Abstract
To test the predictions of competing and mutually exclusive hypotheses for the origin of eukaryotes, we identified from a sample of 27 sequenced eukaryotic and 994 sequenced prokaryotic genomes 571 genes that were present in the eukaryote common ancestor and that have homologues among eubacterial and archaebacterial genomes. Maximum-likelihood trees identified the prokaryotic genomes that most frequently contained genes branching as the sister to the eukaryotic nuclear homologues. Among the archaebacteria, euryarchaeote genomes most frequently harbored the sister to the eukaryotic nuclear gene, whereas among eubacteria, the α-proteobacteria were most frequently represented within the sister group. Only 3 genes out of 571 gave a 3-domain tree. Homologues from α-proteobacterial genomes that branched as the sister to nuclear genes were found more frequently in genomes of facultatively anaerobic members of the rhiozobiales and rhodospirilliales than in obligate intracellular ricketttsial parasites. Following α-proteobacteria, the most frequent eubacterial sister lineages were γ-proteobacteria, δ-proteobacteria, and firmicutes, which were also the prokaryote genomes least frequently found as monophyletic groups in our trees. Although all 22 higher prokaryotic taxa sampled (crenarchaeotes, γ-proteobacteria, spirochaetes, chlamydias, etc.) harbor genes that branch as the sister to homologues present in the eukaryotic common ancestor, that is not evidence of 22 different prokaryotic cells participating at eukaryote origins because prokaryotic “lineages” have laterally acquired genes for more than 1.5 billion years since eukaryote origins. The data underscore the archaebacterial (host) nature of the eukaryotic informational genes and the eubacterial (mitochondrial) nature of eukaryotic energy metabolism. The network linking genes of the eukaryote ancestor to contemporary homologues distributed across prokaryotic genomes elucidates eukaryote gene origins in a dialect cognizant of gene transfer in nature.
Keywords: endosymbiosis, eukaryotes, phylogenomics, lateral gene transfer, horizontal gene transfer, endosymbiotic gene transfer
Introduction
Although the evolutionary details of the prokaryote-to-eukaryote transition are still incompletely resolved (Brown and Doolittle 1997; Koonin 2012), the crucial role that mitochondria played in that transition is becoming increasingly evident (Lane and Martin 2010; Lane 2011). Presently, two main categories of competing hypotheses address the prokaryote-to-eukaryote transition: autogenous and symbiogenic (Maynard-Smith and Szathmáry 1995; Embley and Martin 2006; Pisani et al. 2007; Lane 2009). Autogenous models posit that eukaryotes arose from a single ancestral lineage via mutation in a gradualist type of evolutionary process. Symbiogenic models posit that eukaryotes arose via a symbiotic association of divergent prokaryotic cells, with symbiosis (and gene transfer from endosymbiont to host in some formulations) forging the prokaryote-to-eukaryote transition, with phases of evolutionary innovation marked by distinctly non-gradualist characteristics. Both the autogenous and the symbiogenic categories harbor a number of specific competing alternative hypotheses, respectively, each of which in turn generates testable predictions about the phylogenetic affinities of eukaryotic genes to prokaryotic homologues.
Among the autogenous models, three are currently discussed. The neomuran hypothesis (Cavalier-Smith 1975) argued in its original formulation that eukaryotes arose from cyanobacteria through conventional mutation and selection processes. In more modern formulations, the neomuran hypothesis posits that eukaryotes arose from actinobacteria (Cavalier-Smith 2002); hence, it predicts that eukaryotic genes should uncover detectable affinities to homologues encountered in contemporary actinobacterial genomes. A second and more recent—but far less explicit—autogenous model has it that eukaryotes are descended from planctomycetes or the planctomycete–verrumicrobia–chlamydia (PVC) group (Devos and Reynaud 2010). It predicts eukaryotic genes to uncover widespread sequence similarities to planctomycete homologues, a prediction that is so far unfulfilled (McInerney et al. 2011). A third autogenous theory has it that eukaryotes represent the ancestral state of cell organization and that prokaryotes are derived from eukaryotes via a process that was originally called streamlining (Doolittle 1978) and later called thermoreduction (Forterre 1995). It predicts a three-domain topology for genes shared by eukaryotes and prokaryotes (Forterre and Gribaldo 2010). Common to autogenous theories is the assumption that mitochondria had no role in the prokaryote-to-eukaryote transition, a premise that has become increasingly problematic with data accrued over the last 10 years indicating 1) that mitochondria were present in the eukaryote common ancestor (Embley et al. 2003; van der Giezen 2009) and 2) that, for reasons of bioenergetics, mitochondria were strictly required for the origin of the molecular traits that make eukaryotic cells complex in comparison to their prokaryotic counterparts (Lane and Martin 2010).
Symbiogenic hypotheses can be generally divided into two subcategories. The first subcategory invokes an endosymbiosis to derive a mitochondrion-lacking cell that possess a nucleus, whereby the nuclear compartment is usually viewed as deriving from an endosymbiotic prokaryote. Among current formulations that derive the nucleus from endosymbiosis, the assumed symbiotic partners include 1) a Thermoplasma-like host and a spirochaete endosymbiont (Margulis et al. 2006), 2) a Gram-negative host and a crenarchaeal endosymbiont (Lake and Rivera 1994; Gupta and Golding 1996), 3) a δ-proteobacterial host and a methanogen-like endosymbiont (Moreira and Lopez-Garcia 1998), 4) a γ-proteobacterial host and a Pyrococcus-like endosymbiont (Horiike et al. 2004), and 5) a planctomycete host and a Crenarchaeum-like nucleogenic endosymbiont (Forterre 2011). Like autogenous theories, these models assume that mitochondria had no role in the prokaryote-to-eukaryote transition sensu strictu because a nucleus-forming endosymbiosis is presumed to have generated the eukaryotic lineage, one member of which then acquires the mitochondrion and the other members of which implicitly become extinct because all eukaryotic lineages possess a mitochondrion or did in their evolutionary past (van der Giezen 2009). As discussed elsewhere, there are many serious fundamental problems with the view that the nucleus was ever a free-living prokaryote (Martin 1999a, 2005; Cavalier-Smith 2002).
The second major subcategory among symbiogenic theories invokes endosymbiosis to derive the mitochondrion directly in a prokaryotic host, without any earlier additional symbiotic cell mergers. Because eukaryotes have an archaebacterial genetic apparatus (Langer et al. 1995; Rivera et al. 1998; Cox et al. 2008; Koonin 2009; Cotton and McInerney 2010) and because mitochondria are clearly derived from an endosymbiotic proteobacterium (Gray et al. 1999; Atteia et al. 2009), these theories posit that the host for the origin of mitochondria was a “garden variety” archaebacterium, either related to Thermoplasma (Searcy 1992) or to hydrogen-dependent archaebacteria, with a physiology perhaps similar to methanogens (Martin and Müller 1998; Vellai et al. 1998). In these models, the nucleus arises after the origin of mitochondria and in an autogenous manner that does not require additional endosymbioses (Martin and Koonin 2006).
All of the foregoing theories generate predictions with regard to the branching patterns expected in trees comprising both prokaryotic and eukaryotic genes. Comparatively, few tests of those predictions using alignments for many genes from complete genome data have been reported. Using pairwise sequence similarity matrices, Esser et al. (2004) found that among the 850 yeast genes having homologues among the small prokaryotic sample of 15 archaebacterial and 45 eubacterial genomes, roughly 75% of yeast nuclear-encoded proteins were more similar to eubacterial homologues than to archaebacterial homologues. Using a supertree approach, Pisani et al. (2007) found that when the signal stemming from nuclear genes of cyanobacterial origin in plants is removed from the eukaryote data set, eukaryotes branch among α-proteobacteria, likely reflecting the signal of nuclear genes of mitochondrial origin, and when that signal is removed, eukaryotes then branched with the Thermoplasma lineage among euryarchaeotes. Cox et al. (2008) examined the concatenated phylogeny of genes corresponding to the informational class (information storage and processing) and found evidence linking the eukaryote host lineage to crenarchaeotes, in line with the prediction of the eocyte theory (Lake 1988). Yutin et al. (2008) examined individual phylogenies and obtained conflicting results with respect to the euryarchaeal, crenarchaeal, or ancestral archaebacterial origin of eukaryotic informational genes. Kelly et al. (2011) examined genes that link eukaryotes to archaebacteria and found evidence linking the eukaryotes to the Crenarchaeum/Nitrosopumilus (thaumarchaeal) lineage of archaebacteria.
Autogenous models and symbiogenic models also differ with respect to the predictions that they make about the ancestor of mitochondria and (in some cases) about the nature of eubacterially related genes in eukaryotic genomes. Several recent studies have addressed the origin of mitochondria, but have focused on sequences residing in mitochondrial DNA (mtDNA) (Thrash et al. 2011; Brindefalk et al. 2011; Georgiades and Raoult 2011). Those studies delivered widely conflicting results because of the small sample of mitochondrion-encoded protein available—about 55 at most that can be used to generate trees (Esser et al. 2004)—and the phylogenetic biases introduced by the rapid evolutionary rate and AT richness of mtDNA, which can cause mtDNA-encoded proteins to artefactually group together with homologues from rapidly evolving and AT-rich bacterial lineages, like Rickettsiales (Thrash et al. 2011). Nuclear-encoded proteins should, in principle, be less subject to AT bias and the elevated substitution rate of mitochondrially encoded proteins. They provide a larger gene sample for investigation of mitochondrial origin or of organelle origins in general (Deusch et al. 2008) but that does not mean that they are fundamentally immune to bias or phylogenetic error.
Investigations of mitochondrial origin using many nuclear genes are still scarce. Trees for pyruvate dehydrogenase subunits pointed to Rickettsia-like ancestors (Kurland and Andersson 2000). Trees for Krebs cycle and glyoxylate cycle enzymes (Schnarrenberger and Martin 2002) as well as trees for >200 nuclear-encoded mitochondrial proteins from Chlamydomonas point more frequently to origins from generalist, facultatively anaerobic α-proteobacteria (Atteia et al. 2009), than to Rickettsia-like ancestors, whereby many proteins indicated a eubacterial, but not a specifically α-proteobacterial ancestry. Recent analysis of 86 yeast nuclear-encoded mitochondrial proteins produced a similar result: some point to Rickettsia-like ancestors and some point to facultatively anaerobic Rhodobacter-like ancestors (Abhishek et al. 2011). Although many mitochondria function anaerobically (Tielens et al. 2002; Atteia et al. 2006; Mus et al. 2007), nuclear genes for anaerobic mitochondrial energy metabolism cannot implicate Rickettsia-like ancestors because Rickettsias are strict aerobes that harbor no genes of anaerobic energy metabolism for comparison. Using an automated pipeline for phylogenetic trees, Gabaldon and Huynen (2003) identified 630 nuclear-encoded protein families that trace to the ancestor of mitochondria, although the α-proteobacteria themselves often failed to form a monophyletic group in that study, pointing to the role of LGT in prokaryote evolution.
