | 14.21% | 1.08% | 9.841% | 15.709% | 15.659% |
| Gm12878MusiggMusigg | 26.532% | 15.703% | 25.846% | 27.858% | 27.64% |
| Gm12891InputIggrab | 28.477% | 9.49% | 28.89% | 29.57% | 30.463% |
| Gm12891MusiggMusigg | 23.583% | 11.168% | 22.71% | 25.336% | 25.536% |
| Gm12892InputIggrab | 27.801% | 13.15% | 27.818% | 29.03% | 28.909% |
| Gm12892MusiggMusigg | 30.705% | 13.986% | 31.292% | 31.8% | 29.211% |
| Gm15510InputIggrab | 22.977% | 10.883% | 21.917% | 24.467% | 24.256% |
| Gm15510MusiggMusigg | 20.134% | 4.635% | 19.06% | 21.562% | 21.43% |
| Gm18505InputIggrab | 21.63% | 9.213% | 20.438% | 22.899% | 22.775% |
| Gm18505MusiggMusigg | 27.304% | 15.044% | 27.647% | 28.588% | 28.727% |
| Gm18526InputIggrab | 22.975% | 11.938% | 21.904% | 24.475% | 24.282% |
| Gm18526MusiggMusigg | 25.478% | 11.245% | 25.425% | 26.829% | 26.825% |
| Gm18951InputIggrab | 24.975% | 12.554% | 24.188% | 26.483% | 26.113% |
| Gm19099InputIggrab | 20.983% | 9.691% | 19.188% | 22.744% | 22.456% |
| Gm19099MusiggMusigg | 19.31% | 4.581% | 17.648% | 20.762% | 20.718% |
| Gm19193InputIggrab | 24.35% | 11.492 | 23.484% | 25.86% | 25.521% |
| Gm19193MusiggMusigg | 21.558% | 8.187% | 20.037% | 23.115% | 22.884% |
| Helas3LargeFragment | 19.1% | 4.153% | 16.580% | 21.492% | 21.274% |
| Helas3MnaseV2 | 18.885% | 3.37% | 18.504% | 21.282% | 21.854% |
| Helas3MouseiggV2 | 14.241% | .956% | 10.421% | 16.322% | 16.158% |
| Helas3Nakeddna | 15.007% | 1.895% | 11.678% | 17.094% | 17.125% |
| Hepg2ControlForskln | 32.54% | 15.266% | 32.787% | 33.927% | 34.918% |
| Hepg2ControlInskln | 45.481% | 22.747% | 46.602% | 46.522% | 48.245% |
| Hepg2InputV2 | 48.906% | 36.024% | 48.949% | 50.9% | 50.611% |
| HuvecInputStd | 10.696% | .091% | 5.938% | 12.227% | 12.283% |
| K562InputV3 | 13.664% | .429% | 11.19% | 14.233% | 14.686% |
| K562MusiggMusigg | 13.815% | .456% | 10.269% | 15.126% | 15.114% |
| Mcf7Input | 15.713% | 1.253% | 14.447% | 16.193% | 16.542% |
| Nt2d1InputV2 | 14.945% | 2.775% | 11.624% | 17.56% | 17.546% |
The dispersion parameter φ, as well as all other parameters, are estimated by maximum likelihood. Any nonzero value of φ indicates a departure from the Poisson assumption, which was clearly the case here. Dean’s test and a likelihood ratio test were used to assess the NB model relative to the Poisson model. The likelihood test compares the ratio of model posterior probabilities. Dean’s test tests the sensitivity of the NB model with respect to the dispersion parameter φ (Dean, 1992). Small p-values indicate over-dispersion with respect to the Poisson model and imply the need for a more flexible model of variance. As zero-inflation often masquerades as over-dispersion, we also tested the observed number of zeros against the number of zeros expected under each model. Table 3 compares the observed number of zeros to the number of zeros expected under each of the four regression models. In every dataset, the Poisson model underestimates the number of zeros. Both the ordinary negative binomial and the zero-inflated models capture the number of zeros more accurately, with a modest improvement realized in the zero-inflated models. This analysis implies that the Poisson model is inadequate to capture the biases present in ChIP-seq data.
