Abstract
The functional repertoire of long intergenic noncoding RNA (lincRNA) molecules has begun to be elucidated in mammals. Determining the biological relevance and potential gene regulatory mechanisms of these enigmatic molecules would be expedited in a more tractable model organism, such as Drosophila melanogaster. To this end, we defined a set of 1,119 putative lincRNA genes in D. melanogaster using modENCODE whole transcriptome (RNA-seq) data. A large majority (1.1 of 1.3 Mb; 85%) of these bases were not previously reported by modENCODE as being transcribed. Significant selective constraint on the sequences of these loci predicts that virtually all have sustained functionality across the Drosophila clade. We observe biases in lincRNA genomic locations and expression profiles that are consistent with some of these lincRNAs being involved in the regulation of neighboring protein-coding genes with developmental functions. We identify lincRNAs that may be important in the developing nervous system and in male-specific organs, such as the testes. LincRNA loci were also identified whose positions, relative to nearby protein-coding loci, are equivalent between D. melanogaster and mouse. This study predicts that the genomes of not only vertebrates, such as mammals, but also an invertebrate (fruit fly) harbor large numbers of lincRNA loci. Our findings now permit exploitation of Drosophila genetics for the investigation of lincRNA mechanisms, including lincRNAs with potential functional analogues in mammals.
Keywords: long intergenic noncoding RNAs, modENCODE, transcriptional regulation, evolution, development
Introduction
Large-scale cDNA collections (e.g., Carninci et al. 2005), genome-wide tiling array experiments (Johnson et al. 2005), and whole transcriptome shotgun sequencing (RNA-seq) experiments (Cloonan et al. 2008; Guttman et al. 2010; Cabili et al. 2011) have demonstrated substantial transcriptional activity emanating from sequence lying between protein-coding genes in mammalian genomes. Transcription from these intergenic loci gives rise to several thousand long (>200 bp) intergenic noncoding RNAs (lincRNAs) in mouse, each apparently without protein-coding capability. Mammalian lincRNAs have been shown to regulate gene transcription (reviewed in Ponting et al. 2009; Wilusz et al. 2009) and to contribute to a variety of other cellular functions (reviewed in Prasanth and Spector 2007). For example, the imprinted lincRNA Airn downregulates the expression of the Igf2r gene cluster using a cis-regulatory mechanism (Braidotti et al. 2004), whereas Malat-1 regulates the expression of genes involved in synaptic function (Bernard et al. 2010) and influences alternative splicing through its interaction with splicing factor proteins in the nucleus (Tripathi et al. 2010). Nevertheless, because expression of lincRNA loci is typically low relative to protein-coding genes and because the molecular functions of most lincRNAs remain to be established, there has been considerable debate in the literature concerning their biological importance and molecular mechanisms (Mattick 2003; Hüttenhofer et al. 2005; van Bakel et al. 2010; Clark et al. 2011). Evidence of lincRNA functionality will be most compelling if disruption of loci frequently results in reproducible cellular or organismal phenotypes. However, with mouse as a model organism, only a handful of lincRNA loci, when disrupted, have thus far resulted in overt phenotypes (Ponting and Belgard 2010).
Rapid experimental investigation of lincRNA loci on a more genome-wide scale will require application of a cheaper and more amenable genetic organism than mouse, such as the fruit fly Drosophila melanogaster, which has many benefits for evolutionary and experimental investigations of lincRNA loci. Unlike the large mammalian genomes, which are replete in neutrally evolving and thus functionally inert sequence (Ponting 2008), Drosophila species have a compact 120 Mb genome (Adams et al. 2000), the majority of which appears to be functional (Sella et al. 2009) with half of all noncoding DNA exhibiting evidence of strong purifying selection (Andolfatto 2005). An analysis of D. melanogaster lincRNAs should therefore benefit from substantially greater power to detect evolutionary signatures of functionality than previous analyses in mammals.
Only a handful of lincRNAs have been individually investigated in detail in D. melanogaster, such as roX1, roX2, Hsr, pgc, bxd, αγ-element, iab-4, and bft (Tupy et al. 2005). LincRNAs have long been known to be transcribed from the bithoraxoid region (bxd) of the Ultrabithorax (Ubx) domain (Lipshitz et al. 1987) and have since been suggested to activate Ubx expression by recruiting the epigenetic regulator Ash1 (Sanchez-Elsner et al. 2006), whereas roX1 and roX2 may be analogues of the mammalian Xist transcript (Park et al. 2002). First attempts to identify lincRNAs on a genome-wide scale identified fewer than 150 of such loci, which is likely due to their requirements for lincRNAs to possess either a conserved intron/exon structure (Hiller et al. 2009) or to be supported by full-length cDNA sequence (Inagaki et al. 2005). Nevertheless, up to 5,000 ncRNA loci (of any length, not necessarily >200 bp) have been suggested to be present in the D. melanogaster genome (Li et al. 2009).
The modENCODE consortium recently reported 1,938 new transcribed regions (NTRs), detected using tiling arrays and RNA-seq analysis of total RNA and polyA+ samples, for 30 different developmental time points sampled across the D. melanogaster life cycle (Graveley et al. 2011). The data generated are of greater sequencing depth, and are more comprehensive of diverse developmental stages, than data sets from any other animal species. Large proportions of these NTRs are not linked to previously annotated gene models, but almost 33% contain an open reading frame (ORF) exceeding 100 codons and 42% overlap with previously known genes.
RNA-seq allows the sensitive detection of lowly expressing transcripts (Wang et al. 2009) and does not depend on current gene annotations. It is thus ideal for detecting novel transcripts, including lincRNAs (Wilhelm et al. 2010). Using the large RNA-seq data set produced by modENCODE (Graveley et al. 2011), we adopted a read mapping strategy that specifically enriches for lowly expressed splice junctions to determine the number, expression level, developmental regulation, and genomic complexity of lincRNA loci. Our study did not rely on previously defined loci thereby allowing protein-coding and lincRNA transcripts to be defined using identical criteria, making direct comparisons between them possible.
In this study, we describe the identification of 1,119 D. melanogaster lincRNAs. Only 15% of these lincRNA locus sequences overlap NTRs reported by modENCODE (Graveley et al. 2011). We report that these Drosophila lincRNAs exhibit substantially reduced rates of substitution and insertion–deletion mutations, temporal variations in expression, and a tendency to be transcribed in the vicinity of protein-coding genes involved in development. We also identify 42 pairs of D. melanogaster and mouse lincRNA loci whose locations relative to neighboring orthologous genes are similar. These positional equivalent loci represent the best candidates for lincRNA loci that have been conserved across diverse animal phyla.
Materials and Methods
Data Source
RNA-seq reads, generated from the modENCODE project (http://www.modencode.org/) from 30 developmental time points (Graveley et al. 2011), were acquired from the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/sra?term=srp001065). Each sequencing run was available as a single FASTQ file or as two linked files for paired-end reads. Developmental stages and numbers of reads mapped for each stage are summarized in supplementary table 2 (Supplementary Material online).