Comparative genomics should permit a test of different models for eukaryote origins. Genes suited to such tests are those that are preserved in eukaryotic nuclear genomes that 1) have homologues in prokaryotes and 2) reflect eukaryote monophyly as evidence of their presence in the last eukaryote common ancestor (LECA). Here we have assembled alignments for 712 gene families from 27 eukaryotes, 926 eubacteria, and 68 archaebacteria in order to address the question: given the present (limited) genome sample, how many eukaryotic genes with prokaryotic homologues actually reflect a single origin? Those that trace to the eukaryote common ancestor allow us to furthermore ask: in which prokaryotic lineages are those genes currently found? We then contrast the results with the predictions generated by competing theories for eukaryote origins, but do so in a modern context taking into account the circumstance that free-living prokaryotes have been undergoing LGT during the time since eukaryotes arose, such that the collection of genes that eukaryotes acquired from the ancestor of mitochondria reflects a sample of ancient prokaryote gene diversity, not a collection of genes that we would expect to find in any contemporary free-living prokaryote (Martin 1999b; Esser et al. 2007; Richards and Archibald 2011). In that sense, the concept of prokaryotic lineages is poorly defined when it comes to the phylogeny of individual genes (Doolittle and Bapteste 2007; Bapteste et al. 2009), a circumstance that figures prominently in the interpretation of the results.
Methods
Data
Proteomes of 27 eukaryotes and 994 prokaryotes were retrieved from the public databases. The following proteomes were downloaded from RefSeq database (Pruit et al. 2005): Hydra magnipapillata, Ciona intestinales, Caenorhabditis elegans, Physcomitrella patens, all 994 prokaryotes (11/2009 version), Oryza sativa, and all fungi and animal sequences (02/2008 version). Additional proteomes were retrieved from the JGI database (http://www.jgi.doe.gov/): Populus trichocarpa (v1.1), Ostreococcus tauri and Chlamydomonas reinhardtii (v3.1). The Arabidopsis thaliana proteome was downloaded from TAIR project (Swarbreck et al. 2008). The Cyanidoschyzon merolae proteome was downloaded from the genome project Web site (Matsuzaki et al. 2004). The complete list of genomes used is given in supplementary Table S1, Supplementary Material online.
Clusters of Homologous Proteins
For the reconstruction of eukaryotic protein families, a reciprocal best Blast (v2.2.20; Altschul et al. 1997) hit procedure was used (Tatusov et al. 1997). Only BBH having an e-value ≤1 × 10−10 were retained. Pairs of reciprocal BBHs were aligned using the Needleman–Wunsch algorithm by the needle program included in the EMBOSS package (Rice et al. 2000). Homologous pairs having ≤40% identical amino acids were excluded from the data set. All remaining eukaryotic homologues were clustered into protein families with MCL (v1.008 Enright et al. 2002) using the default parameters. This analysis yielded 37,101 protein families comprising 165,329 proteins. Excluding protein families comprising less than 4 members in total (18,116) or less than 3 main eukaryotic groups (animals, fungi, algae, plants) resulted in 1,122 protein families retained for further analysis. To find prokaryotic homologues to all clustered eukaryotic proteins, these proteins were BLASTed against 994 prokaryotic proteomes. A total of 367 clusters had no homologues within the prokaryotic genomes. Prokaryotic homologues were added to the clusters using a reciprocal BBH procedure applying an e-value threshold of ≤1 × 10−10 and ≥30% identical amino acids. All prokaryotic hits per eukaryotic query protein were added to the respective protein family, and redundant prokaryotic proteins were omitted. In case of multiple prokaryotic homologues in different strains of the same species, only one homologue having the highest sequence similarity was included in the protein family. The analysis resulted in 755 protein families of eukaryotic sequences and their prokaryotic homologues.
The functional classification of all protein families is based on the KOG database (Tatusov et al. 2003). A total of 626 protein families that overlapped with KOG clusters were annotated to have the same function as the matching KOG. The remaining protein families were manually classified by sequence similarity to known KOGs using the KOGnitor tool (http://www.ncbi.nlm.nih.gov/COG/grace/kognitor.html). A total of seven protein families had no homologues in the KOG database and were annotated as unknown function.
Phylogenetic Analysis
The protein families were aligned with MAFFT (v6.832b; Katoh et al. 2002) using Blosum62 substitution matrix (Henikoff and Henikoff 1992). Alignment quality was tested using the HoT procedure (Landan and Graur 2007) and 20 alignments having SPS <70% were excluded from the data set. Phylogenetic trees were calculated from the alignments using maximum-likelihood approach with RAxML (v7.0.4; Stamatakis 2006). Substitution rate per site was estimated from a gamma distribution with four discrete rate categories and the WAG substitution matrix (Whelan and Goldman 2001). The proportion of invariable sites was estimated from the data. Eukaryotic monophyly within the reconstructed trees was tested using an in-house PERL script. A group is considered as monophyletic if there exists a bipartition (branch) in the tree that includes only species from that group. Thus in trees testifying eukaryotic monophyly, there exists a branch that splits between eukaryotes and prokaryotes.
Single eukaryotic sequences branching within the prokaryotic clade were manually tested for possible bacterial contaminations using BLAST at NCBI Web site (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against the nr database. We manually excluded sequences that were obvious bacterial contaminations where possible. For example, 39 genes annotated as belonging to the Hydra genome actually belong to the genome of the eubacterial endosymbiont of Hydra, Curvibacter spec. (Chapman et al. 2010). Another eight genes in the Populus genome are >90% identical at the amino acid level to prokaryotic proteins and were classified as a putative bacterial contamination, as were two sequences from the Bos taurus genome. In several cases, the bacterial contamination “Hydra” sequence was the only representative from the metazoa, such that in total 24 alignments were excluded from the analysis, leaving 712 alignments for analysis, which are available upon request.
To specify the sister group to the eukaryotes, there are several possibilities. In each tree, the branch connecting the monophyletic eukaryotic clade to the prokaryotes serves to splits the prokaryote clade into two groups, one of which is selected as the sister group. We tested several criteria to decide which is the sister group: the group with the smaller number of sequences, the group with the smaller average distance to the eukaryotes, and the group that does not include the root after midpoint rooting. The sister group frequencies inferred are robust to the three different criteria (supplementary fig. S1, Supplementary Material online). For simplicity, we used the criterion of the smaller clade to specify the sister group, use of other criteria would not alter the results.
We did not initiate exhaustive topology searches or likelihood optimization efforts searches beyond those performed by RAxML in order to find more or fewer cases of eukaryote monophyly in the data. For those genes that did reflect eukaryote monophyly, we were interested in the identity and nature of the genomes harboring the nearest prokaryotic neighbors.
Network and Reference Tree Reconstruction
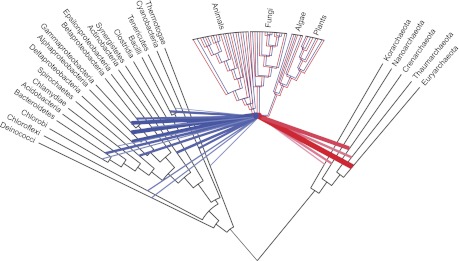
The prokaryotic clade of the universal reference tree was retrieved from Popa et al. (2011) that reconstructed it from the ribosomal RNA operon sequences within a taxonomic framework. The tree was reconstructed for prokaryotic main taxa by using the consensus sequences of the ribosomal RNA genes for each bacterial group. The groups correspond to the phyla of the bacterial species or the class in the case of Proteobacteria and Firmicutes. Three archaebacterial groups including the Nanoarchaeota, Thaumarchaeota, and Korarchaeota that were missing in the Popa et al. (2011) tree were included according to their phylogenetic position in Makarova et al. (2010). The eukaryotic clade is a consensus tree reconstructed from 12 gene trees that include all eukaryotic species in our analysis (excluding the highly reduced genome of Encephalitozoon cuniculi). The network in figure 8 combines 571 sister group specifications as lateral edges connecting vertical edges of the reference tree. The edge weight is the frequency in which species in the given prokaryotic taxon branched within the sister group to the eukaryotic clade. The universal tree with lateral edges signifying prokaryotic contributions was depicted with a MatLab© script.
Fig. 8.—
Network linking apparent prokaryotic donors to the eukaryote common ancestor according to the present findings. This network based on a traditional phylogenetic tree to which lateral edges were added. Color intensity and width of the lateral edges reflect the frequencies with which these groups appear as sisters to the eukaryotic clade.
Results
Eukaryotic Genes Reflect Single Origins
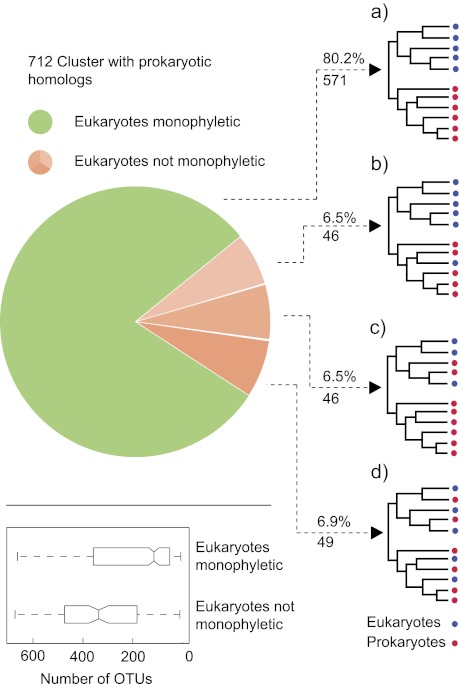
Clustering the eukaryotic and prokaryotic proteins by sequence similarity yielded 712 inter-kingdom families of homologous proteins. All protein families include at least three of the main eukaryotic groups: animals, fungi, algae, and plants. Phylogenetic trees were reconstructed from the protein families by a maximum-likelihood approach and rooted on the branch that maximizes the ratio between eukaryotes and prokaryotes in the resulting clades. The resulting rooted trees were classified into four categories according to the branching pattern of the eukaryotic species within the tree (fig. 1).