Our study also revealed that the pattern of variance in count appeared to be consistent between technical replicates, but differed across different cell lines. This was evidenced by an inter-replicate spectral analysis of Input data, presented in the next section, which showed a strong correlation between the frequency responses of technical replicates at all frequencies (Pearson correlation coefficients 0.7–0.99). In contrast, an inter-cell line spectral analysis of Input data (data not shown) demonstrated a wide range of correlations (Pearson correlation coefficients 0.08–0.99) at different frequencies. This observation suggests that a model of cell line-specific biases in tag density can be constructed. In the next section, we describe how spectral analysis can extract reproducible trends in tag density. We will then improve our regression models by accounting for such cell line-specific biases.
4 Model-based Bias Correction
4.1 Spectral analysis
In this section we employ a technique from signal processing known as spectral analysis. This method decomposes a function into components with given characteristic length scales known as the function’s spectrum. Wavelets provide one of the most powerful tools for spectral analysis (Daubechies, 1992). Wavelets allow us to decompose the alignment density into its component peaks of various amplitudes and fluctuation scales. We can then determine what percentage of the enrichment profile is composed of peaks of a given size, and we can correlate this information across datasets to formulate models of cell line-specific bias. Toward this end, we decomposed the Input alignment densities from ENCODE Yale TF ChIP-seq by using Coiflet wavelets (Daubechies, 1992) of order 1. We used a 15 level decomposition. Level 1 captures changes in alignment density occurring over length scales of 1.25 kb. Each subsequent level increases this scale by a factor of 2. Thus, a level 15 decomposition will allow us to classify changes in alignment density ranging over a spectrum of 1.25 kb to 40.96 Mb. We refer the reader to the appendix for a brief overview of wavelets and additional details of our decomposition methods.
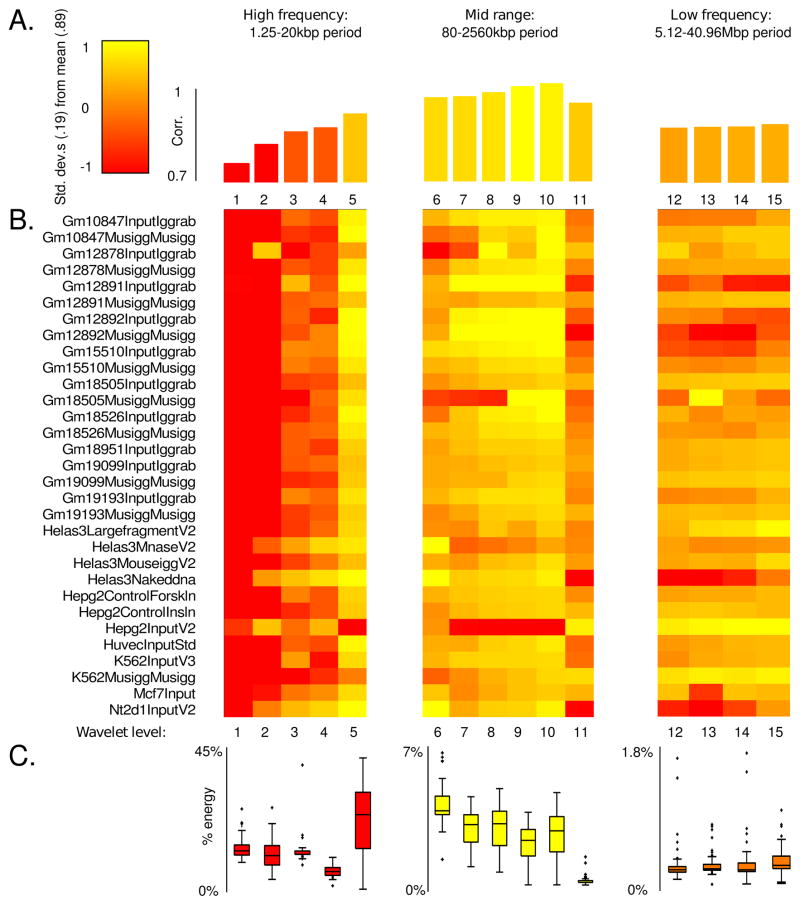
We analyzed the Pearson correlation between the wavelet coefficients of decomposed replicate data. We compared these correlations to the distribution of spectral energy. Formally the spectral energy is the sum of the squares (Euclidean length squared or L2-norm squared) of the wavelet coefficients at a particular level. Consequently, the spectral energy gives a measure of the magnitude of the component of the alignment density oscillating with a given period. The purpose of this analysis is to determine the frequencies which simultaneously capture the predominant trend in an individual dataset (high spectral energy) and are reproducible across datasets (high correlation). Figure 7 A summarizes the correlation between technical replicates as a function of characteristic length scale described by level. Each correlation value is the average over all datasets, at that level, and Figure 7 B is a heat map showing the individual correlations between replicates by level. The correlation between replicates is generally strong with a mean of 0.89. Replicate Input data