Short-Read Assembly
We mapped these sequences onto the D. melanogaster reference genome assembly (build 5.3) separately for each developmental time point data set. These sequences were then assembled into gene models using a procedure summarized in supplementary figure 1 (Supplementary Material online).
Both pairs of each paired-end read were mapped separately using Bowtie (Langmead et al. 2009). This allowed the mean and standard deviation of the insert size for paired-end reads to be calculated for each sequencing run. This information was required for later mapping stages using TopHat (Trapnell et al. 2009).
The 5′ and 3′ positions of splice junctions were mapped separately for each sequencing run (whether single- or paired-ended) using TopHat. This program was provided with D. melanogaster splice junctions from FlyBase release 5.27 gene annotations (Tweedie et al. 2009) and from a set of candidate lincRNAs previously defined using publicly available intergenic D. melanogaster expressed sequence tag (EST) sequences (Young RS, unpublished data). To exclude putative intergenic transcripts that represent unannotated exons of proximal protein-coding genes, we defined raw junctions (option j for TopHat) as the adjacent end points of neighboring EST-defined lincRNA loci and FlyBase genes. This directs TopHat to seek reads that span 5′ and 3′ positions of previously unannotated splice junctions. All other options were left at default values. RPKM (reads per kilobase of exon model per million mapped reads) values were calculated for each FlyBase-defined gene model for each sequencing run. This was achieved by dividing the number of reads mapping to a particular gene by the length of the gene and the total number of reads mapped in that run. Splice junctions that were newly identified from one or more sequencing runs but the same cDNA library were collated and appended to the previous raw junctions prior to a second remapping of reads using TopHat (with all other parameters held constant). This allowed TopHat to identify reads in one sequencing run which supported a splice junction found in a separate run but which previously had insufficient reads to be called. A single RPKM value was then calculated for each FlyBase gene model using reads from all sequencing runs for that cDNA library. Splice junctions called for each cDNA library and for each individual developmental time point were collected together and added to the raw junctions defined by neighboring FlyBase genes and EST-defined lincRNA loci. All reads from this time point were then mapped for a third and final time using TopHat. This allowed reads in one cDNA library to now support a splice junction found in a separate library. The consistency of this mapping procedure 1) across sequencing runs from the same cDNA library and 2) across cDNA libraries from the same tissue is illustrated in supplementary figure 2 (Supplementary Material online). This final collection of mapped reads was assembled into a set of time point–specific transcripts using the Cufflinks program (Trapnell et al. 2010). Here, the mean mate-pair insert size and standard deviation supplied to the program were calculated from all paired-end reads mapped for the cDNA library.
Comparative Transcriptomics
We used Cuffcompare (Trapnell et al. 2010) to build a consensus transcript set using transcript models from all 30 developmental time points. The mate-pair insert size and standard deviation were calculated from all paired-end reads mapped across all stages. Differential expression of these transcripts across time points was then estimated using Cuffdiff (Trapnell et al. 2010), where the maximum number of iterations for maximum likelihood estimation was increased from the default 5,000 to 25,000. As Cuffdiff allows only pairwise comparisons, developmental time points were analyzed sequentially and then separately for males and females when appropriate. Also, differences between age-matched male and female samples were investigated, with the parameters set as above. Here, instead of using RPKM values as above, individual transcript expression levels were quantified using FPKM values (fragments per kilobase of exon per million fragments mapped) as reported by Cuffdiff. The use of this quantity is appropriate for paired-end reads as it reports on the concomitant mapping of the two read ends of the cDNA fragment rather than on the mapping of individual reads. We used the Cufflinks-reported FPKM values, rather than RPKM values, because this allows overlapping transcripts to be quantified separately, depending on to which transcript individual fragments had been assigned. These FPKM values were log2-transformed to produce an approximately normal distribution from which standard analysis could be applied. When considering stage-specific expression (embryo, larva, pupa, and adult), a gene was considered to be expressed in a stage if it was associated with an FPKM value of at least 1 (Mortazavi et al. 2008) for at least one of the time points contained within that stage. A male- or female-specific gene model was defined if it was expressed with an FPKM value of at least 1 in at least one stage in one sex but was not expressed in all stages in the other sex.
Transcript and Gene Annotation
To ensure that our results were not influenced by genomic DNA contamination in the cDNA libraries, we only considered transcripts longer than 200 bp that were either:
Multiexonic or
Unspliced and expressed in multiple tissue samples, where the transcript contained sufficient reads for Cuffdiff to test for differential expression in at least one comparison.
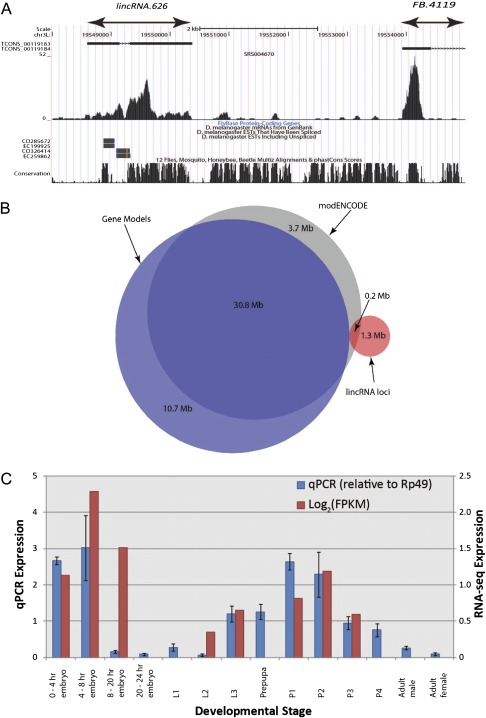
We define a gene model as a cluster of one or more transcripts, which are connected through shared exonic or intronic bases, as shown in figure 1A. Note that not all pairs of transcripts in a gene thus need overlap.
FIG. 1.—
(A) Definition of genomically adjacent protein-coding gene model (FB.4119) and a novel putative lincRNA locus (lincRNA.626). The black boxes denote exons called by Cufflinks for this tissue, with arrowed lines representing introns separating exons within the same transcript. A histogram of read counts that support these models’ sequences is shown below (from embryonic tissues, 4–6 h after egg laying). Note that only Cufflinks transcripts >200 bp are displayed. At the foot of this UCSC genome browser snapshot (Kent et al. 2002) is the FlyBase annotation corresponding to FB.4119, supporting messenger RNAs and ESTs, and a PhastCons track showing genome sequence conservation across multiple arthropods. (B) Venn diagram showing strong overlap between modENCODE (Graveley et al. 2011) and gene model exons and a low degree (13%) of overlap between the lincRNA exons defined in this study and modENCODE exons. (C) Concordance of qRT–PCR data with stage-matched log2(FPKM) expression values from RNA-seq analysis for lincRNA.626. Mean log2(FPKM) values are calculated and plotted for qRT–PCR experiments which cover more than one modENCODE developmental time point. Error bars represent 95% confidence intervals for qRT–PCR.
FlyBase Models
Models overlapping a known FlyBase gene by at least one base on either strand were associated with that gene. Those transcript models that lay in the intergenic regions thus represent putative lincRNA loci.