Fig. 1.—
A distribution of topologies among 712 inter-kingdom trees. The schematic trees on the right symbolize the branching patterns of eukaryotic and prokaryotic species observed in each category. (a) Eukaryotes and prokaryotes form monophyletic clades. (b) Eukaryotes are polyphyletic (≤6 eukaryotic species branch within the prokaryotic clade) and prokaryotes are paraphyletic. (c) Eukaryotes are paraphyletic (between 1 and 22 prokaryotic species branch within the eukaryotic clade) and prokaryotes are polyphyletic. (d) A mixed topology of eukaryotes and prokaryotes. The boxplots in the lower panel show the distribution of the number OTUs in trees where the eukaryotes are 1) monophyletic or 2) not monophyletic.
Most of the tree topologies (571/712, 80.2%) recovered the eukaryotic genes as a monophyletic clade. The remaining trees fell into three different categories in similar shares. In polyphyletic trees (46, 6.5%), there exists a branch that splits the tree into a eukaryotes-only clade and a prokaryotic clade that includes a few eukaryotes (fig. 1b). The frequency of eukaryotes in the prokaryotic clade ranged between 1 and 6 species. In 12 of those polyphyletic trees, the eukaryotes branching within the prokaryotic clade were photosynthetic eukaryotes branching as the nearest neighbors of cyanobacteria, the expected result for genes that were transferred from the ancestor of plastids into the nuclear genome of photosynthetic eukaryotes (Timmis et al. 2004). Genes in this group include the ClpB heat shock protein Kda100 (C. reinhardtii, C. merolae, and O. tauri) and phosphoglycerate kinase (Brinkmann and Martin 1996) from C. reinhardtii, C. merolae, and A. thaliana (supplementary Table 2, Supplementary Material online). Most (34) of the remaining trees in this category included a single eukaryote within the prokaryotic clade, with many proteins being annotated as “predicted protein” or “hypothetical protein.” Genes of the red algae C. merolae branched frequently within the prokaryotic clade with 18 tree topologies placing this species within the prokaryotic clade next to a noncyanobacterial nearest neighbor (supplementary Table 2, Supplementary Material online).
Paraphyletic trees are the mirror image of polyphyletic trees, as they include a branch that splits the tree into a prokaryotes-only clade and a eukaryotic clade that includes several prokaryotes (fig. 1c). The number of prokaryotes in the latter clade ranged between 1 and 22 (see distribution in supplementary fig. S2, Supplementary Material online). In 29 of the 46 paraphyletic trees, prokaryotes branching within the eukaryotic clade included one or more eukaryote-associated microbes (e.g., human pathogens and plant endosymbionts). These could be prokaryote acquisitions of eukaryotic homologues, as has been previously observed, for example for tubulin (Pilhofer et al. 2007). In six trees, all of the prokaryotes that branched within the eukaryotic clade were cyanobacterial species next to plants or algae. The mixed prokaryotic–eukaryotic branching pattern of the remaining 49 (6.9%) trees did not enable a clear cut rooting and classification of the trees into one of the above categories (fig. 1d). In trees with eukaryote monophyly, operational genes and informational genes are present in a ratio of 362:209, in the trees where eukaryotes are nonmonophyletic, operational genes are significantly enriched (129:12, P < 0.0001).
LECA Genes with Prokaryotic Homologues
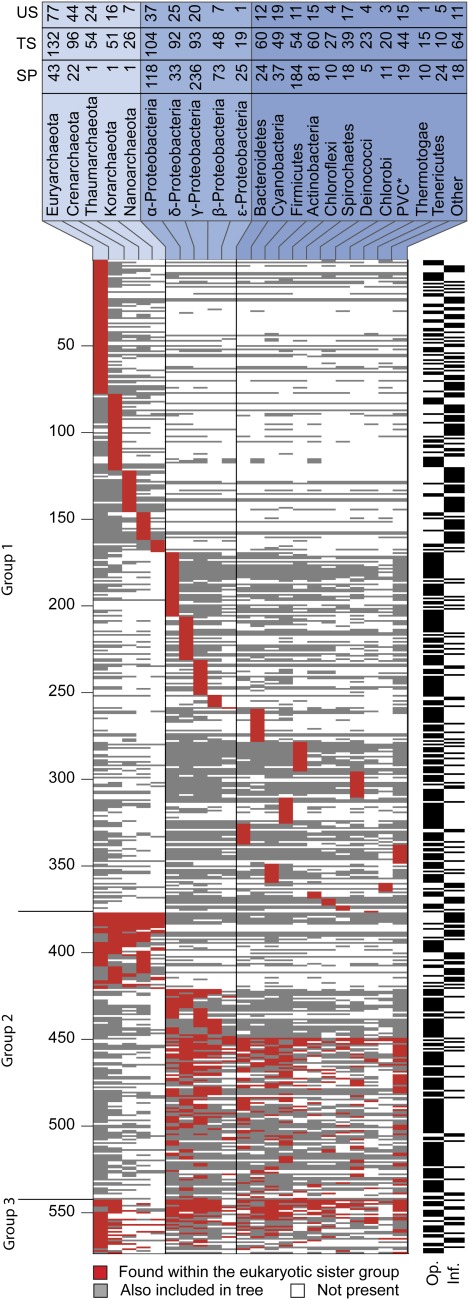
For 571 protein families, ML trees indicate eukaryote monophyly. Because we only considered trees that spanned the opisthokont/archaeplastida divide, and because of current views for the placement of the eukaryotic root (Hampl et al. 2009), eukaryote monophyly indicates their presence in LECA. The distribution across prokaryotic genomes of genes that appear as sisters to the eukaryotic nuclear copy is of interest because their phylogenetic affinities can, in principle, help to discriminate between competing theories for eukaryote origins. An overview of the results is shown in figure 2. For simplicity, the topologies can be divided into three general categories with respect to the taxonomic distribution of eukaryote sister genes among prokaryotes. Group 1: The sister genes occur only among genomes of one of the higher prokaryotic taxa shown in figure 2, for example, euryarchaeotes, thaumarchaeotes, α-proteobacteria, firmicutes, or the like. Among the 571 LECA gene families, 375 yield this result. Group 2: The sister genes are not restricted to a particular class or phylum but occur only among members of the archaebacteria or eubacteria, 165 alignments and trees yield this result. Group 3: The sister group to the eukaryotic nuclear gene includes genes that occur among members of both the archaebacteria and the eubacteria, 31 alignments and trees deliver this result.
Fig. 2.—
A presence–absence pattern (PAP) of the bacterial taxonomic groups in trees supporting the eukaryotic monophyly. The rows correspond to 571 trees in which the eukaryotes were monophyletic, the columns correspond to 22 bacterial groups. A cell i,j in the matrix is colored if tree i included a homologue from bacterial group j. Taxonomic groups harboring a gene that branches as a nearest neighbor to the eukaryotic clade in each tree are marked by a red cell. Taxonomic groups that are also present in the tree are marked by a gray cell. An asterisk indicates that species from the Chlamydiae, Verrumicrobia, and Plancomycetes taxa were combined into one group (PVC) (Wagner and Horn 2006). Group 1 included only trees where exactly one bacterial group was found, Group 2 included trees where 1) only Archaea were found, 2) only proteobacteria were found, and 3) were only eubacteria were found. Group 3 included all other trees. The black and white bars on the right indicate whether the KOG underlying the tree belongs to an informational (Inf.) or to an operational (Op.) class (Rivera et al. 1998). The numbers in the top panel indicate the following. US: The number of times that the given taxon was the only taxon in the sister group to the eukaryotic sequence (unique sisterhood, US). TS: the number of times that the taxon was either included in a unique sister group or in a sister group consisting of mixture of prokaryotic taxa total sisterhood (TS). SP: the number of sequenced strains from that taxon in our genome sample.
The trees in Group 3 contain the least information and are also the easiest to interpret. Monophyletic eukaryotes nested within and as the sister to clades in which the archaebacteria and the eubacteria are interleaved indicate that the eukaryotic gene reflects a single origin, but that during the time subsequent to the origin of eukaryotes, the prokaryotic gene has undergone so much sequence divergence and/or LGT among prokaryotic groups that it is not possible to generate a strong inference about the source of that gene in LECA through the vantage point of phylogenetic trees. Many phylogenetic artefacts affecting the prokaryote topology might also rest in this category, distinguishing between LGT and phylogeny or alignment artefacts is not straightforward (Roettger et al. 2009). These 31 trees thus are equivocal about gene origins in LECA.
The 165 trees in Group 2 show the eukaryote nuclear genes branching as the sister to groups containing homologues present in several different archaebacterial or several different eubacterial higher taxa. These genes tend to reflect archaebacterial or eubacterial ancestries for the eukaryotic gene, respectively, without implicating a specific higher-level taxon as the donor lineage. Among these genes, 44 reflect an archaebacterial ancestry, whereas 121 reflect a eubacterial ancestry. Of the 44 archaebacterial-derived genes in the LECA, 28 belong to the informational class (involved in information storage and processing), whereas 103 out of the 121 eubacterial-derived LECA genes belong to the operational class (involved in biochemical and biosynthetic processes). Thus, the informational and operational classes of eukaryotic genes well-established in analyses of the yeast genome (Rivera et al. 1998; Cotton and McInerney 2010) as well as the preponderance of eubacterial-derived over archaebacterial-derived genes in eukaryotic genomes (Esser et al. 2004; Pisani et al. 2007) are also evident for these 165 genes present in LECA. However, for these 165 trees, the sister group relationship to the eukaryotic gene appears more or less as a bucket of mixed pickles, but archaebacterial or eubacterial pickles. Although the proteobacteria are clearly the most frequently represented among the 121 trees indicating a eubacterial ancestry of the eukaryote nuclear genes, all eubacterial groups are ultimately represented.
The 375 trees that we classified as Group 1 show one of the higher prokaryotic taxa sampled as harboring the sister gene of the eukaryote common ancestor homologue. That is, the sister of the eukaryotic nuclear gene contained only members of one of the 21 higher prokaryotic taxa (22 including the category “other”) shown in figure 2. The most frequent taxon uniquely harboring sister genes to genes present in the eukaryote common ancestor were the euryarchaeotes (77 genes), followed by the crenarchaeotes (44 genes), the α-proteobacteria (37 genes), the δ-proteobacteria (25 genes), the thaumarchaea (24 genes), the γ-proteobacteria (20 genes), the cyanobacteria (19 genes), the spirochaetes (17 genes), the korarchaeote (16 genes), the actinobacteria (15 genes), the PVC group (15 genes), the bacteriodetes (12 genes), etc. In fact, all of the higher taxa sampled harbor a gene with a sister group relationship to the eukaryotic nuclear homologue. Some might conclude from this that all prokaryote lineages sampled donated genes to LECA but that is a much too simplistic inference that entails unrealistic assumptions about the nature of prokaryotic lineages and the affects of LGT over geological timescales as outlined in the Discussion.