demonstrate a stratified correlation profile with weak correlation at high frequency (corresponding to changes in enrichment occuring over a 1.25–10 kb interval), and increased correlation in mid to low frequencies (length scales of 20 kb to 40.96 Mb). In contrast, Figure 7 C shows that spectral energy is almost completely concentrated in high frequencies, consistent with Input data being dominated by small scale noise. Interestingly, the frequency band corresponding to a 20 kb period is simultaneously high energy and highly correlated. We interpret this band as containing cell line-specific sonication bias. ChIP-seq Input datasets derived from the same cell line thus exhibit scale-dependent correlation in both spacial and frequency domains. This analysis suggests that wavelet decompositions can be used to formulate models of cell line-specific biases by designing filters targeting frequencies correlated amongst Input datasets from the same cell type. We describe this approach in the next subsection.
Figure 7. Frequency correlation and spectral energy for ChIP-seq replicates.
Level 15 Coiflet wavelet decompositions were performed on the alignment densities for each replicate experiment pair in the ENCODE Yale TF Input dataset. At each level we computed the Pearson correlations of the detail coefficients, as well as their spectral energy. (A) The average correlation over all datasets, at a given wavelet level. Over all datasets, over all levels, the mean correlation was 0.89 with a standard deviation of 0.19. (B) Correlation between replicates at a given wavelet decomposition level. (C) Percentage energy allocated to a given level, averaged over all experiments. Each box summarizes the distribution of spectral energies across datasets, at a given level.
4.2 Bias correction
We will use the H3K4me3 ChIP-seq dataset in NSC to illustrate the use of nonlinear regression to generate null models of alignment density (reads/kb). We combine the above spectral analysis with insights gained from studying over-dispersion and zero-inflation to formulate the following zero-inflated negative binomial regression model of the SES normalized Input channel, designed to account for the functional dependence of read density on GC content, mappability, and sonication bias. This model is a mixture of a point mass concentrated at zero with probability δ and a negative binomial probability function, NB(y|μ, φ), where μ is the mean and φ a genome-wide estimate of dispersion. The variance of the NB component is given by μ + φμ2. Zero counts can be produced by either the point mass or by the negative binomial component. Under this mixture model, the probability of observing a zero count is P(k = 0) = δ + (1 − δ)NB(0|μ, φ), while the probability of a nonzero count y is P(k = y) = (1 − δ)NB(y|μ, φ). The model mean μ is chosen to be log linear in the regressors. The point mass probability δ is modeled as logistic in the regressors. We fit the model using the SES normalized Input channel alignment density as a response to mappability, GC content, and a truncated wavelet expansion of Input data as regressors using maximum likelihood estimation. The details of the model, how the regressors were aggregated, and how the wavelet expansion was constructed are found in the appendix.