LincRNA Loci
We calculated the coding potential of all putative lincRNA loci using the Coding Potential Calculator (CPC) (Kong et al. 2007). The exonic bases for each transcript in a model were analyzed separately and in both orientations (forward and reverse strand). A transcript was deemed to be noncoding if the coding potentials of both strands scored less than zero. Benchmarking of the CPC algorithm demonstrated its efficacy in distinguishing known protein-coding from noncoding genes. A total of 1.3% of genes annotated as protein-coding by FlyBase are designated as being noncoding by CPC (score >0), whereas 2.8% of annotated noncoding genes were predicted to be coding by CPC (score <0). If all transcripts within an intergenic model were considered to be noncoding, only then was it defined as a lincRNA locus.
We also examined the evolutionary signatures of lincRNAs to determine the likelihood of their representing unannotated protein-coding genes using the phyloCSF program (Lin et al. 2011). A multiple-species alignment between D. melanogaster, D. simulans, and D. yakuba was submitted for each transcript and the maximum scoring transcript (i.e., that most likely to be protein-coding) within each gene model recorded.
Intergenic Regions
All intervals between gene models (of either type) were annotated as “intergenic sequence.”
The numbers of each category of gene, their lengths, and their expression profiles are summarized in table 1.
Table 1.
Characteristics of Gene Models and Putative LincRNA Loci
| Gene Type | Structure/Expression | Number of Gene Loci | Median Gene Length (bp) | Median Number of Alternative Transcripts | Median Number of Tissues in Which Expressed | Median log2(FPKM) | Standard Error log2(FPKM) |
| Gene model | Multiexonic | 7,414 | 1,700 | 2 | 30 | 1.93 | 1.61 |
| Single exon, expressed in multiple tissues | 126 | 873 | 1 | 18.5 | N/A | N/A | |
| LincRNA loci | Multiexonic | 1,049 | 443 | 1 | 11 | −1.52 | 1.54 |
| Single exon, expressed in multiple tissues | 70 | 235 | 1 | 2 | 0.30 | N/A |
Reverse Transcription and Quantitative Polymerase Chain Reaction Validation
RNA was extracted from different stages of fly development using a miRNeasy kit (Qiagen), including additional DNAse I digestion. Total RNA (1 μg) was reverse transcribed with Quantiscript reverse transcriptase (Qiagen) using random hexamer primers. Gene expression was determined by quantitative polymerase chain reaction (qPCR) from cDNA with SYBR green (Sigma) on an ABI 7500 thermocycler. Oligonucleotides for amplification of lincRNA cDNAs were designed using E-RNAi (http://www.dkfz.de/signaling/e-rnai3/). Sequences were as follows for lincRNA626 F—5′-TCAAAACTGTACCAGCTGCCTGGT-3′, R—5′-TGGTCGCTTGTGCTCGGATCG-3′; Rp49 F—5′-TACAGGCCCAAGATCGTGAA-3′, R—5′-TCTCCTTGCGCTTCTTGGA-3′. The delta-delta-Ct method was used to calculate messenger RNA abundance, using Rp49 expression as the reference.
Evolutionary Analyses
Nucleotide Substitution Rate
Pairwise genomic sequence alignments for D. melanogaster against D. yakuba or D. simulans (http://hgdownload.cse.ucsc.edu/downloads.html#fruitfly) were used to obtain alignments of all exonic bases for each gene model. Positions were removed if they contained a gap in either of the aligned species or bordered a gap in the alignment as these are known to bias substitution rate estimations (Lunter et al. 2008).
Substitution rates were estimated for each D. melanogaster gene (using the exonic sequence only) when aligned to D. yakuba (or D. simulans) using the baseml program from PAML (Yang 2007) and the HKY85 substitution model. Genes with an estimated substitution rate greater than 1 were discarded because their genomic alignments were likely to be between nonorthologous sequences.
The significance of individual lincRNA substitution rates was estimated by comparison to that of putatively neutrally evolving short (≤86 bp) intron sequence (Haddrill et al. 2005). D. melanogaster intronic sequences which mapped uniquely to the D. yakuba (or D. simulans) genome using BLAT (Kent 2002) were aligned and the sites required for correct intron splicing (6 bp at the 5′-end and 16 bp at the 3′-end of all introns) were then removed. These were then concatenated into a single alignment of presumed neutrally evolving sequence. One thousand such alignments were then generated for the exonic sequence of each lincRNA by sampling aligned positions with replacement from the concatenated alignment and their substitution rates were similarly estimated using baseml. A lincRNA was considered to be significantly constrained if fewer than 25 of the 1,000 neutral values were less than that of the lincRNA (i.e., P < 0.025). The false discovery rate (FDR) was estimated by partitioning the estimated P-values into 2.5% bins and then calculating the mean number of entries in the neutral bins (P > 0.025; P < 0.975) and then calculating the mean number of entries in the neutral bins. The ratio of this number to the number of constrained lincRNAs is then the FDR.
Population Genetics
In addition to the reference genome, we used data described in Rogers et al. from 37 genomes of North Carolina strains sequenced as part of the Drosophila Population Genomics Project (DPGP, www.dpgp.org). Using bases with a quality score of at least 20, we were able to collect a total of 74,042 polymorphic sites within both lincRNA exons and introns as well as within small protein-coding introns. We determined the derived and ancestral state for 48,374 of these sites using the UCSC genome alignments of D. melanogaster with D. simulans and with D. yakuba. We implemented a modified McDonald–Kreitman test (McDonald and Kreitman 1991) comparing the ratio of polymorphic to divergent sites between D. melanogaster and D. simulans within small introns to lincRNA exons and lincRNA introns. Differences between sequence categories were assessed using a chi-square test.
Comparison with Mouse LincRNAs
Mouse and fruit fly reference genomes (mouse NCBIv37 and FlyBase release 5.27) were partitioned into protein-coding gene territories. For each genome, we determined the mid-distance, i, between each known protein-coding gene’s terminus and its closest upstream and downstream protein-coding neighbors i − 1 and i + 1 (Ponjavic et al. 2009). A gene’s territory is defined as the interval delimited by the genomic co-ordinates i − 1 to i + 1. LincRNA loci lying within each territory were associated with the corresponding protein-coding gene. D. melanogaster orthologous protein-coding genes in Mus musculus were defined by the InParanoid database (Berglund et al. 2007). D. melanogaster lincRNAs were associated with mouse lincRNAs defined using the FANTOM3 cDNA collection (Marques and Ponting 2009) if they were found within a protein-coding gene territory in D. melanogaster whose orthologue’s protein-coding gene territory also contained a lincRNA locus.
Genome-Wide Association
The significant association of lincRNAs with a variety of genomic features was assessed as previously, using a randomization procedure (Ponjavic et al. 2007). In this context, protein-coding genes and lincRNA loci are referred to as “annotations.” The instances of a particular feature, whose enrichment or deficit is being tested, are referred to as “segments.” The number of nucleotides shared between these two sets is recorded and compared by simulation to the overlap expected if segments were to be randomly distributed across a background workspace. Here, the workspace represents all regions in the genome in which it is possible to find a particular set of annotations; for the protein-coding genes, this is the completely sequenced regions of the genome, whereas for the lincRNA loci and intergenic regions, this is the portion of the sequenced genome that lies between the gene models defined here. The segments that were tested for association with these annotations are as follows:
Indel-purified segments defined between D. melanogaster and D. simulans at a 10% FDR (Meader et al. 2010).