Functional Categories
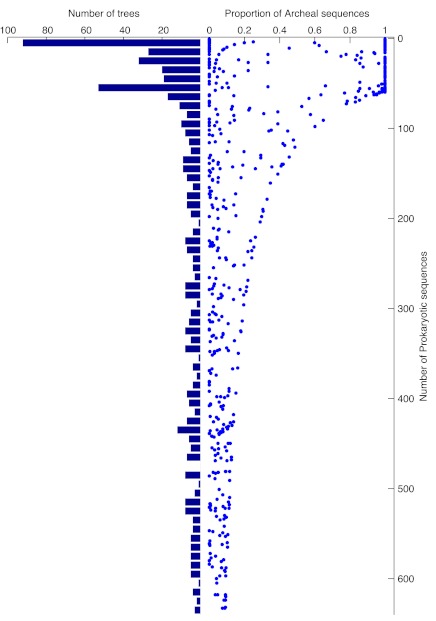
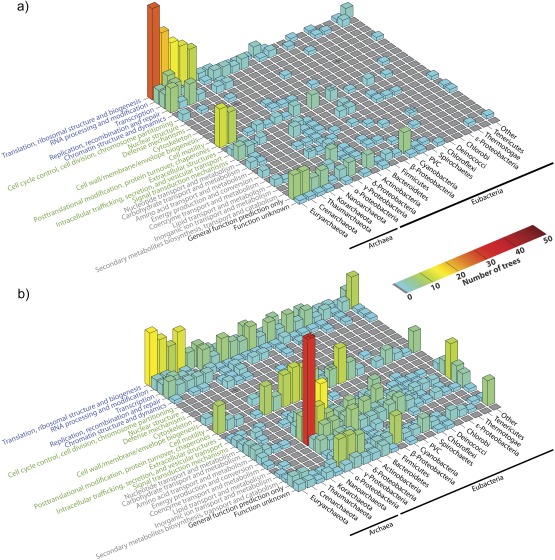
There are only 68 archaebacteria in our genome sample, and it is evident in figure 2 that among those alignments and trees where eukaryotic genes branch with archaebacteria as sisters, the eubacteria are underrepresented. We plotted the frequency distribution of the number of Operational Taxonomic Units (OTUs) in the protein family and for the same distribution the proportion of archaebacterial sequences in each alignment, or tree, as shown in figure 3. Among trees having 68 prokaryotic taxa or fewer, there are 47 that contain only archaebacterial homologues and 61 in which over 90% of the OTUs are archaebacterial. With increasing numbers of prokaryotes in the trees, the proportion of archaebacteria declines quickly, and there is clearly a bimodal distribution with regard to the presence and frequency of archaebacterial sequences in the protein families containing fewer than, or more than, 68 prokaryotic sequences (fig. 3). Because this bimodality is not independent of the informational–operational gene dichotomy, we plotted functional category assignments against sister group relationship for the 571 genes showing eukaryote monophyly in two bins, that is, trees containing fewer than 68 prokaryotic sequences (fig. 4a) or more than 68 prokaryotic sequences (fig. 4b).
Fig. 3.—
Proportion of archaeal sequences per alignment in the data set. The left bar graph shows the distribution of bacterial sequences in all trees where the eukaryotes form a monophyletic clade in bin intervals of 10. The plot on the right indicates the proportion of archaeabacterial sequences in each tree. There were only 68 archaea in the data, hence a skew distribution of trees containing many or mostly archaeabacterial sequences versus eubacterial sequences in alignments with more than 68 OTUs (see also fig. 4).
Fig. 4.—
Three-dimensional bar graphs of prokaryotic groups found as sister groups to the eukaryotes distributed across functional categories according to KOG groups. The four main groups are information storage and processing (classes colored in blue), cellular processes and signaling (classes colored in green), metabolism (classes colored in gray), and poorly characterized proteins (classes colored in black). (a) Including the data from trees with 68 or fewer prokaryotic sequences. (b) Including the data from trees with more than 68 bacterial sequences. Bar height and color indicate how often a certain group was found in a tree belonging to a certain category.
In figure 4a, the archaebacterial nature of the eukaryotic genetic apparatus, ribosome biogenesis in particular, stands out. Figure 4b summarizes the eukaryote sisterhood frequencies for the trees with stronger eubacterial representation. In addition to the archaebacterial informational signal, the most notable feature of figure 4b is the frequency with which proteobacteria branch as sister to the eukaryotes in operational genes, in particular energy metabolism. We note that the frequencies in figure 4 have not been normalized with respect to the number of species or genes per category. For example, the high frequency of euryarchaeotal sisterhood observed is not completely independent of the heavier taxon sampling for euryarchaeotes, which are twice as frequent in the data (43 genomes) as crenarchaeotes (22 genomes). In the same vein, the appreciable frequency of γ-proteobacterial sisterhood in energy metabolism category or firmicute sisterhood in the category posttranslational modification and chaperones (fig. 4) is not independent of the large number of genomes sampled for these groups in our data, which is, for obvious reasons, strongly skewed toward pathogens: the 236 γ-proteobacterial and 184 firmicute genomes in our data (fig. 2, top). However, normalization is not as easy as it might seem because many of the elements on both matrices (fig. 4a and b) are empty and because the genomes within the higher taxa indicted are extremely diverse with respect to genome size and frequencies of various functional categories. We plotted sisterhood occurrence for how often a gene from the taxon was found in the sister clade normalized by the frequency of genes in that taxon that are present in the trees, hence capable of appearing as the eukaryote sister. Although there are sufficient number of observations to normalize at the level of taxa, when normalization is extended to functional categories, spurious results are obtained, even when the empty or nearly empty elements of the matrix are removed (supplementary fig. S3, Supplementary Material online).
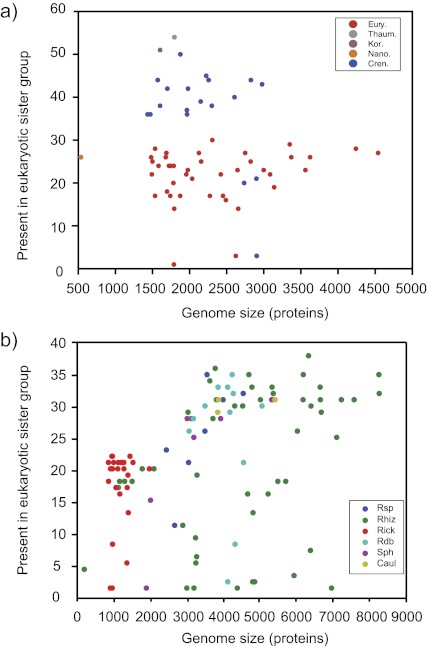
The apparent strong contributions of euryarchaeotes and α-proteobacteria are notable and robust. Among the archaebacteria, the crenarchaeotes are, on a per gene basis, more frequently found in the sister group than the euryarchaeotes (fig. 5a). Among the α-proteobacteria, there is a positive correlation (ρ = 0.0437, P = 2.8 × 10−6) between genome size and eukaryote sisterhood frequency (fig. 5b), indicating in the simplest interpretation, that the ancestor of mitochondria had a large genome. If we reduce the result of the functional category analysis to its most basic statement, the data reveal clear evidence for the archaebacterial nature of the eukaryotic genetic apparatus and the eubacterial nature of eukaryotic energy metabolism.
Fig. 5.—
Correlation between genome size and strain presence in the eukaryotic sister clade. (a) All archaebacterial species that were found as eukaryotic sisters plotted against their genome size. Correlation was measured using the spearman rang correlation, resulted in ρ = –0.1106, P = 0.3882. (b) All α-proteobacterial species that were found as eukaryotic sisters plotted against their genome size. Correlation was measured using the Spearman rang correlation, resulted in ρ = 0.4371 and P = 2.8 × 10−6.
Genes Dispersed (Widely) from the Ancestor of Mitochondria
Theories on the origin of eukaryotes differ with respect to the role of mitochondria therein. Some theories view the origin of mitochondria as distinct from and mechanistically irrelevant to the origin of eukaryotes (Kurland et al. 2006; Forterre and Gribaldo 2010). Others view the origin of mitochondria as coinciding with the origin of eukaryotes (Martin and Müller 1998; Embley and Martin 2006), as having precipitated the origin of the nucleus (Martin and Koonin 2006), and as an energetic conditio sine qua non for the origin of eukaryote-specific gene families that underpin eukaryotic cell and cell cycle complexity (Lane and Martin 2010). Most studies aiming to identify the sister group to mitochondria have focused on genes encoded in mtDNA. But mtDNA-encoded proteins are often highly divergent or rapidly evolving and phylogenetic problems thus arise with their tendency to branch with proteins from other rapidly evolving lineages such as Rickettsias (Bridefalk et al. 2011; Georgiades and Raoult 2011; Thrash et al. 2011). In phylogenetics, the problem is well-known and called long-branch attraction (Lockhart et al. 1994). The most slowly evolving prokaryotic homologues of eukaryotic nuclear-encoded proteins should be least affected by long-branch attraction, and nuclear genome data have not been examined from that standpoint so far. Hence we examined all 571 trees showing eukaryote monophyly to find the prokaryotic homologues that had the least total distance (the shortest path) in the ML tree to the eukaryotic nuclear genes. The result (not shown) was similar to figure 4a and b in that the highest frequencies of sister group occurrence were observed in the euryarchaeotal category for ribosome biogenessis and the α-proteobacterial category energy metabolism.