As a point of comparison we use a MACS style variable rate Poisson model, with SDS and model mean μ = max(μgw, μ1kbp, μ5kbp, μ10kbp), where μgw is the genome-wide average tag count in the SDS normalized Input, and μ*kbp is an average Input tag count over a given interval width. For the remainder of this subsection we will simply refer to this as the Poisson model.
To compare the ZINB and Poisson models of the Input channel, we first assessed model fit. As in the ENCODE dataset, the ZINB regression model shows significant improvement both in its ability to handle over-dispersion and zero inflation when compared to the Poisson model. Since the MACS Poisson model is not obtained via maximization of a likelihood function and since it is not a special case of the ZINB, as is the case for P vs. NB, we cannot use Dean’s test or the likelihood ratio test to formulate a comparison. However, as in (Cameron, C. A., Tricedi, 1998), we can estimate dispersion by an ordinary least squares fit α of , where the regression coefficient α gives a type of coefficient of variation and estimates genome-wide dispersion, μi the Poisson model mean estimates for the i-th bin computed as above, and yi the Input alignment density counts. 0 ≤ α ≤ 1 indicates mild over-dispersion and anything over 1 indicates significant over-dispersion. For the H3K4me3 dataset, this dispersion estimate is on the order of 104, which is significant. Even after removing possible outliers of yi > 1000, the dispersion estimate drops to 19.73, but is still highly over-dispersed. Due to the nature of the Poisson model, μ is never less than the global average (≈ 10); and, as a consequence, the model vastly underestimates the number of zero counts. In fact, the expected number of zeros under the Poisson model is < 10−7%. The observed number of zeros is actually 10.52%, much closer to the ZINB prediction of 11.62%.
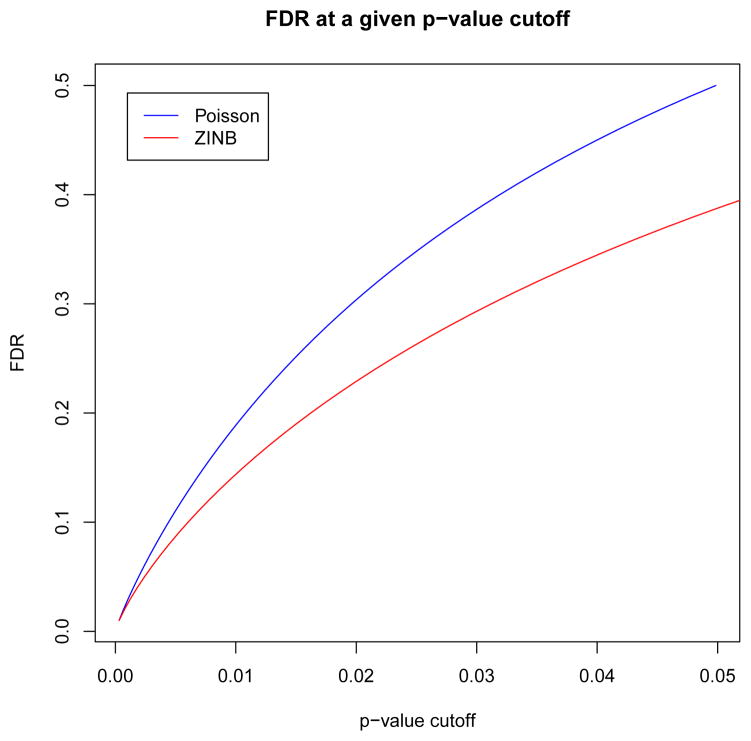
In addition to accessing model fit, we also compared false discovery rates (FDRs) between the ZINB and Poisson models when used to assess H3K4me3 enrichment in NSC. We computed the statistical significance of the observed IP alignment density with respect to both null models for each nucleotide on chromosome 1. The observed count was given by the IP alignment density based on a 1 kb window centered at each nucleotide. Likewise the GC content, mappability, and sonication (wavelet approximation) predictor values were averaged in the same window when computing the probabilities with respect to the ZINB model. This generated approximately 1.9 × 108 hypothesis tests for which each model assigned a p-value. The probability density