PhastCons regions of deep conservation across the Drosophila phylogeny (Siepel et al. 2005).
MicroRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), and endogenous small interfering RNAs (esiRNAs). We downloaded miRNA (Ruby et al. 2007) and esiRNA (Czech et al. 2008) sequences and aligned them to the D. melanogaster genome using Bowtie and BLAT, respectively. We considered only esiRNAs that produced a unique BLAT hit with 100% identity. We downloaded piRNA cluster annotations (Yin and Lin 2007) and removed any mapped esiRNAs that were found within these clusters. The coordinates of each set of short RNAs were clustered to produce a nonoverlapping set of genomic intervals.
Gene Ontology (GO) Analysis. We annotated each of the protein-coding gene territories defined above with the GO terms (Ashburner et al. 2000, release date 28 March 2008) associated with the protein-coding gene in the territory. An annotation was then created for each GO term. Those annotations with an expected lincRNA density of less than 1% were removed to reduce the number of false positives associated with very small overlaps.
Chromatin domain types obtained from Filion et al. 2010.
For each annotation, each set of segments was repeatedly sampled 10,000 times to generate an empirical distribution from which the P-value and significance of the observed over- or under-representation can be calculated. A P-value < 0.025 was considered to be significant.
Results
1,119 Putative LincRNA Loci in the D. melanogaster Genome
We used 4,054,717,403 sequencing reads of poly(A)+-selected RNA-seq evidence collected by the modENCODE consortium (Graveley et al. 2011) to define 1,119 putative lincRNA locus models in the D. melanogaster genome; of these, only 156 (14%) previously had EST support, defined as at least one base overlap between a previously reported EST and a lincRNA model. Transcripts were initially assembled separately at each of 30 developmental time points for which RNA-seq data were available and subsequently merged to produce a single consensus transcript set (see Materials and Methods). In order to discard genomic DNA contaminants, single exon models were only retained when supporting evidence from multiple developmental time points was available (see Materials and Methods). A gene was then defined as the set of transcripts that share at least one intronic or exonic base on either strand, as the RNA-seq data lacked strand information (fig. 1A). We recorded 7,414 gene models which overlap known FlyBase (Release 5.27, www.flybase.org) genes and which are hereafter labeled “gene models.” Transcriptional evidence was available for 13,463 (90.8%) FlyBase gene models, including 441 non–protein-coding genes. Those intergenic regions between gene models and lincRNA loci, for which there is no evidence of transcription, were annotated as “intergenic sequences.”
LincRNAs from 1,119 loci were identified and defined as transcripts longer than 200 bp which did not overlap any FlyBase gene model, and whose transcripts lacked evidence of significant protein-coding ability, as recorded by the CPC (Kong et al. 2007). CPC uses six features of putative ORFs to define transcripts as protein-coding whose conceptual translations are relatively long and/or that are sequence-similar to known proteins. The remaining transcripts that do not show these characteristics were defined as being noncoding. The proportion of transcripts predicted by CPC to be ncRNAs that are instead protein-coding was estimated to be only 1.3% (Materials and Methods). Furthermore, of 51,343 peptide sequences in the Peptide Atlas (Deutsch et al. 2008), none could be mapped to conceptual translations on either strand for the 1,119 lincRNA sequences, whereas 16,842 could be mapped to 2,692 (35.7%) of our gene models. This will reflect the low expression level of these putative lincRNA transcripts and also the strong likelihood that a high proportion of these transcripts’ sequence is, indeed, non–protein-coding. An additional approach, PhyloCSF (Lin et al. 2011), broadly validated the distinction of protein-coding from noncoding loci (supplementary fig. 3, Supplementary Material online). Only 17% of the protein-coding gene models, but 95% of the lincRNAs, have phyloCSF scores lower than 0. An upper bound estimate is thus that 17% of the set of 1,119 candidate lincRNA loci are, instead, protein-coding genes. Taken together, although recognizing that some transcripts may encode short polypeptides of low sequence similarity to known proteins, we will refer to these sequences simply as “lincRNAs” because our three approaches support the majority of these lincRNAs’ sequence as being noncoding.
We then adopted a two-stage strategy to consider the novelty and validity of this set of 1,119 lincRNA loci: We first compared the set with the 1,938 NTRs reported by modENCODE (Graveley et al. 2011) and then used reverse transcription and quantitative PCR (qRT–PCR) to validate a large number of these lincRNAs (see below). The most important distinction between our RNA-seq read mapping protocol and that of the modENCODE consortium relates to our use of three rounds of splice site junction detection which resulted in our mapping of approximately 200 million additional reads. As a result, we predicted three times more lincRNA loci (1,119 lincRNA loci; 1.3 Mb) than modENCODE (333 loci; 0.2 Mb) (fig. 1B). Only 73 (7%) of our lincRNA loci are completely covered by modENCODE transcripts. In contrast, most (largely protein-coding) gene models’ exons defined by us were also identified by modENCODE (fig. 1B). Of the 3.7 Mb of transcribed sequence found only by modENCODE, 1.3 Mb is intronic to our models and much of the remaining 2.4 Mb likely reflects poly(A)− transcripts detected by modENCODE total RNA and microarray experiments that were not considered in our analyses. The differences in approach to lincRNA identification are likely to explain why some of our putative lincRNA loci which overlap NTRs may have been incorrectly annotated by Graveley et al. as being protein-coding (e.g., see fig. 4A). In contrast to our approach described above, Graveley at al. use a sole criterion to define transcripts that contain an ORF longer than 100 amino acids as being protein-coding (Graveley et al. 2011).
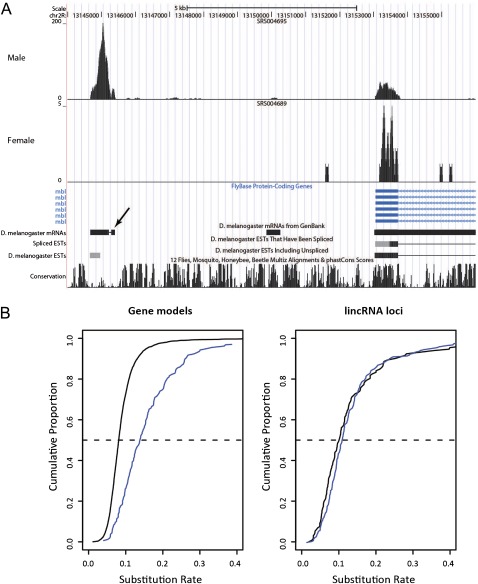
FIG. 4.—
(A) Example UCSC genome browser view of a spliced putative lincRNA locus in the vicinity of the mbl protein-coding gene which has read support for expression in one sex but not the other. The small exon at the right of the lincRNA (indicated by an arrow) is supported by messenger RNA but not EST evidence and has been annotated by Graveley et al. as a protein-coding gene (CG43108). Note that this annotation has not been added to the UCSC genome browser. (B) Cumulative distributions of the nucleotide substitution rate for gene models (left) and lincRNA loci (right) with different sex-specific expression profiles. Blue—male-specific; solid black—no sex specificity. The dashed line indicates the 50th percentile.