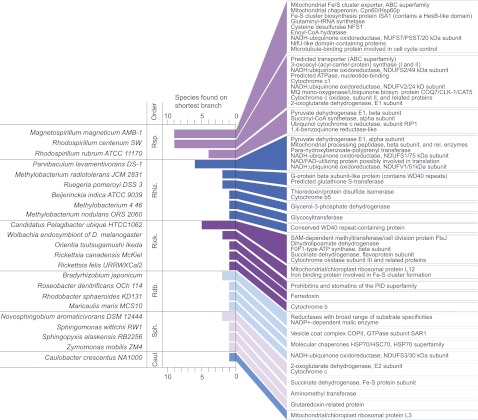
The α-proteobacterial genomes harboring the slowly evolving genes were mainly facultative anaerobes from the Rhodospirilliaceae (Magnetospirillum and Rhodospirillum) with the spectrum of functions represented being metabolic and thereby very distinct from the ribosomal proteins and respiratory chain components that are typically encoded in mitochondrial genomes (fig. 6). Thus, the result that one obtains for the inferred nature of the ancestor of mitochondria depends strongly upon which genes one considers: The fast-evolving genes in mtDNA often point to a fast-evolving mitochondrial ancestor related to Rickettsias (Bridefalk et al. 2011; Thrash et al. 2011; but see also Esser et al. 2004 and Abhishek et al. 2011 for different results), whereas the proteins encoded in nuclear DNA point to facultative anaerobic generalist α-proteobacteria as the mitochondrial ancestor (Atteia et al. 2009)—the most slowly evolving proteins in particular—as seen in figure 6.
Fig. 6.—
Frequency of single α-proteobacterial species found on the shortest path to the eukaryotic clade, by summing up the branch lengths. For each of the respective sequences, the functional annotation is also given. Abbreviations refer to α-proteobacterial families. Rsp: Rhodospirillales, Rhiz: Rhizobiales, Rick: Rickettsiales, Rdb: Rhodobacterales, Sph: Sphingomonadales, and Caul: Caulobacterales.
There are 106 α-proteobacteria in our sample, about one-fourth of which are intracellular pathogens belonging to the Rickettsiales. Figure 7 plots the frequency of proteins from 106 α-proteobacterial genomes appearing in the sister group to the eukaryotic genes (dark blue fields), how often each genome harbors a protein that does not branch as the eukaryote sister (light blue fields), or whether the gene is missing in the genome altogether (white fields). The α-proteobacterial strains with the highest frequency of occurrence in the sister group were Rhizobium NGR 234 (Rhizobiales, 38 times), Beijerinckia indica ATCC 9039 (Rhizobiales, 36 times), Acidiphilium cryptum JF-5 (Rhodospirillales, 35 times), Ruegeria pomeroyi DSS 3 (Rhodobacterales, 35 times), Sinorhizobium meliloti (Rhizobiales, 35 times), Azorhizobium caulinodans ORS 571 (Rhizobiales, 35 times), and Methylobacterium nodulans ORS 2060 (Rhizobiales, 35 times; for the complete list see supplementary Table S3, Supplementary Material online). Clearly, among those genes where an α-proteobacterial homologue resides in the eukaryote sister group, different genes implicate different ancestors of mitochondria within the α-proteobacteria, and each of the genomes is implicated as the sister of a eukaryotic nuclear gene at least once. It has been suggested that such patterns could reflect multiple origins of mitochondria (Georgiades and Raoult 2011). It is more likely however, in our view, that such patterns reflect a single origin of mitochondria followed by subsequent LGT among free-living prokaryotes (Martin 1999b; Richards and Archibald 2011).
Fig. 7.—
Distribution of α-proteobacterial groups found as sister group to the eukaryotic clade. These presence absence matrix gives the functional description of all 104 trees (as rows) where the α-proteobacteria (106 different species, as columns) were found as the eukaryotic sister clade. The color indicates whether a group was found as sister clade (deep blue) or was just present in the tree (light blue). Abbreviations of α-proteobacterial families as given in the legend to figure 6.
A Network Linking LECA to Prokaryotes
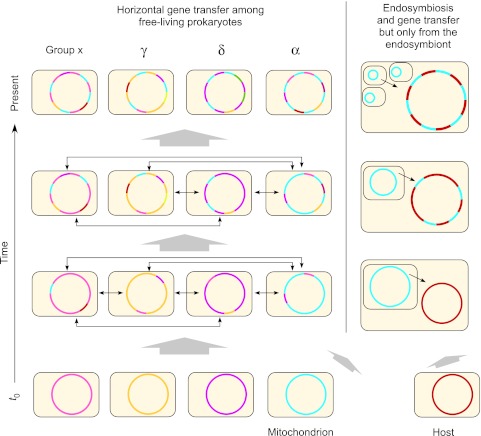
Based on the current sample of 994 genomes, 571 trees implicate many different prokaryotes as gene donors to the eukaryote common ancestor (fig. 7). In fact, the trees implicate all 22 prokaryote higher taxa sampled here are gene donors to LECA (figs. 2 and 4). Figure 8 summarizes those results in a network in which the weight of the edges connecting prokaryotes to LECA reflects the relative frequency of gene contribution to LECA by the respective lineage in the current sample. The eubacterial contributions are shaded blue, the archaebacterial contributions are shaded red, and the retention of genes from both sources in diversifying eukaryotic lineages is indicated accordingly. As in earlier studies (Esser et al. 2004; Rivera and Lake 2004; Dagan and Martin 2006; Pisani et al. 2007), the eubacterial contribution to eukaryotic genomes is quantitatively predominant, a crucial circumstance that is still too often overlooked (Gribaldo et al. 2010). These distinct contributions from archaebacteria and eubacteria require a network-based framework, rather than a tree-based framework, for addressing eukaryote origins. Under the simplest working hypothesis, the eubacterial and the archaebacterial contributions stem from only one cellular donor each, the eubacterial ancestor of mitochondria and its archaebacterial host, respectively. The (erroneous, in our view) implication of several different donor lineages stems merely from the natural workings of LGT among free-living prokaryotes subsequent to the origin of eukaryotes, as sketched in figure 9.
Fig. 9.—
Lateral gene transfer between free-living prokaryotes subsequent to the origin of organelles requires that we think at least twice when interpreting phylogenetic trees for genes that were acquired from mitochondria (or chloroplasts, not shown). Genes that entered the eukaryotic lineage via the genome of the mitochondrial endosymbiont represent a genome-sized sample of prokaryotic gene diversity that existed at the time that mitochondria arose. The uniformly colored chromosomes at t0 indicate that at the time of mitochondrial origin, there existed for individual prokaryotes specific collections of genes in genomes, much like we see for strains of Escherichia coli today. If an E. coli cell would become an endosymbiont today, it would not introduce an E. coli pangenome’s worth of gene diversity (some 18,000 genes) into its host lineage, rather it would introduce some 4,500 genes or so. The free-living relatives of that endosymbiont would go on reassorting genes across chromosomes via gene transfer at the pangenome (species and strain) level, at the genus level, at the family level, and at the level of proteobacteria, the environment, and so forth. After 1.5 billion years, it would be very unreasonable to expect any contemporary prokaryote to harbor exactly the same collection of genes as the original endosymbiont did. Instead, the descendant genes of the endosymbiont (labeled blue in the figure) would be dispersed about myriad chromosomes, and we would eventually find them one at a time through genome sequencing of individuals from different groups. Though not shown here, for reasons of space limitation, exactly the same process also applies, in principle, for the host’s genome. Redrawn from Martin (1999b) and from figure 5 of Rujan and Martin (2001).
In order for that explanation to be tenable, a considerable amount of LGT must have occurred for the genes under study among the ancestors of the groups sampled here. To see if there is evidence for that, we looked to see how often the prokaryotic groups in question were monophyletic across the 571 trees for which the eukaryotes were monophyletic. The result is shown in Table 1. The worst “LGT offenders” were the δ-proteobacteria, the firmicutes, and the γ-proteobacteria, which each were monophyletic groups in less than 10% of the trees studied. Aside from archaebacteria and α-proteobacteria, these three groups were also the largest apparent contributors to the functional classes in figure 4b.
Table 1.
Prokaryote Monophyly in Eukaryote Monophyly Trees
| Degree of Prokaryote Monophyly | |||
| Group | Stricta | Outerb | Innerc |
| Chlamydiae | 0.844 | 0.856 | 0.962 |
| Chlorobi | 0.672 | 0.695 | 0.893 |
| Deinococcus | 0.654 | 0.693 | 0.882 |
| Thermotogae | 0.579 | 0.635 | 0.851 |
| ε-Proteobacteria | 0.534 | 0.583 | 0.785 |
| Cyanobacteria | 0.473 | 0.557 | 0.760 |
| Crenarchaeota | 0.341 | 0.598 | 0.660 |
| Chloroflexi | 0.286 | 0.364 | 0.665 |
| Spirochaetes | 0.249 | 0.298 | 0.641 |
| β-Proteobacteria | 0.237 | 0.415 | 0.501 |
| Bacteroidetes | 0.214 | 0.334 | 0.642 |
| Euryarchaeota | 0.200 | 0.476 | 0.505 |
| α-Proteobacteria | 0.194 | 0.398 | 0.499 |
| Actinobacteria | 0.170 | 0.359 | 0.534 |
| Archaea, otherd | 0.092 | 0.159 | 0.486 |
| γ-Proteobacteria | 0.090 | 0.382 | 0.325 |
| Firmicutes | 0.080 | 0.310 | 0.345 |
| δ-Proteobacteria | 0.056 | 0.152 | 0.361 |
| Bacteria, other | 0.038 | 0.099 | 0.292 |
The proportion of trees in which the group is monophyletic.
The proportion of the members of the given group that are present in the tree and contained within the smallest clade containing all members of the group (and members of other groups); ngroup/nclade, where ngroup is the number of members of the group in the clade and nclade is the number of OTUs in that clade. Value shown is the mean across all trees.
The proportion of the members of the given group that are present in the tree and contained within the group’s largest monophyletic clade; ngroup{clade}/ngroup{tree}, where ngroup{clade} is the number of members of the group in the clade and ngroup{tree} is the number of group members in the tree. Value shown is the mean across all trees.
Designates a grouping of Nanoarchaea, Thaumarchaea, and Korarchaeota lumped together, the individual samples of which are either one or too small to consider monophyly.
Discussion
Single Ancient Acquisition, Not Continuous Influx
We identified and investigated 712 eukaryotic protein families that have prokaryotic homologues. Of those trees, 571 (80%) reflect a single origin for the eukaryotic gene. For the remaining 141 genes whose trees do not directly reflect a single origin, there are causes other than LGT from prokaryotes to eukaryotes that can readily account for the lack of observed eukaryote monophyly, the two most obvious of which are computational (phylogenetic parameters) and biological (differential loss). Examining alignment characteristics that are correlated with the inference of LGT from discordant branching, Roettger et al. (2009) found that the number of OTUs in the alignment (tree) was among the most highly correlated with LGT inference: the more OTUs in the alignment, the more likely the inference of LGT. The median number (± standard error) of OTUs in our 141 trees that did not recover eukaryote monophyly is 337 ± 179, which is significantly higher (P < 10−11) than the median value of 115 ± 193 for the 571 trees in which the eukaryotes comprised a monophyletic group. The large number of OTUs is potentially a biased source of alignment and phylogeny artefacts that could disrupt eukaryote monophyly.