governing the p-values p of large ensembles of multiple hypothesis tests has been well studied in the context of microarray analysis (Dudoit et al., 2003) and is typically modeled as a two component mixture density f(p) = π0fn(p) + π1fa(p), such that π0 + π1 = 1, with fn(p) = 1 and fa(p) governing the null and alternate hypotheses, respectively (Storey, 2003). By Bayes theorem the probability of a true null hypothesis (non-enriched locus) given a positive test (observed p-value less than a given cutoff p*) is , where F(p) is the cumulative density function of f(p). Following (Pounds and Morris, 2003), we estimate the density under the alternate hypothesis of H3K4me3 enrichment as a beta distribution apa−1 on observed p-values p ∈ [0,1]. The parameter a ∈ [0,1], as well as the proportion π0 of true null hypotheses, were computed via maximum likelihood estimation using the BUM class of the ClassComparison library in the OOMPA R language library suite (oompa, 2010). FDRs were also computed via the same package which estimates false discovery rate at a given p-value cutoff p* as , where π̂0 is an upper bound on π0 obtained via a confidence interval (Pounds and Morris, 2003, Casella and Berger, 1990). Figure 8 shows that the ZINB model demonstrates a lower FDR at a given p-value cutoff than the Poisson model.
Figure 8. FDRs for the Poisson and ZINB models.
FDR as a function of p-value cutoff was estimated from the distribution of p-values produced by both the Poisson and ZINB models using a beta-uniform mixture model. The ZINB model exhibits a lower FDR than the Poisson model at a given p-value cutoff.
4.3 Comparison with other methods
To further validate our approach, we analyzed the ENCODE c-Myc and RNA polymerase II (Pol II) ChIP-seq data in the leukemia cell line K562. A recent study shows that c-Myc plays a critical role in releasing Pol II from promoter-proximal pausing and that the majority of c-Myc-bound genes undergo active transcriptional elongation (Rahl et al., 2010). We thus expect functional c-Myc ChIP-seq peaks to co-localize with Pol II peaks. We called peaks using the default parameters at 1% FDR using MACS, CCAT, and PeakSeq and compared the results to our ZINB model with SES at 1% FDR (Storey, 2003). The results of this comparison are shown in Table 4.
Table 4.
A comparison of Pol II and c-Myc peaks called by MACS, CCAT, Peak-Seq, and the ZINB model. A. Number of peaks called by each method and the percentage of c-Myc peaks overlapping with Pol II. B. The percentages of peaks called by other methods overlapping with ZINB peaks. C. The percentage of ZINB peaks that overlap with peaks detected by other methods.