Previously unknown loci would be expected to be expressed at low levels. Indeed, the novel lincRNAs we identified tended to be expressed at reduced levels than those present in both the modENCODE and our data sets (Mann–Whitney test on maximum FPKM values, P < 2.2 × 10−16). Consequently, we sought to verify their expression using qRT–PCR for a similarly diverse range of developmental time points. Of the 66 lincRNAs tested, expression was validated for 58 (87.9%) (e.g., see fig. 1C), which is more than double the validation rate seen in previous studies of Drosophila lincRNAs defined by cDNA evidence (Inagaki et al. 2005; Tupy et al. 2005). Seventeen qPCR products were validated by sequencing, while all 58 products were of the expected sizes, as shown by gel electrophoresis. All primer sequences were designed not to amplify nonspecific sequences and they did not target repeat elements. We were able to reliably detect expression, using qRT–PCR, of lincRNAs associated with a maximum FPKM value of only 0.23 in the RNA-seq data, although this represents only a conservative lower limit of detection. The eight transcripts whose expression was not validated could be false positives; however, we note that these, and even more lowly expressed lincRNAs, may yet be detected upon closer inspection and examination of more restricted tissue samples. This value of 0.23 is lower than the minimum of 1 FPKM cited as being required for convincing expression in RNA-seq studies of this type (Mortazavi et al. 2008) and is likely due to the much greater sequencing depth within this data set. From 274 qRT–PCR experiments, a highly significant relationship was observed between these qRT–PCR data and stage-matched FPKM values (log2(qRT-PCR) versus log2(FPKM) linear correlation coefficient = 0.54, P < 2.2 × 10−16). This provides independent experimental evidence that our novel lincRNA transcripts, including those expressed at low levels, are indeed transcribed into RNA.
Further details and annotations of our lincRNA locus models, together with whether these are validated by qRT–PCR, are provided in supplementary table 1 (Supplementary Material online).
It follows from our definition of a lincRNA loci that each is distinct, with no evidence either from pre-existing or modENCODE data that they represent alternative transcripts of genomically adjacent protein-coding genes. Inspection of individual loci (e.g., fig. 1A) shows that most often, there is clear separation between adjacent gene models and lincRNA loci with intervening regions showing little or no evidence of transcription. Indeed, lincRNA loci frequently lie in gene-poor regions: They tend to be further from gene models than these models are from one another (median 2,269 bp for gene-lincRNA intervals vs. 452 bp for gene–gene intervals; Mann–Whitney P < 2.2 × 10−16).
LincRNA loci tend to be less complex than gene models, as was also observed for the modENCODE noncoding NTRs. As summarized in table 1 and as expected for previously unrecognized transcripts, they are shorter and have fewer transcripts contained within each locus. These differences further support the distinction of our lincRNA set from protein-coding genes. Most (94%) contained multiple exons. Only a minority of these lincRNAs appear to be the precursors of previously identified short RNA species, as shown in supplementary figure 4 (Supplementary Material online). Three hundred and five lincRNA loci (27%), and 5,961 gene models (80%), overlap one or more regions from which these short RNA species are transcribed. LincRNA loci are significantly (P < 2.4 × 10−3) depleted in microRNAs (Ruby et al. 2007), piRNAs (Yin and Lin 2007), and esiRNAs (Czech et al. 2008) relative to random expectations (miRNAs, −13.1%; piRNAs, −98.6%; esiRNAs, −56.7%, respectively). Rather, and as expected, miRNA sequences are significantly enriched within gene models (17.4%, P < 1.0 × 10−4), and esiRNA and piRNA sequences are significantly enriched in intergenic genomic regions (3.7%, P < 1.0 × 10−4 and 6.5%, P < 1.0 × 10−4, respectively). EsiRNAs and piRNAs do not possess poly(A)+ tails, being transcribed by RNA Polymerase III (Miyoshi et al. 2010); hence, we would not expect them to be found within gene models or lincRNA loci defined using poly(A)+-selected transcriptome data.
LincRNAs Exhibit Signatures of Evolutionary Constraint
If these 1,119 lincRNA loci express functional transcripts in D. melanogaster and/or its close relative D. yakuba, then their sequences will have purged deleterious substitutions or insertions and deletions (indels) since these species’ last common ancestor. Indeed, we found these loci to be associated with substantially and significantly lower rates of nucleotide substitution (median rate 0.11) compared with either untranscribed intergenic sequence or neutrally evolving short introns (Haddrill et al. 2005) (median rates of 0.18 and 0.25, respectively); surprisingly, their substitution rates are similar to those for the gene models (median rate of 0.10) (fig. 2A). Similar results were obtained for alignments of D. melanogaster and D. simulans sequences (supplementary fig. 5, Supplementary Material online).
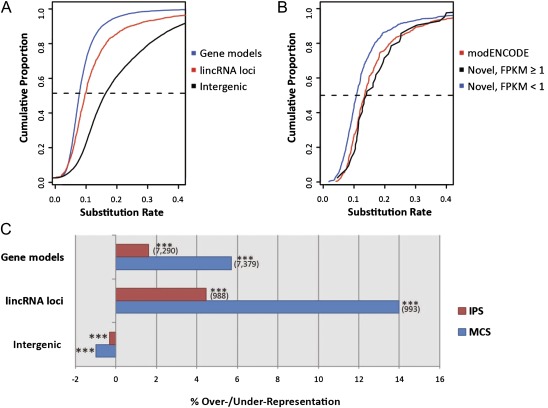
FIG. 2.—
Evidence for substantial purifying selection acting on putative lincRNA sequences. (A) Cumulative frequency distributions of exonic nucleotide substitution rates when aligned between Drosophila melanogaster and D. yakuba: Substitution rates of gene models are indicated in blue, and those for lincRNA loci are in red. The black line plots the cumulative substitution rates for untranscribed intergenic regions. The dashed line indicates the 50th percentile. (B) Cumulative frequency distributions of exonic nucleotide substitution rates when aligned between D. melanogaster and D. yakuba for lincRNA loci identified by modENCODE (red), novel lincRNAs with a maximum FPKM ≥1 (black), and novel lincRNAs with a maximum FPKM <1. (C) Enrichments or deficits of conserved sequence (indel-purified segments, in red, and MCS, in blue) within exonic sequences from gene models and lincRNA loci, and intergenic space, relative to genome-wide random expectations (***P < 0.001). Numbers of gene models and lincRNA loci overlapping each conserved sequence type are displayed in brackets
If lowly expressed lincRNAs are often “biological noise,” and thus lack function, or if our set of novel lincRNAs contained large numbers of such transcripts, then we expect their sequence substitution rates to be relatively high. By contrast, we found the opposite trend: lowly expressed (maximum FPKM < 1) and novel lincRNAs—those not sharing any overlap with modENCODE transcript models—tended to have significantly lower substitution rates than those also overlapping modENCODE models (Mann–Whitney test, P = 7.6 × 10−11; fig. 2B).