Another mechanism that could readily produce the 141 cases of eukaryote non-monophyly is differential loss among paralogous gene families that were inherited as paralogs from the mitochondrial ancestor in LECA. About 50% of the genes in an average contemporary prokaryotic genome have duplicates within the genome (Hooper and Berg 2003). For our present considerations, it is immaterial whether the source of the duplicated prokaryotic gene is from within the chromosome or via lateral acquisition, although genome data argue in favor of the latter (Treangen and Rocha 2011). If the mitochondrial ancestor had a typical genome with about 4,000 genes, it would have then harbored about 2,000 genes existing within paralogous families, like Escherichia coli or Bacillus subtilis do (D’Antonio and Ciccarelli 2011). Transfer from a single source, for example the host or the mitochondrial ancestor, followed by differential loss within such gene families during eukaryote evolution (Zmasek and Godzik 2011), would produce non-monophyletic eukaryote trees in a manner that does not involve LGT.
At the same time, there are limits as to how many such patterns can be explained with differential loss only. If differential loss (instead of LGT) is invoked to explain the presence/absence patterns of all nonuniversally distributed genes, the Genome of Eden problem (Doolittle et al. 2003) ensues: inferred ancestral genome sizes become orders of magnitude larger than any observed contemporary prokaryotic genome, an untenable proposition (Dagan and Martin 2007). But for a mere 140 genes families the situation is less severe, especially given that the genome of the mitochondrial ancestor probably harbored on the order ∼1,000 gene families. Furthermore, at least eight ancient gene duplicate pairs (16 gene families) have been universally, or nearly so, conserved across prokaryotic genomes (Dagan et al. 2010). Thus, it would be unreasonable to assume that there was neither paralogy nor differential loss in the 20% fraction of trees where eukaryotes appeared non-monophyletic, especially given that 50% of a typical prokaryotic genome falls into intragenomic gene families.
Given eukaryote age, there have been ∼1.8 billion years (Parfrey et al. 2011) of opportunity for these eukaryote lineages to reacquire these 571 genes from prokarytes via LGT. But that has not happened, indicating that LGT from prokaryotes to eukaryotes—outside the context of endosymbiotic organelle origins—is rare in eukaryote evolution. It is certainly far more rare than LGT among prokaryotes in evolution. This is consistent with the lack of functional gene acquisition by aphids from Buchnera endosymbionts (Nikoh et al. 2010), despite more than 100 million years of intracellular coevolution. By contrast, at the origins of chloroplasts and mitochondria, gene transfers from the genomes of the respective endosymbionts and functional integration of those genes into the metabolism of the resulting cell were abundant (Timmis et al. 2004; Lane and Archibald 2008).
Much current thinking on eukaryote origins is still focused on debating the branching orders in alternative trees (Gribaldo et al. 2010): a tree x versus tree y debate. But a spectrum of alternatives that consider only trees is not broad enough. A considerable amount of evidence indicates that the process of eukaryote origins was not tree-like to begin with. Eukaryote genome evolution entails many non–tree-like processes, and these non–tree-like events (endosymbiosis and gene transfer) could be the decisive events in eukaryote evolution (Lane and Martin 2010; Koonin 2012). Gene origin and evolution in the eukaryotic tree of life has many tree-like components (Bapteste et al. 2009). But when the overall process of eukaryote (genome) evolution is set in the context of a realistic model of prokaryotic genome evolution, with abundant gene transfer among prokaryotes and occasional major influxes into eukaryote genomes via endosymbiosis and gene transfers from organelles, the non-treelike evolutionary events (Lane and Archibald 2008; McInerney et al. 2008) stand out—they are components of eukaryote genome evolution that do not fit on a tree. They require network approaches.
Many Theories and Many Trees
Among the 571 trees that recovered eukaryote monophyly, the higher prokaryotic taxa harboring a gene with a sister group relationship to the eukaryotic nuclear homologue are shown in figure 2. These genes could provide evidence to discriminate between different current theories for eukaryote origin. We start with theories that received the least support.
One theory has it that the eukaryotic lineage is of equal age as the two prokaryotic domains (Kurland et al. 2006); it predicts that we should mainly obtain a topology of three monophyletic domains among our trees. The three-domain tree was however observed in only three cases out of 571 (0.5% of all trees): the 60s ribosomal protein L2/L8 (KOG2309), the 40S ribosomal protein S16 (KOG1753), and the large subunit of RNA polymerase III (KOG0261). Gribaldo et al. (2010) argued that the three-domain tree is correct, but reported no new analyses to test that view and considered only a specific subset of genes—those that specifically link eukaryotes and archaebacteria and hence fit the metaphor of a tree. In doing so, they disregared the eubacterial majority of genes in eukaryotic genomes. Among theories considered here, the three-domain tree received the least support. The next lowest rung on the ladder of support is occupied by the theory that the eukaryotic nucleus arose within an endospore forming Gram-positive bacterium (Gould and Dring 1979), in which case eukaryotes should branch with firmicutes, which was observed in only 11 trees (fig. 2).
That is followed by theories that entail the planctomycetes (the PVC group) as intermediate steps in the prokaryote-to-eukaryote transition (Devos and Reynaud 2010) or as the host for an endosymbiotic origin of the nucleus (Forterre 2011). PVC sisterhood is observed in 15 trees (2.6%). That is the same level of support as current versions of the neomuran theory receive (Cavalier-Smith 2002) because actinobacteria branched as eukaryote sisters in 15 trees. The theory of the late Lynn Margulis that spirochaetes were crucial to eukaryote origin via the simultaneous origin of eukaryotic flagella and the nucleus (Margulis et al. 2006) fared incrementally better, with 17 trees pegging spirochaetes as eukaryotic sisters (fig. 2). Better still fared the original version of the neomuran theory (Cavalier-Smith 1975) with eukaryotes viewed as direct descendants of cyanobacteria (19 trees).
Some theories have it that the nucleus arose as an archaebacterial endosymbiont in a Gram-negative host (Gupta and Golding 1996), in some formulations a γ-proteobacterial host (Horiike et al. 2004). The corresponding γ-proteobacterial sisterhood is observed in 20 trees. Related theories have it that the host for an endosymbiotic origin of the nucleus was a δ-proteobacterium (Moreira and Lopez-Garcia 1998), a topology that is observed for 25 genes (fig. 2). Although an endosymbiotic theory for the origin of the nucleus belongs to the very first formulations of endosymbiotic theory (Mereschkowsky 1905, 1910), there are a number of fundamental and serious problems with the view that the nucleus was ever free-living prokaryote (Martin 1999a; Cavalier-Smith 2002).
Several modern formulations of endosymbiotic theory that posit only two cells at eukaryote origin, an archaebacterial host and a mitochondrial endosymbiont (Searcy 1992; Martin and Müller 1998; Vellai et al. 1998). One formulation of endosymbiotic theory entailing a prokaryotic host posits mass transfer of genes from the genome of the mitochondrial endosymbiont to the chromosomes of the host, while directly accounting for the common ancestry of mitochondria and hydrogenosomes (Martin and Müller 1998), and an autogenous origin of the nucleus in a mitochondrion-bearing cell, mechanistically precipitated via the invasion of Group II introns from the symbiont into the host’s chromosomes and their transition there to spliceosomal introns (Martin and Koonin 2006). This is supported by the most frequent class of eubacterial sisterhood observed was for α-proteobacteria (37 genes; fig. 2).
The archaebacterial genomes sampled revealed some cases of eukaryotic sisterhood for Nanoarchaea (Huber et al. 2002) and Korarchaeota (Elkins et al. 2008), which have so far not been implicated in eukaryote origins, as well as more frequent sisterhood for mesophilic crenarchaeotes currently called thaumarchaeotes (24 trees), crenarchaeotes (44 trees), and euryarchaeotes (77 trees), which have (Embley and Martin 2006; Cox et al. 2008; Kelly et al. 2011). Because of imbalanced lineage sampling, the data do not speak unambiguously in favor on any particular theory. Nevertheless, the distribution of signals in figure 4 is more in line with the prediction of an “archaebacterial nature of the eukaryotic genetic apparatus and a eubacterial ancestry of eukaryotic energy metabolism” (Martin and Müller 1998) than with the predictions of other theories.
Some might take the sisterhood frequency of δ-proteobacterial genes to eukaryotic homologues as evidence in favor of a participation of δ-proteobacteria at eukaryote origins, but the same logic would then have to be applied to γ-proteobacterial genes, actinobacterial genes, cyanobacterial genes, spirochaete genes, and so forth; the various theories for the origin of eukaryotes that generate those predictions cannot all be simultaneously correct. The simplest interpretation in our view is that shown in figure 9. Particularly with regard to δ-proteobacterial sisterhood, we point out that in gene sharing networks of proteobacteria, the frequency of lateral gene sharing between δ-proteobacteria and α-proteobacteria is higher than within α-proteobacteria themselves (Kloesges et al. 2011), such that gene transfer among prokaryotes prior to, and subsequent to, the origin of mitochondria could readily account for the observation. In that sense, the mitochondrion remains a plausible alternative as the sole biological source of oddly branching eubacterial genes in the genome of the eukaryotic ancestor, one requiring little in the way of corollary assumptions—all we have to assume is the gene transfer among prokaryotes has always been more or less like it is today, and we do not have to assume additional cellular partners whose ribosomes disappear. Eukaryotes (that lack plastids) possess only two kinds of ribosomes: archaebacetrial ribosomes in the cytosol and eubacterial ribosomes in the mitochondrion. Theories that posit cellular partners other than the mitochondrion and its host have to account for the disappearance of the additional genomes and ribsomes, and why the data should tend to support one partner (a δ-proteobacterium for example) over another (a γ-proteobacterium or spirochaete) even though the competing alternative signals are more or less equally strong.