| A. Number of peaks | ||||
|---|---|---|---|---|
| MACS | CCAT | PeakSeq | ZINB | |
| c-Myc | 25676 | 13022 | 15749 | 13550 |
| Pol II | 18688 | 41979 | 21631 | 33138 |
| Pol II-bound c-Myc | 50% | 80% | 65% | 77% |
| B. Peaks from other methods found by ZINB | ||||
| MACS | CCAT | PeakSeq | ||
| c-Myc | 45% | 76% | 68% | |
| Pol II | 99% | 92% | 98% | |
| Pol II-bound c-Myc | 67% | 81% | 78% | |
| C. Peaks from ZINB found by other methods | ||||
| MACS | CCAT | PeakSeq | ||
| c-Myc | 82% | 67% | 73% | |
| Pol II | 48% | 68% | 47% | |
| Pol II-bound c-Myc | 79% | 75% | 72% | |
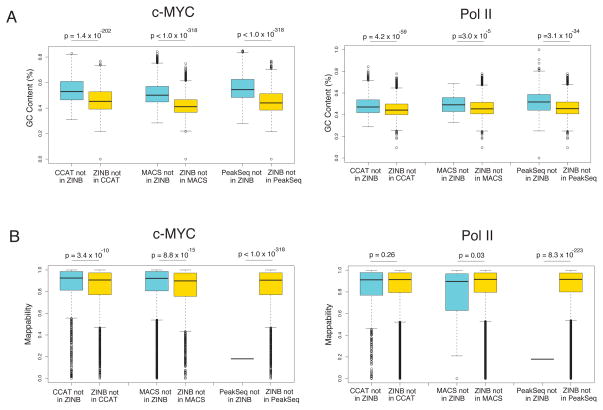
We found considerable overlap with other methods for Pol II-bound c-Myc peaks. Note that our method and CCAT which both employ a normalization scheme based on low order bins produce the greatest enrichment of Pol II for c-Myc peaks of 77% and 80%, respectively. We also tried MOSAiCS (Kuan et al., 2009), but the software could not complete analysis on chromsomes 20, 21, 22, X and Y for c-Myc and on chromosomes X and Y for Pol II. At 1% FDR, MOSAiCS found only 1963 c-Myc peaks on chromsomes 1–19 and 11505 Pol II peaks on chromosomes 1–22, and 99% of the MOSAiCS peaks were found by our method. This analysis suggests that MOSAiCS may have many false negatives, at least for the dataset that we have analyzed. In addition, we analyzed the mappability and GC content of the peaks that did not overlap between our method and others. We found that peaks unique to ZINB were significantly lower in GC content than those unique to all other methods for both Pol II and c-Myc, as shown in Figure 9. Mappability was also lower for our method’s unique peaks with respect to MACS and CCAT on the c-Myc dataset and was comparable on the Pol II dataset. The peaks called only by PeakSeq had an extremely low mappability, consistent with the fact that the algorithm also adjusts for mappability. We ascribe this difference in GC content and mappability in differential peaks to the adjustment made by our regression model to compensate for GC content and mappability bias. For example, read count is correlated with both GC content and mappability (Aird et al., 2011, Li et al., 2010), so that regions of higher GC content and mappability tend to have higher read counts in both IP and input channels. Consequently, the fact that peaks unique to other methods have higher GC content is consistent with their use of a Poisson (MACS) or binomial (CCAT/PeakSeq) null model, since we have shown that a simultaneous scaling of the IP and Input read densities causes the Poisson p-values to drop artificially. Conversely, our method seems to be more sensitive in detecting peaks where lower GC content and poor mappability may allow only modest IP enrichment.
Figure 9. Comparison of the distributions of (A) GC content and (B) mappability in peaks unique to individual algorithm.
Peaks unique to our method generally show lower GC content and mappability. The statistical significance of the difference in distribution was assessed by the Wilcoxan rank sum test.
5 Conclusions
The proposed signal extraction scaling provides an effective approach to normalizing paired sequencing data in background genomic regions that give rise to non-specific DNA not directly targeted by antibodies. The statistical significance of the enrichment of IP over Input post normalization can be then assessed, relative to a null hypothesis of no enrichment, via a divergence test. A high divergence test p-value can rapidly identify a failed ChIP-seq experiment, and the proposed approach thus provides a powerful quality control method. It should be noted that the resulting scaling factor is in practice quite aggressive and should be implemented in the context of additional bias corrections and a null distribution with a flexible variance model, as described here. The Poisson distribution, for example, is highly sensitive to scaling due to the fact that its variance is equal to its mean (as opposed to a negative binomial model where the variance is quadratic in the mean). Consequently, SES must be complemented with an accurate estimation of the Poisson rate in order to control for false discovery. We have shown that regression models provide a framework for modeling the biases inherent to ChIP-seq data. As more control data become available, the accuracy of the regression will greatly increase in the future.