Ninety-six percent of lincRNA loci (with a FDR of 0.1%) individually show a suppressed substitution rate, relative to putative neutrally evolving short intron sequence, which is indicative of a significant degree of purifying selection (see Materials and Methods). None of the remaining 45 lincRNA loci individually exhibited evidence for a significantly elevated substitution rate above neutrality in comparisons of D. melanogaster with D. simulans and D. yakuba. LincRNAs also were shown to tolerate fewer insertion–deletion (indel) mutations, as shown in figure 2C by a significant 4.5% (P < 1 × 10−4) enrichment in their indel-purified segments between D. melanogaster and Drosophila simulans (Meader et al. 2010). When considering greater phyletic distances across all 12 Drosophila species whose genomes have been sequenced, and Anopheles mosquito, honeybee and Tribolium beetle, lincRNAs are also significantly enriched (14.0%, P < 1 × 10−4) in multispecies conserved sequence (MCS) regions (fig. 2C; Siepel et al. 2005). Ninety-five percent (1,063 of 1,119) of these lincRNAs contain such MCSs. These observations are consistent with these lincRNA locus sequences being constrained, and thus functional, both between these three fruit fly species and among others across the Drosophila and insect clades.
LincRNA loci exhibit evidence for constraint not just over the long periods of evolution separating these species but also within the shorter time since the coalescence of the modern D. melanogaster population. A detailed analysis requires data from an ongoing population genetics study (DPGP, www.dpgp.org), but preliminary findings from 48,374 variants detected in 37 individuals (Rogers et al. 2010) support purifying selection on substitutions in transcribed lincRNA sequence. This is because we find a significantly higher polymorphism/divergence ratio within both lincRNA exons (0.3801) and lincRNA introns (0.2246) in comparison to small introns (0.1613, two-tailed chi-squared test, P < 1 × 10−3 after a Bonferroni correction for multiple testing for both comparisons).
Developmental Expression of lincRNAs
We first examined the contributions of lincRNA transcription to the transcriptome of each of the 30 developmental time points. As for mammals (Guttman et al. 2010; Cabili et al. 2011), lincRNA expression levels in D. melanogaster tend to be substantially lower than those of gene models; this is apparent from the two very different scales on which their summed log2(FPKM) values are plotted in figure 3A. Across the different samples, the total gene model expression was, on average, 253-fold higher than for lincRNA loci. As observed by the modENCODE consortium (Graveley et al. 2011), expression levels of gene models increase during later developmental stages, with significant increases above embryonic expression in the pupal stage (1.1-fold difference in log2 expression values, P = 6.3 × 10−5) and for adult males (1.2-fold, P = 9.6 × 10−6). By contrast, the total expression of all lincRNA loci was more variable over these developmental stages. A significant decrease in lincRNA expression occurs during the pupal stage (−1.7-fold, P = 1.5 × 10−2), whereas they were significantly upregulated in males (1.5-fold, P = 4.9 × 10−5).
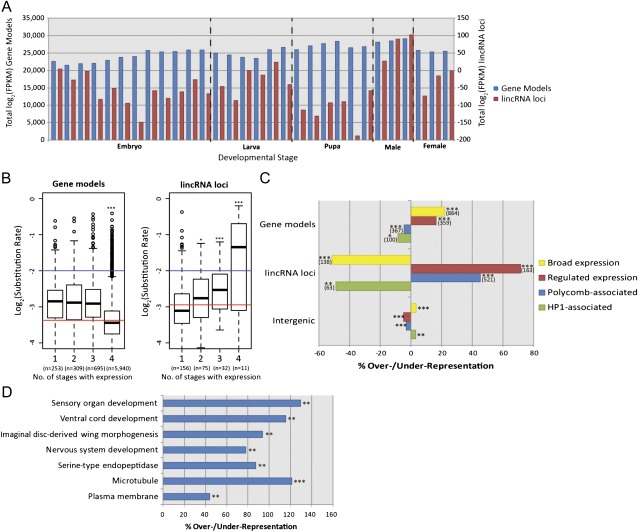
FIG. 3.—
(A) Expression levels of gene models and putative lincRNA loci across 30 developmental time points. Summed log2(FPKM) values for each time point are plotted for gene models (left vertical axis) and lincRNA loci (right axis). (B) Box and whiskers plots of log2(substitution rates) for gene models (left) and lincRNA loci (right) for increasing breadth of expression across one or more of four developmental stages (linear regression, ***P < 0.001). Red lines indicate log2(mean substitution rate) for the genes examined here. Blue lines indicate the log2(mean substitution rate) for presumed neutrally evolving short introns. Note that only genes and lincRNAs that are expressed at greater than 1 FPKM in at least one developmental stage are graphed here. (C) Enrichments or deficits of different chromatin types within gene models, lincRNA loci, and untranscribed intergenic sequence relative to genome-wide random expectations (*P < 0.05, **P < 0.01, and ***P < 0.001). Numbers of gene models and lincRNA loci overlapping each chromatin type are displayed in brackets. Repressive (“Black”) chromatin is depleted approximately 8% for both lincRNAs and gene models and modestly (0.6%) enriched in intergenic regions. (D) GO terms with associated protein-coding gene territories, which contain a significantly greater than expected density of lincRNA loci using a genome-wide association test (P < 0.01, FDR < 0.6). The top two terms are “cellular component” terms, whereas “serine-type endopeptidase activity” is a “molecular function” term and remaining terms are drawn from the “biological process” ontology.
Next, we considered whether the evolutionary rate of transcribed gene model or lincRNA locus sequence is influenced by the number of developmental stages during which it is expressed. In D. melanogaster, it was previously found that proteins expressed during early-to-mid development tend to have evolved the slowest (Davis et al. 2005; Artieri et al. 2009), whereas in previous studies of mammalian protein–coding genes, it was found that those which are broadly expressed (“housekeeping genes”) tend to evolve more slowly than those expressed in few tissues (Duret and Mouchiroud 2000; Winter et al. 2004). As expected, gene models that are broadly expressed in all four developmental stages (i.e., housekeeping genes) evolve the slowest since they exhibit the lowest nucleotide substitution rates (fig. 3B). By contrast, lincRNA loci that are expressed in three or four developmental stages (those that are “broadly expressed”) have a significant tendency to have evolved more rapidly than those expressed in only one or two stages (fig. 3B). More broadly expressed lincRNA loci thus appear to be less constrained not just in their expression but also in their sequence. Thirty-three percent of the 43 broadly expressed lincRNA loci exhibit substitution rates that exceed the expected neutral rate, estimated from short intron sequences (Haddrill et al. 2005). However, as we noted previously, none show statistically significant evidence for positive selection (see Materials and Methods).