Too Many Inferred Cells and Donor Lineages
In the literature on endosymbiosis and gene transfer from organelles to the nucleus, it is commonplace to speak about “eukaryotic genes of α-proteobacterial origin.” But the taxonomic or lineage designation “α-proteobacterial” is in fact very problematic, and perhaps even more arbitrary that problematic. In the context of eukaryote gene origins, most readers will associate “α-proteobacterial” with “mitochondrial,” and attribution of a gene origin to a cellular partner is uncontroversial; the existence of a donor cell is inferred from an observation in a phylogenetic tree. The implicit reasoning is: per donor lineage identified, add one cellular partner. That is seemingly unproblematic for α-proteobacteria (or cyanobacteria for plastids), but by that measure, we would infer 22 different prokaryotic cells (including a cell from the phylum “other”) at the origin of eukaryotes on the basis of the present data. The participation of 22 different cells to construct LECA and then no subsequent additions for the next ∼1.8 billion years for the genes and lineages sampled here are not likely in our view, though not prohibitively complex as an idea, if we think openly. But the more subtle problem lies elsewhere: the arbitrary level at which we define a lineage from which the cell is to be inferred.
Namely, if we alter the level of taxonomic specificity with which we describe in figure 2, we will infer fewer or more numerous cells participating at eukaryote origin as endosymbionts and hosts. For example, if we were to increase the taxonomic resolution for our designation/definition of a “donor lineage” to the level of prokaryotic families, then we would infer 148 different donor lineages (i.e., how many families there are in our sample whose genes populate eukaryote sister groups in our trees) and hence 148 different prokaryotic cells in symbiotic association at eukaryote origin, given the present sample. Or we can take things one step further: at the level of genera and species, the numbers increase to 349 cells and 768 cells participating in the origin of the eukaryote common ancestor, respectively. And as the sample of sequenced genomes grows over time, so will the number of inferred donors to LECA.
Thus, that avenue of interpretation (one cell per lineage) is clearly problematic and leads to chaos because of the arbitrariness of choosing or defining the taxonomic level at which to seek or find a donor lineage. One solution is to simply zoom out in terms of taxonomic resolution, and conceptually operate at the level of domains, in which case we would conveniently have one eubacterium (the mitochondrial endosymbiont) and one archaebacterium (the host) implicated at eukaryote origins. That would solve the “one cell inferred per lineage identified” conundrum, but it is only half of the problem. The other half concerns the concept of a prokaryotic “lineage” in the context of the amount of geological time (about 1.8 billion years) that the fossil record implores us to keep in mind when considering eukaryote origins.
In terms of how genes behave in chromosomes over time, there are two ways to think about prokaryote lineages: they have static chromosomes that are immune to LGT and differences in gene content across members of a lineage are generated only by gene loss or they have fluid chromosomes with genes coming into exiting genomes of members of the “lineage” over time. The latter fluid chromosome view has recently been presented in more generalized form as the public goods hypothesis for prokaryotic genes (McInerney, Pisani, et al. 2011). Readers familiar with prokaryote chromosome evolution will immediately complain that the static chromosome model is unrealistic and outdated, but it is very real and manifest—but usually implicit—in literature concerning eukaryote gene origins. Clearly, what we consider to be a “donor lineage” at eukaryote origin depends on the level of taxonomic resolution chosen to represent a “lineage” in our prokaryotic survey.
For example, if we go to the level of species or strains, and adhere to the concept of a static prokaryote chromosome model (that is if we neglected LGT among prokaryotes over geological time, which we do not), we would conclude that the most potent donor of genes to the common ancestor of the eukaryotic lineage was the creanarchaeon (thaumarchaeote) Nitrosopumilus maritimus strain SCM1, which appears in the clade adjacent to the eukaryote gene 54 times, 3 time more than the next most potent apparent donor, the korarchaeon Candidatus korarchaeum cryptofilum strain OPF8. But the origin of eukaryotes occurred some ∼1.8 billion years ago (Parfrey et al. 2011). If we were assuming a static chromosome model and thus claiming (which we are not) that specific strains of prokaryotes served as donors of eukaryotic genes ∼1.8 billion years ago, then we would be assuming (which we do not) that Nitrosopumilus maritimus SCM1, defined by the specific collection of 1,795 genes in its genome, existed ∼1.8 billion years ago. Implicitly, we would then also be assuming (which we do not) that all the other species and strains in the present study also existed in their modern form ∼1.8 billion years ago.
The cogent reader would immediately, and rightly, protest that no contemporary prokaryotic strain with its current specific collection of genes could have existed ∼1.8 billion years ago. This is all the more evident in light of gene content differences among E. coli strains, a well-studied case. In 61 sequenced E. coli genomes, there are about 18,000 genes, only about 4,500 of which occur in any individual strain (Lukjancenko et al. 2010). Each new sequenced strain uncovers a new combination of genes present in the E. coli pan-genome and about 200 ORFs new to the species pan-genome. The E. coli core genome (present in all 61 sequenced strains) is currently ≤1,000 genes, and no E. coli genome harbors more than about 25% of all 18,000 genes found in the species (Lukjancenko et al. 2010). If we move up the taxonomic scale to the level of γ-proteobacteria, the problem gets worse: A sample of 157 γ–proteobacteria (subclass) was found to harbor 40,327 different gene families (Kloesges et al. 2011), and a sample of 329 proteobacteria (class or phylum) was found to harbor about 75,000 different genes (Kloesges et al. 2011). An average individual proteobacterium only has about 3,000–4,000 genes and none has more than 9,703, the number found in Sorangium cellulosum So ce 56. The difference between ∼75,000 genes present in proteobacteria and ∼3,000 present in a given strain is not attributable to differential loss from a proteobacterial ancestor that had 75,000 genes, but it is readily attributable to gene flow (LGT) among individuals and individual strains of proteobacteria and among proteobacteria with other prokaryotes (Doolittle 1999; Doolittle et al. 2003; Dagan et al. 2008).
Because of the foregoing considerations, reasoning along the lines of “one cell inferred per donor lineage identified” is misleading, mainly because if we are talking about genes in genomes (which we are), the word “lineage” does not have a well-specified meaning in a context where we try to relate collections of genes present in individual prokaryote genomes that existed ∼1.8 billion years ago to genes present in individual modern sequenced genomes. Nonetheless, there is a current trend in the literature of inferring cells where a few genes give unexpected trees, the invisible “chlamydial” sidekick at plastid origins being perhaps the most prominent example (Huang and Gogarten 2007; Becker et al. 2008; Moustafa et al. 2008). But that line of argumentation will necessarily subside at some point, for two reasons. First, it will not lead to any form of convergence as more genomes become sampled. On the contrary, with increased sampling of prokaryotic genomes for reference comparisons, the phylogenetic “identity” of donor lineages to the eukaryotic common ancestor will continue to change and will furthermore continue to spread out across more prokaryotic genomes. Second, and more severely, if one looks for a few genes that suggest a chamydial partner one will find them, but if one looks for a few genes that suggest a spirochaete partner, one will find them too, and a clostridial partner to boot, and so forth. Because eukaryotes have so many genes, if we look for a particular phylogenetic pattern, the chances are that we will find it at a low frequency in eukaryote genomes by chance alone (Stiller 2011), but we need to look at all the genes in eukaryote genomes, not just ad hoc gene samples that support a particular story. In phylogenomics, we need to keep random phylogenetic error in mind (Stiller 2011), and the interpretations of the data need to encompass gene transfer among prokarytes because it is a very real component of microbial evolution.
All things being equal, if our present considerations are approximately on target, as sampling improves the end result might tend to asymptotically approach one apparent prokaryotic donor chromosome per eukaryotic gene, even if—as we maintain—only two cells, a mitochondrial endosymbiont and its archaebacterial host, each with a discrete and specific collection of genes, participated at eukaryote origin. As sampling improves, a more realistic, temporally dynamic prokaryote lineage concept with fluid genomes and genes as public goods will figure more widely into thinking on eukaryote gene origins. Because genes that contributed to the eukaryote common ancestor lineage have different individual histories, networks, rather that trees alone, are integral to the study of eukaryote origins, and the explanatory context needs to recognize the importance of endosymbiosis and gene transfer in evolution.
Supplementary Material
Supplementary tables and figures are available at Genome Biology Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank the European Research Council for financial support (NETWORKORIGINS grant number 232975) and James O. McInerney for helpful comments on the text.