Acknowledgments
We would like to thank Henrik Bengtsson, Adam Olshen, Ritu Roy, Taku Tokuyasu, Mark Segal, Saunak Sen, Barry Taylor, Yuanyuan Xiao, and Hao Xiong for helpful discussions. This project was in part supported by a Sontag Foundation award and NIH DP2OD006505 to DAL and by grants from the PhRMA Foundation, UCSF RAP, UCSF Academic Senate, the Sontag Foundation, and the National Cancer Institute (R01CA163336) to JSS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health
6 List of abbreviations
- NGS
Next-generation DNA Sequencing
- TF
transcription factor
- ChIP-seq
chromatin immunoprecipitated DNA sequencing
- MeDIP-seq
Methylated-DNA immunoprecipitation sequencing
- SDS
sequencing depth scaling
- SES
signal extraction scaling
- NCS
neural stem cells
- P
Poisson
- NB
negative binomial
- ZIP
zero-inflated Poisson
- ZINB
zero-inflated negative binomial
- FDR
false discovery rate
7 Appendix
7.1 Wavelet decomposition
An excellent introduction to wavelets is (Daubechies, 1992). In section 4.1, the raw alignment counts were binned into 1kb non-overlapping windows. We then performed a level 15 Coiflet-1 wavelet decomposition by using the MATLAB command wdencmp. The first 4 components corresponding to high-frequency noise (< 20 kbp) were removed in the regression models.
7.2 Regression models
In this paper we consider 4 regression models: 1. a variable rate Poisson model (P), 2. a variable rate negative binomial model (NB), 3. a zero-inflated Poisson model (ZIP), and 4. a zero-inflated negative binomial model (ZINB). While the Poisson models assume that the model mean μ is equal to the variance σ2, the negative binomial models relax the condition with a quadratic model of variance σ2 = μ + φμ2, where φ is a constant dispersion parameter estimated from the data (Hilbe, 2011). The model definitions for P and NB are:
| (3) |
| (4) |
ZIP and ZINB generalize P and NB respectively to mixture models with two components. The first component is one of the above probability masses and the second component, in both cases, is a point mass concentrated at zero with probability δ. Given x ∈ ℝn×r, a model matrix of n observations of the tag count at each of the r chosen regressors we model δ as a logistic function . The model mean μ is chosen to be log linear in the regressors, i.e., log (μ) = xβ. β, γ ∈ ℝr×1 and φ ∈ ℝ are constants determined via maximum likelihood estimation (MLE) as implemented in the the R language packages glm (Davies, 1992), used to fit the Poisson, and pscl (Jackman, 2010), used to fit NB, ZINB, and ZIP models.
We fit the models using the Input channel alignment count as a response to mappability and GC content in our analysis of the Poisson assumption in section 3. We added a truncated wavelet expansion of Input data as a regressor in section 4.2. We choose not to add this regressor in section 3 since our purpose was to study zero-inflation and over-dispersion in the raw data and provide motivation for adding the wavelet regressor as a form of bias correction in section 4.2. The mappability was obtained from the UCSC 36-mer CRG Alignability track (Kent et al., 2002) which measures how uniquely 36-mer sequences align. The GC content regressor was determined as the percentage of G and C bases in 5-base windows. The count, mapability, and GC content were measured in non-overlapping 1kb windows. The wavelet regressor was determined in several steps as follows. As in our spectral analysis, genome-wide alignment counts in the Input channel were binned into 1kb non-overlapping windows, a first order Coiflet wavelet decomposition of the alignment density was performed using MATLAB and components with characteristic length scales less than 20 kbp were then dropped. This has the effect of removing the component of the alignment density profile corresponding to high frequency noise and leaving only the component corresponding to bias that is highly correlated across replicate data, as shown in section 4.1. The log fold-change of the wavelet regression with respect to the genome-wide mean tag count in the Input channel was then used as the regressor. The log scale was chosen since the model mean is log linear in the regressors. Consequently, in the absence of GC content and mappability bias, the model mean will be proportional to the wavelet approximation.
