Abstract
The chemical stability of phosphodiester bonds of some oligoribonucleotides in the presence of a cofactor like polyvinylpyrolidine (PVP) is sequence dependent. It was found that pyrimidine-A (YA) and pyrimidine-C (YC) are especially susceptible to hydrolysis. The hydrolyzability of this same phosphodiester bond is dependent on its position in the oligomer. The presence of 3' and 5'-adjacent nucleotides enhances hydrolysis of the UA phosphodiester bond. The acceleration of the hydrolysis of UA by a 5'-adjacent nucleotide is not base dependent. However, a 3'-adjacent purine increases hydrolysis of a UA phosphodiester bond more than a 3'-pyrimidine. The presence of the exoamino group on the 3'-side base (on 6 and 4 position for adenosine and cytidine, respectively) of YA or YZ phosphodiester bond is required for hydrolysis.
Full text
PDF
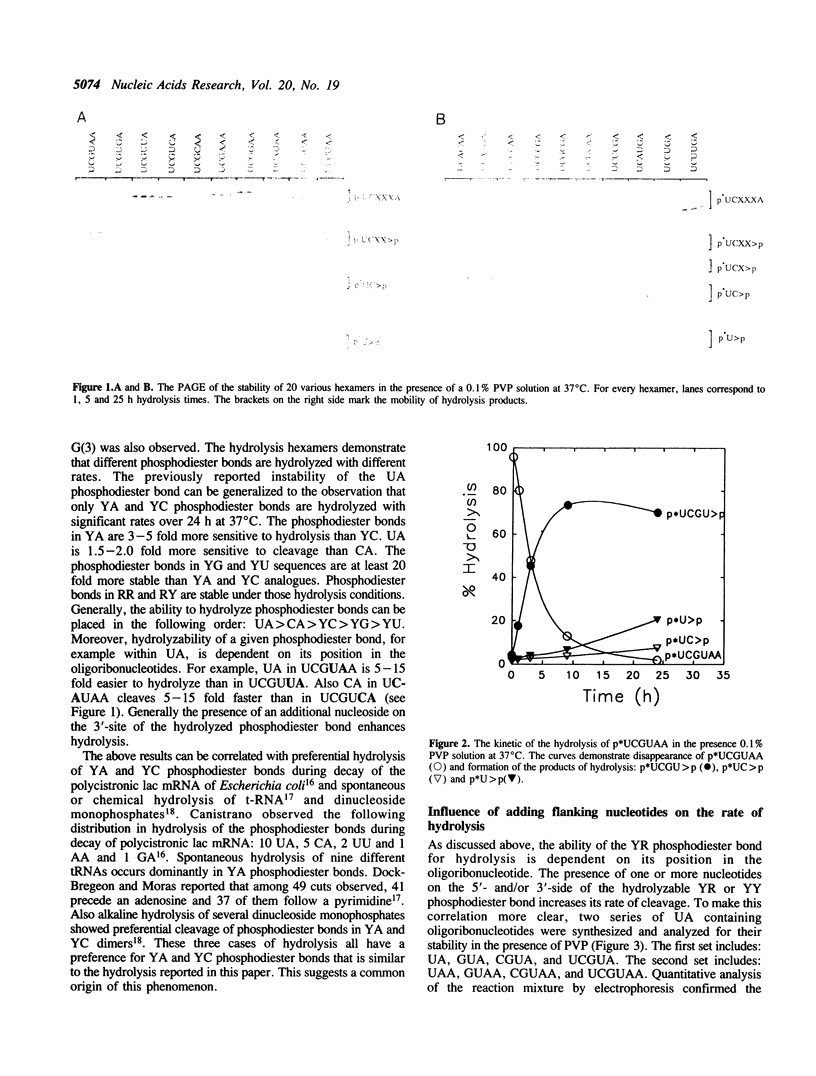
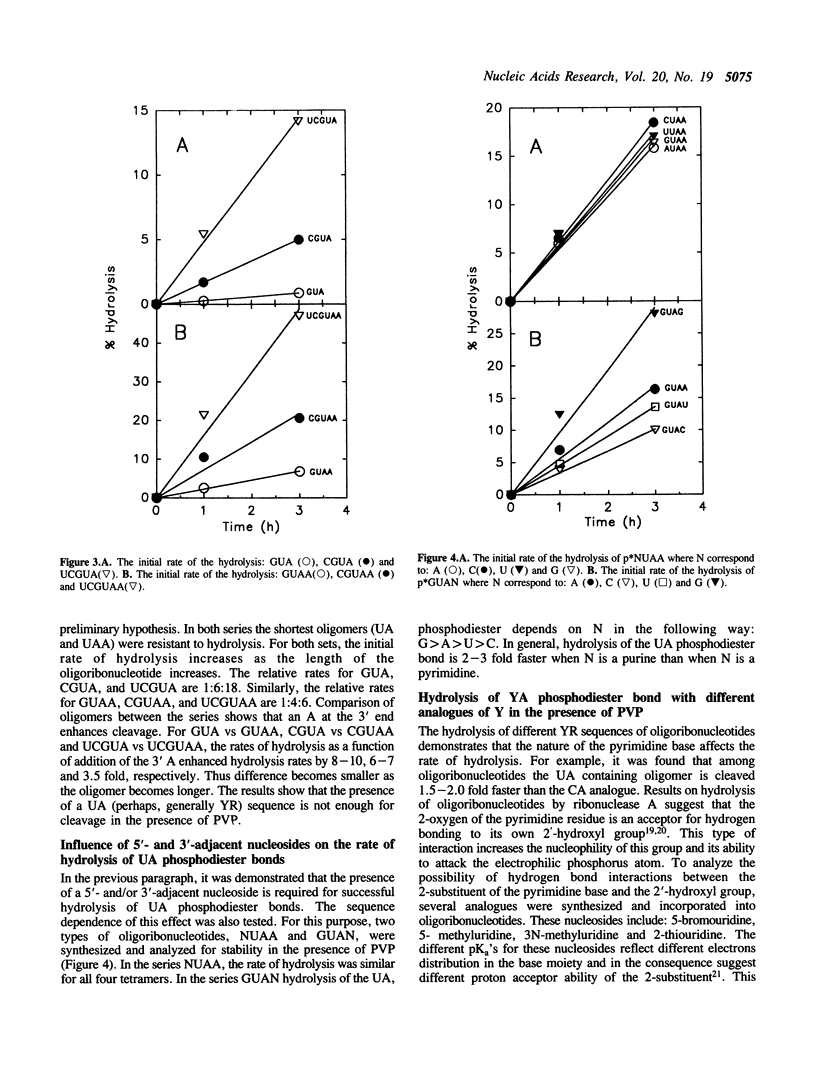


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Chowrira B. M., Burke J. M. Binding and cleavage of nucleic acids by the "hairpin" ribozyme. Biochemistry. 1991 Sep 3;30(35):8518–8522. doi: 10.1021/bi00099a003. [DOI] [PubMed] [Google Scholar]
- Ezra F. S., Lee C. H., Kondo N. S., Danyluk S. S., Sarma R. H. Conformational properties of purine-pyrimidine and pyrimidine-purine dinucleoside monophosphates. Biochemistry. 1977 May 3;16(9):1977–1987. doi: 10.1021/bi00628a035. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Kierzek R., Caruthers M. H., Longfellow C. E., Swinton D., Turner D. H., Freier S. M. Polymer-supported RNA synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry. 1986 Dec 2;25(24):7840–7846. doi: 10.1021/bi00372a009. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Quigley G. J., Suddath F. L., McPherson A., Sneden D., Kim J. J., Weinzierl J., Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973 Jan 19;179(4070):285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- Kole R., Altman S. Reconstitution of RNase P activity from inactive RNA and protein. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3795–3799. doi: 10.1073/pnas.76.8.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H., Tinoco I., Jr Conformation studies of 13 trinucleoside diphosphates by 360 MHz PMR spectroscopy. A bulged base conformation. I. Base protons and H1' protons. Biophys Chem. 1980 Apr;11(2):283–294. doi: 10.1016/0301-4622(80)80031-7. [DOI] [PubMed] [Google Scholar]