References
- Abhishek A, Bavishi A, Choudhary M. Bacterial genome chimaerism and the origin of mitochondria. Can J Microbiol. 2011;57:49–61. doi: 10.1139/w10-099. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atteia A, et al. Pyruvate formate-lyase and a novel route of eukaryotic ATP-synthesis in anaerobic Chlamydomonas mitochondria. J Biol Chem. 2006;281:9909–9918. doi: 10.1074/jbc.M507862200. [DOI] [PubMed] [Google Scholar]
- Atteia A, et al. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-proteobacterial mitochondrial ancestor. Mol Biol Evol. 2009;29:1533–1548. doi: 10.1093/molbev/msp068. [DOI] [PubMed] [Google Scholar]
- Bapteste E, et al. Prokaryotic evolution and the tree of life are two different things. Biol Direct. 2009;4:34. doi: 10.1186/1745-6150-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Hoef-Emden K, Melkonian M. Chlamydial genes shed light on the evolution of photoautotrophic eukaryotes. BMC Evol Biol. 2008;8:203. doi: 10.1186/1471-2148-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindefalk B, Ettema TJG, Viklund J, Thollesson M, Andersson SG. A phylometagenomic exploration of oceanic alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade. PLoS One. 2011;6:e24457. doi: 10.1371/journal.pone.0024457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann H, Martin W. Higher-plant chloroplast and cytosolic 3-phosphoglycerate kinases: a case of endosymbiotic gene replacement. Plant Mol Biol. 1996;30:65–75. doi: 10.1007/BF00017803. [DOI] [PubMed] [Google Scholar]
- Brown JR, Doolittle WF. Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The origin of nuclei and of eukaryotic cells. Nature. 1975;256:463–468. doi: 10.1038/256463a0. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton JA, McInerney JO. Eukaryotic genes of archaebacterial origin are more important than the more numerous eubacterial genes, irrespective of function. Proc Natl Acad Sci U S A. 2010;107:17252–17255. doi: 10.1073/pnas.1000265107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci U S A. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc Natl Acad Sci U S A. 2007;104:870–875. doi: 10.1073/pnas.0606318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Roettger M, Bryant D, Martin W. Genome networks root the tree of life between prokaryotic domains. Genome Biol Evol. 2010;2:379–392. doi: 10.1093/gbe/evq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Antonio M, Ciccarelli DF. Modification of gene duplicability during the evolution of protein interaction network. PloS Comp Biol. 2011;7:e1002029. doi: 10.1371/journal.pcbi.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deusch O, et al. Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol Biol Evol. 2008;25:748–761. doi: 10.1093/molbev/msn022. [DOI] [PubMed] [Google Scholar]
- Devos DP, Reynaud EG. Evolution. Intermediate steps. Science. 2010;330:1187–1188. doi: 10.1126/science.1196720. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Genes in pieces: were they ever together? Nature. 1978;272:581–582. [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Bapteste E. Pattern pluralism and the tree of life hypothesis. Proc Natl Acad Sci U S A. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, et al. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos Trans R Soc Lond B Biol Sci. 2003;358:39–58. doi: 10.1098/rstb.2002.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins JG, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci U S A. 2008;105:8102–8107. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Embley TM, et al. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life. 2003;55:387–395. doi: 10.1080/15216540310001592834. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Martin W, Dagan T. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol Lett. 2007;3:180–184. doi: 10.1098/rsbl.2006.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, et al. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- Forterre P. Thermoreduction, a hypothesis for the origin of prokaryotes. C R Acad Sci III. 1995;318:415–422. [PubMed] [Google Scholar]
- Forterre P. A new fusion hypothesis for the origin of Eukarya: better than previous ones, but probably also wrong. Res Microbiol. 2011;162:77–91. doi: 10.1016/j.resmic.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S. Bacteria with a eukaryotic touch: a glimpse of ancient evolution? Proc Natl Acad Sci U S A. 2010;107:12739–12740. doi: 10.1073/pnas.1007720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, Huynen MA. Reconstruction of the proto-mitochondrial metabolism. Science. 2003;301:609–609. doi: 10.1126/science.1085463. [DOI] [PubMed] [Google Scholar]
- Georgiades K, Raoult D. The rhizome of Reclinomonas americana, Homo sapiens, Pediculus humanus and Saccharomyces cerevisiae mitochondria. Biol Direct. 2011;6:55. doi: 10.1186/1745-6150-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould GW, Dring GJ. Possible relationship between bacterial endospore formation and the origin of eukaryotic cells. J Theoret Biol. 1979;81:47–53. doi: 10.1016/0022-5193(79)90079-1. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. The origin of eukaryotes and their relationship with the Archaea: are we a phylogenomic impasse? Nat Rev Microbiol. 2010;8:743–752. doi: 10.1038/nrmicro2426. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Golding GB. The origin of the eukaryotic cell. Trends Biochem Sci. 1996;21:166–171. [PubMed] [Google Scholar]
- Hampl V, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups.”. Proc Natl Acad Sci U S A. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89:10915–101919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SD, Berg OG. On the nature of gene innovation: duplication patterns in microbial genomes. Mol Biol Evol. 2003;20:945–954. doi: 10.1093/molbev/msg101. [DOI] [PubMed] [Google Scholar]
- Horiike T, Hamada K, Miyata D, Shinozawa T. The origin of eukaryotes is suggested as the symbiosis of Pyrococcus into γ-proteobacteria by phylogenetic tree based on gene content. J Mol Evol. 2004;59:606–619. doi: 10.1007/s00239-004-2652-5. [DOI] [PubMed] [Google Scholar]
- Huang J, Gogarten JP. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 2007;8:R99. doi: 10.1186/gb-2007-8-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, et al. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, Wickstead B, Gull K. Archaeal phylogenomics provides evidence in support of a methanogenic origin of the Archaea and a thaumarchaeal origin for the eukaryotes. Proc R Soc Lond B Biol Sci. 2011;278:1009–1018. doi: 10.1098/rspb.2010.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloesges T, Martin W, Dagan T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 2011;28:1057–1074. doi: 10.1093/molbev/msq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009;37:1011–1034. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. The logic of chance: the nature and origin of biological evolution. Upper Saddle River (NJ): FT Press; 2012. [Google Scholar]
- Kurland CG, Andersson SG. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 2000;64:786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryotic cells. Science. 2006;312:1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- Lake JA. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 1988;331:184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- Lake JA, Rivera MC. Was the nucleus the first endosymbiont? Proc Natl Acad Sci U S A. 1994;91:2880–2881. doi: 10.1073/pnas.91.8.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landan G, Graur D. Heads or tails: a simple reliability check for multiple sequence alignments. Mol Biol Evol. 2007;24:1380–1383. doi: 10.1093/molbev/msm060. [DOI] [PubMed] [Google Scholar]
- Lane CE, Archibald JM. The eukaryotic tree of life: endosymbiosis takes its TOL. Trends Ecol Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Lane N. Life ascending: the ten greatest inventions of evolution. London: Profile Books; 2009. 344 p. [Google Scholar]
- Lane N. Energetics and genetics across the prokaryote-eukaryote divide. Biol Direct. 2011;6:e35. doi: 10.1186/1745-6150-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- Langer D, Hain J, Thuriaux P, Zillig W. Transcription in Archaea: similarity to that in Eukarya. Proc Natl Acad Sci U S A. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart PJ, Steel MA, Hendy MD, Penny D. Recovering evolutionary trees under a more realistic evolutionary model. Mol Biol Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- Lukjancenko O, Wassenaar TM, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microbiol Ecol. 2010;60:708–720. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol. 2010;8:731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, Chapman M, Guerrero R, Hall J. The last eukaryotic common ancestor (LECA): acquisition of cytoskeletal motility from aerotolerant spirochetes in the Proterozoic Eon. Proc Natl Acad Sci U S A. 2006;103:13080–13085. doi: 10.1073/pnas.0604985103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Martin W. A briefly argued case that mitochondria and plastids are descendants of endosymbionts, but that the nuclear compartment is not. Proc R Soc Lond B Biol Sci. 1999a;266:1387–1395. [Google Scholar]
- Martin W. Mosaic bacterial chromosomes—a challenge en route to a tree of genomes. BioEssays. 1999b;21:99–104. doi: 10.1002/(SICI)1521-1878(199902)21:2<99::AID-BIES3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Martin W. Archaebacteria (Archaea) and the origin of the eukaryotic nucleus. Curr Opin Microbiol. 2005;8:630–637. doi: 10.1016/j.mib.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Martin W, Koonin EV. Introns and the origin of nucleus–cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red algae Cyanidioschizon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- Maynard-Smith J, Szathmáry E. The major transitions in evolution. Oxford: Oxford University Press; 1995. 346 p. [Google Scholar]
- McInerney JO, Cotton JA, Pisani D. The prokaryotic tree of life: past, present, and future? Trends Ecol Evol. 2008;23:276–281. doi: 10.1016/j.tree.2008.01.008. [DOI] [PubMed] [Google Scholar]
- McInerney JO, Pisani D, Bapteste E, O'Connell MJ. The public goods hypothesis for the evolution of life on Earth. Biol Direct. 2011;6:41. doi: 10.1186/1745-6150-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney JO, et al. Planctomycetes and eukaryotes: a case of analogy not homology. BioEssays. 2011;33:810–817. doi: 10.1002/bies.201100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereschkowsky C. Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol Centralbl. 1905;25:593–604. [English translation in Martin W, Kowallik KV. 1999. Eur J Phycol. 34:287–295.] [Google Scholar]
- Mereschkowsky C. Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis, einer neuen Lehre von der Entstehung der Organismen. Biol Centralbl. 1910;30 278–288, 289–303, 321–347, 353–367. [Google Scholar]
- Moreira D, Lopez-Garcia P. Symbiosis between methanogenic archaea and δ-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J Mol Evol. 1998;47:517–530. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- Moustafa A, Reyes-Prieto A, Bhattacharya D. Chlamydiae has contributed at least 55 genes to plantae with predominantly plastid functions. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. Anaerobic acclimation in Chlamydomonas reinhardtii—anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem. 2007;282:25475–25486. doi: 10.1074/jbc.M701415200. [DOI] [PubMed] [Google Scholar]
- Nikoh N, et al. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 2010;6:e1000827. doi: 10.1371/journal.pgen.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey WP, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, Rosati G, Ludwig W, Schleifer KH. Coexistence of tubulin and ftsZ in different Prosthecobacter species. Mol Biol Evol. 2007;24:1439–1442. doi: 10.1093/molbev/msm069. [DOI] [PubMed] [Google Scholar]
- Pisani D, Cotton JA, McInerney JO. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol. 2007;24:1752–1760. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal barriers and bypasses to lateral gene traffic among sequenced prokaryote genomes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruit KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richards TA, Archibald JM. Gene transfer agents and the origin of mitochondria. Curr Biol. 2011;21:R112–R114. doi: 10.1016/j.cub.2010.12.036. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci U S A. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- Roettger M, Martin W, Dagan T. A machine-learning approach reveals that alignment properties alone can accurately predict inference of lateral gene transfer from discordant phylogenies. Mol Biol Evol. 2009;26:1931–1939. doi: 10.1093/molbev/msp105. [DOI] [PubMed] [Google Scholar]
- Rujan T, Martin W. How many genes in Arabidopsis come from cyanobacteria? An estimate from 386 protein phylogenies. Trends Genet. 2001;17:113–120. doi: 10.1016/s0168-9525(00)02209-5. [DOI] [PubMed] [Google Scholar]
- Schnarrenberger C, Martin W. Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants: a case study of endosymbiotic gene transfer. Eur J Biochem. 2002;269:868–883. doi: 10.1046/j.0014-2956.2001.02722.x. [DOI] [PubMed] [Google Scholar]
- Searcy DG. Origins of mitochondria and chloroplasts from sulphur-based symbioses. In: Hartman H, Matsuno K, editors. The origin and evolution of the cell. Singapore: World Scientific; 1992. pp. 47–78. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stiller J. Experimental design and statistical rigor in phylogenomics of horizontal and endosymbiotic gene transfer. BMC Evol Biol. 2011;11:259. doi: 10.1186/1471-2148-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic respective on protein families. Science. 1997;24:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep. 2011;1:e13. doi: 10.1038/srep00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielens AG, Rotte MC, van Hellemond JJ, Martin W. Mitochondria as we don’t know them. Trends Biochem Sci. 2002;27:564–572. doi: 10.1016/s0968-0004(02)02193-x. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Rocha EPC. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PloS Genet. 2011;7:e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J Eukaryot Microbiol. 2009;56:221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs K, Vida G. A new aspect to the origin and evolution of eukaryotes. J Mol Evol. 1998;46:499–507. doi: 10.1007/pl00006331. [DOI] [PubMed] [Google Scholar]
- Wagner M, Horn M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotech. 2006;17:41–49. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. The deep archaeal roots of eukaryotes. Mol Biol Evol. 2008;25:1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek CM, Godzik A. Strong functional patterns in the evolution of eukaryotic genomes revealed by the reconstruction of ancestral protein domain repertoires. Genome Biol. 2011;12:R4. doi: 10.1186/gb-2011-12-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]