Contributor Information
Aaron Diaz, University of California, San Francisco.
Kiyoub Park, University of California, San Francisco.
Daniel A. Lim, University of California, San Francisco
Jun S. Song, University of California, San Francisco
References
- Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, Gnirke A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biology. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows PM. Expected selection differentials for directional selection. Biometrics. 1971;28:2091–2110. [PubMed] [Google Scholar]
- Cameron CA, Tricedi PK. Regression Analysis for Count Data. Cambridge; 1998. [Google Scholar]
- Casella G, Berger R. Statistical Inference. Pacific Grove, CA: Wadsworth and Brooks/Cole; 1990. [Google Scholar]
- Chui CK. Multivariate Splines. SIAM; 1987. [Google Scholar]
- Daubechies I. Ten Lectures on Wavelets. Philadelphia, PA: Society for Industrial and Applied Mathematics; 1992. [Google Scholar]
- Davies S. Fitting generalized linear models. 1992 URL http://stat.ethz.ch/R-manual/R-devel/library/stats/html/glm.html.
- Dean CB. Testing for Overdispersion in Poisson and Binomial Regression Models. Journal of the American Statistical Association. 1992;87:451. [Google Scholar]
- Dudoit S, Schaffer J, Boldrick J. Multiple hypothesis testing in microarray experiments. Statistical Science. 2003;18:71–103. [Google Scholar]
- Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Research. 2010;38:1–7. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbe J. Negative Binomial Regression. Cambridge, UK: Cambridge University Press; 2011. [Google Scholar]
- Jackman AS. Pscl:political science computational laboratory. 2010 URL http://cran.r-project.org/web/packages/pscl/index.html.
- Jaynes ET. Information theory and statistical mechanics. Phys Rev. 1957;106:620–630. [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The Human Genome Browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nature Biotechnology. 2008;26:1351–9. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan PF, Chung D, Pan G, Thomson JA, Stewart R, Keles S. A Statistical Framework for the Analysis of ChIP-Seq Data. Technical Report. 2009 doi: 10.1198/jasa.2011.ap09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jiang H, Wong WH. Modeling non-uniformity in short-read rates in RNA-Seq data. Genome Biology. 2010;11:R50. doi: 10.1186/gb-2010-11-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- oompa. Object-oriented microarray and proteomic analysis. 2010 URL http://bioinformatics.mdanderson.org/Software/OOMPA.
- Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics (Oxford, England) 2007;23:2881–7. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nature Biotechnology. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Majumder T, Kalyanaraman A, Pande PP. Hardware Accelerators for Biocomputing: A Survey. Electrical Engineering. 2010:3789–3792. [Google Scholar]
- Shannon CE. The mathematical theory of communication. 1963. MD computing: computers in medical practice. 1948;14:306–17. [PubMed] [Google Scholar]
- Spyrou C, Stark R, Lynch AG, Tavaré S. BayesPeak: Bayesian analysis of ChIP-seq data. BMC Bioinformatics. 2009;10:299. doi: 10.1186/1471-2105-10-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. The positive false discovery rate: A Bayesian interpretation and the q-value. The Annals of Statistics. 2003;31:2013–2035. [Google Scholar]
- Teytelman L, Ozaydin B, Zill O, Lefrançois P, Snyder M, Rine J, Eisen MB. Impact of chromatin structures on DNA processing for genomic analyses. PLoS One. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28:516–520. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Handoko L, Wei X, Ye C, Sheng J, Wei CL, Lin F, Sung WK. A signalnoise model for significance analysis of chip-seq with negative control. Bioinformatics. 2010;26:1199–1204. doi: 10.1093/bioinformatics/btq128. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]