To further investigate the developmental regulation of lincRNA loci, we considered their genomic locations within five principal chromatin types recently delineated in a Drosophila embryonic cell line (Filion et al. 2010). LincRNA loci showed substantially greater specificity for such chromatin types than the gene models, being greatly overrepresented in euchromatin containing genes whose transcription is specific to a few embryonic stages and tissues (“red”, fig. 3C) and in Polycomb group protein–associated chromatin (“blue”, fig. 3C). Polycomb regions frequently regulate genes with developmental functions (Sparmann and van Lohuizen 2006), and this result is consistent with recent studies suggesting a role for lincRNAs in regulation of Polycomb group protein recruitment (Sanchez-Elsner et al. 2006; Rinn et al. 2007; Zhao et al. 2010). As might be expected from their frequent narrow expression specificity, lincRNA loci are substantially depleted in broadly expressed euchromatin (“yellow”, fig. 3C) and in HP1-associated heterochromatin (“green”, fig. 3C).
In mammals, transcription in the vicinity of enhancer sites can generate a class of ncRNAs termed enhancer RNAs (eRNAs; Ørom et al. 2010). Expression levels of eRNAs and of transcripts from genomically adjacent protein-coding genes appear to be positively correlated (Ponjavic et al. 2009; Ørom et al. 2010). To consider correlated expression between noncoding and protein-coding transcripts, we first asked whether Drosophila lincRNA loci are enriched in the genomic vicinity of protein-coding genes associated with particular GO term annotations (see Materials and Methods). This analysis adopts a simplifying conservative assumption that lincRNA loci are more likely to regulate transcription of the most proximal gene than of other nearby genes.
We identified lincRNA loci as being significantly enriched in the vicinity of genes annotated as being involved in nervous system development, imaginal disc–derived wing morphogenesis, sensory organ development, ventral cord development, serine-type endopeptidase activity, the microtubule-associated complex, and the plasma membrane (fig. 3D). All results are significant (P < 0.01) and are associated with a low FDR (less than 0.6 false annotations are expected for each ontology we considered). Next, we sought evidence that the expression levels of protein-coding genes with these specific functional annotations are correlated with the expression levels of their adjacent lincRNA loci. Forty lincRNAs (25.6%) were found to be positively correlated with their neighboring protein-coding gene, whereas 4 (2.56%) were negatively correlated. This represents a significant increase in the number of correlations observed when comparing the expression of these lincRNAs to their other flanking protein-coding gene that lacked such specific functional annotations (two-tailed chi-squared test, P < 3 × 10−2). These findings are consistent with a minority of the 1,119 lincRNAs being either eRNAs that enhance the expression of genomically neighboring protein-coding genes or RNAs whose expression is coregulated with adjacent protein-coding genes.
Sex-Specific Expression of LincRNAs
One hundred and fifty-one lincRNAs were expressed in only one sex at one or more of the three adult time points for which sex-specific data are available (fig. 4A); these loci outnumber sex-specific gene models (151 vs. 121), despite there being overall seven times fewer lincRNA loci than gene models. Of these 151, 139 are specific to males with 110 being expressed in the testis or accessory gland. Male-specific protein-coding gene models show an increased substitution rate (median increase 1.5-fold, Mann–Whitney test, P < 2.2 × 10−16), relative to those that show no specificity, a result which is consistent with their roles in sexual selection (Haerty et al. 2007). In contrast, male-specific lincRNAs show no such bias (Mann–Whitney test, P < 0.21). Rather than participating in conspecific selection, male-specific lincRNAs are thus likely to contribute to male-specific, perhaps testis-specific, developmental processes.
Positionally Equivalent LincRNAs between Drosophila and Mouse
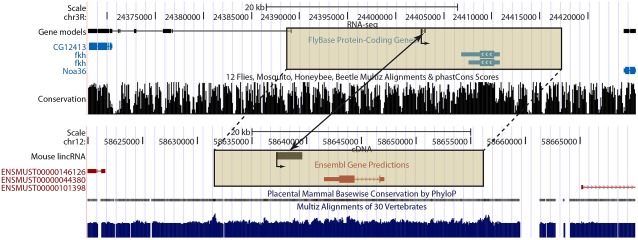
Finally, we considered whether lincRNA loci can be identified in two diverse animal species, D. melanogaster and mouse, that may act analogously in cis (Engström et al. 2006) on genomically neighboring protein-coding genes that are predicted by InParanoid as being orthologues in the two species (fig. 5). If so, then these lincRNA loci could have arisen either independently, by functional convergence, or else from a common ancestor approximately 700 million years ago but whose sequences have diverged to such an extent that resemblance to the ancestral sequence has been eroded. Certainly, noncoding sequence similarity is not expected to be retained between these species across such a considerable evolutionary time (Woolfe et al. 2004).
FIG. 5.—
An example of positionally equivalent putative lincRNA loci in both Drosophila melanogaster and Mus. musculus. The arrows within the protein-coding gene models and originating at the lincRNA transcriptional start sites indicate the shared orientation of transcription in both species. The boxed genomic regions indicate the orthologous protein-coding gene neighborhoods for D. melanogaster (fkh) and M. musculus (Foxa1). Note that only multiexonic transcripts are shown for the D. melanogaster gene models. The positionally equivalent lincRNA loci are indicated by the two-headed arrow.
We sought orthologous protein-coding genes that, in both species, are in the genomic vicinity of a lincRNA locus. Using a genomic association test (see Materials and Methods), we then observed that D. melanogaster lincRNA loci were significantly (P < 2.4 × 10−2) and substantially (57% increase) more likely to lie in the vicinity of genes whose mouse orthologues were also in the genomic vicinity of one or more lincRNA loci. The 42 orthologous gene neighborhoods that contain a lincRNA locus represent a significant 34% increase (two-tailed chi-squared test, P < 4.5 × 10−2) on the expected number of such loci. The mouse genes whose orthologous territories also contain a lincRNA in Drosophila are significantly (1.9- to 3.8-fold; P ≤ 1.1 × 10−4) enriched for annotations related to, among others, developmental regulation, including multicellular organismal development, cell differentiation, nucleic acid binding, and transcriptional regulator activity. These lincRNAs may therefore be eRNAs involved in developmental pathways conserved between these two diverse organisms.
To our knowledge, this study has provided the first evidence that lincRNA transcription is especially concentrated near to orthologous genes in species that are separated by such a long evolutionary distance. Further experimental investigation in D. melanogaster and mouse should determine whether these lincRNA loci are not only conserved in genomic position but are also conserved in cis-regulatory mechanism.
Discussion
We report the first genome-wide and deep sequencing study in which intergenic noncoding expression has been followed throughout an animal’s life cycle. We report a set of 1,119 candidate lincRNA loci, of which only 17%, at most, are predicted to represent previously undiscovered protein-coding genes. Our results show that lincRNA loci are commonplace and should now prompt experimental investigations into whether they represent an important component of the functionality of the Drosophila genome. We were able to validate, using qRT–PCR, expression from 87% of our lincRNA loci which we assayed, even for loci with maximum FPKM values as low as 0.23. The number of annotated loci in D. melanogaster found in FlyBase now increases by 7.5% (from 14,833 to 15,952) with many of these novel loci, as expected, being expressed at low levels and in restricted numbers of tissues and developmental stages. As lincRNAs are generally shorter than protein-coding transcripts, this increase in the number of loci is not matched by a corresponding increase in the number of bases covered by these annotations (2% increase, from 91 to 93 Mb). Despite the greater range of developmental time points used for these RNA-seq data, the number of D. melanogaster lincRNAs is already exceeded by known mouse lincRNA loci (Carninci et al. 2005; Guttman et al. 2009; Guttman et al. 2010; Cabili et al. 2011), a set that will clearly increase upon further RNA-seq interrogation of the mouse transcriptome (Marques and Ponting 2009). The increased complexity of the mouse over the fruit fly therefore appears to be matched by increases in the number of lincRNA loci, as well as protein-coding genes.