- MARKHAM R., SMITH J. D. The structure of ribonucleic acid. I. Cyclic nucleotides produced by ribonuclease and by alkaline hydrolysis. Biochem J. 1952 Dec;52(4):552–557. doi: 10.1042/bj0520552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. Cleavage of oligoribonucleotides by a ribozyme derived from the hepatitis delta virus RNA sequence. Biochemistry. 1992 Jan 14;31(1):16–21. doi: 10.1021/bi00116a004. [DOI] [PubMed] [Google Scholar]
- Ray J., Manning G. S. Theory of delocalized ionic binding to polynucleotides: structural and excluded-volume effects. Biopolymers. 1992 May;32(5):541–549. doi: 10.1002/bip.360320510. [DOI] [PubMed] [Google Scholar]
- Seela F., Ott J., Franzen D. Poly(2-methylthio-7-deazainosinic acid)--hydrophobic stabilization of polynucleotide secondary structure by the 2-methylthio group. Nucleic Acids Res. 1983 Sep 10;11(17):6107–6120. doi: 10.1093/nar/11.17.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Suddath F. L., Kim J. J., Rich A. RNA double-helical fragments at atomic resolution. I. The crystal and molecular structure of sodium adenylyl-3',5'-uridine hexahydrate. J Mol Biol. 1976 Jun 14;104(1):109–144. doi: 10.1016/0022-2836(76)90005-x. [DOI] [PubMed] [Google Scholar]
- Stone M. P., Winkle S. A., Borer P. N. 13C-NMR of ribosyl ApApA, ApApG and ApUpG. J Biomol Struct Dyn. 1986 Feb;3(4):767–781. doi: 10.1080/07391102.1986.10508460. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Seeman N. C., Kim S. H., Berman H. M. Crystal structure of a naturally occurring dinucleoside phoaphate: uridylyl 3',5'-adenosine phosphate model for RNA chain folding. J Mol Biol. 1972 May 28;66(3):403–421. doi: 10.1016/0022-2836(72)90423-8. [DOI] [PubMed] [Google Scholar]
- Thedford R., Fleysher M. H., Hall R. H. Polynucleotides. 3. Synthesis of (3'-5')-linked diribonucleoside phosphates containing 3- and 5- methyluracil. J Med Chem. 1965 Jul;8(4):486–491. doi: 10.1021/jm00328a016. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Usher D. A. On the mechanism of ribonuclease action. Proc Natl Acad Sci U S A. 1969 Mar;62(3):661–667. doi: 10.1073/pnas.62.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]