If lincRNA loci in Drosophila were not to impart function, then their sequence evolution would not be expected to differ from untranscribed intergenic sequence, their transcript levels would not vary over developmental stages, and their genomic positions would occur randomly with respect to chromatin domains and neighboring protein-coding gene classes. Instead, we have shown that these lincRNA loci are almost as intolerant of substitution mutations as gene models and are considerably less tolerant than intergenic sequence for which we have no evidence of transcription. Ninety-five percent of lincRNA loci contain an MCS, arguing for their long-lasting functionality across the entire Drosophila phylogeny.
Our data suggest a major biological role for lincRNAs in transcription regulation during development. This is implied by their more prominent expression at the earlier embryonic and larval stages and their loci being enriched in Polycomb protein–associated domains which are known to harbor developmentally relevant genes. LincRNAs with the highest sequence constraint, which might be expected to convey the most fundamental roles, are expressed preferentially during single developmental stages, rather than over multiple stages, and represent the best candidates for further experimental scrutiny into their contributions to developmental processes.
Like other molecule types, lincRNAs are expected to possess many diverse molecular roles. Nevertheless, a substantial minority of lincRNAs (155 of 1,119, 14%) are transcribed in the vicinity of protein-coding genes from particular functional classes, which is approximately 2-fold more than expected by chance (fig. 3D). Expression of genes from these classes is also significantly more likely to be positively correlated with transcription from genomically adjacent lincRNA loci. These biases suggest this fraction of RNAs first as eRNAs that actively promote transcription of genomically adjacent protein-coding genes and second as RNAs with roles in development. Specifically, the role of this fraction of lincRNAs may be in the development of the nervous system. Similar findings were reported previously for mouse lincRNA loci (Ponjavic et al. 2009). LincRNAs have previously been shown to be important in the mammalian nervous system (Mercer et al. 2008) and their brain expression patterns can be conserved between diverse vertebrates (Chodroff et al. 2010). Our findings in D. melanogaster, an invertebrate, suggest a role for lincRNAs in regulating developmental processes and in the development of the nervous system more generally across the animal kingdom. The 255 pairs of D. melanogaster lincRNA and protein-coding loci that contribute to these enrichments represent a rich resource for future investigations of the molecular mechanisms of transcriptional regulation during development.
The availability of lincRNA loci from both D. melanogaster and mouse allowed us to identify lincRNAs in each species that lie in the genomic vicinity of orthologous protein-coding genes. Such lincRNAs, through the potential cis-regulation of orthologous genes, may possess analogous, or even homologous, functional roles, which our results suggest would most likely be in developmental processes. We observed an increased frequency of D. melanogaster lincRNAs in the genomic vicinities of genes whose mouse orthologues also neighbored a lincRNA locus. As discussed above, mouse lincRNA catalogues remain incomplete and so the true enrichment may be higher than reported here. Similar positionally equivalent lincRNA loci were previously identified between human and mouse (Engström et al. 2006). To our knowledge, there have been only two previous reports of analogous lincRNA action between such distantly related species as mammals and Drosophila (Deng and Meller 2006; Jolly and Lakhotia 2006). In both instances, lincRNAs from both species are seen to participate in chromatin remodeling, through dosage compensation or the heat shock response but otherwise exhibit little else in common. These species’ high divergence disallows sequence similarities, and thus distinction between analogy and homology, to be discerned between paired lincRNAs; hence, this issue will, in the future, require experimental resolution.
Whether these lincRNAs function to regulate these protein-coding genes through a purely cis-acting mechanism could be tested by introducing genetic lesions, such as a premature transcriptional termination signal, to these sequences. Transfecting short hairpin RNAs (shRNAs) constructs (Guttman et al. 2011), which only target the mature lincRNA molecule, preferentially reveal trans-acting functions of the lincRNAs.
The data presented here for D. melanogaster and elsewhere for mouse and other species (Yazgan and Krebs 2007) suggest that the genomes of diverse animals contain large numbers of lincRNA loci that can confer biological function. The 1,119 D. melanogaster lincRNA loci provide excellent experimental candidates for testing the functional hypotheses advanced by this study, such as sex-specific regulation, regulation by chromatin states, the analogous activity of lincRNAs between D. melanogaster and mouse, and the cis-regulation of neighboring protein-coding genes. In all, 632 (56.5%) of our lincRNAs can be tested for at least one of these four functions. Genetic transformation techniques are available for D. melanogaster, which allow these hypotheses to be addressed. For example, 117 of our lincRNA loci contain a P-element for which it is already possible to obtain a mutant stock. Preliminary results (data not shown) reveal that several such P-element insertion lines exhibit a lethality phenotype, and these will be reported elsewhere. Clearly, the powerful genetic toolkit of D. melanogaster can now be applied to determine the molecular deficits that underlie such phenotypic changes for these, and many other, lincRNA loci.
Supplementary Material
Supplementary figures 1–5 and tables 1 and 2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We are grateful to T. Grant Belgard for assistance and scripts for the short-read assembly pipeline and to members of the C.P.P. and J.L.L. groups for valuable discussions. We thank the modENCODE consortium, in particular Brenton Graveley, for their community spirit in generating and sharing the RNA-seq data set. This work was supported by the Biotechnology and Biological Sciences Research Council (grant BB/F007590/1), European Research Council (DARCGENs), and by the UK Medical Research Council. A.C.M. was funded by a Marie Curie Intra-European Fellowship.
References
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437(7062):1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Artieri CG, Haerty W, Singh RS. Ontogeny and phylogeny: molecular signatures of selection, constraint, and temporal pleiotropy in the development of Drosophila. BMC Biol. 2009;7:42. doi: 10.1186/1741-7007-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund AC, Sjolund E, Ostlund G, Sonnhammer ELL. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 2007;36 doi: 10.1093/nar/gkm1020. (Database):D263–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29(18):3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braidotti G, et al. The air noncoding RNA: an imprinted cis-silencing transcript. Cold Spring Harb Symp Quant Biol. 2004;69:55–66. doi: 10.1101/sqb.2004.69.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chodroff RA, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11(7):R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, et al. The reality of pervasive transcription. PLoS Biol. 2011;9(7):e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5(7):613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453(7196):798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JC, Brandman O, Petrov DA. Protein evolution in the context of Drosophila development. J Mol Evol. 2005;60(6):774–785. doi: 10.1007/s00239-004-0241-2. [DOI] [PubMed] [Google Scholar]
- Deng X, Meller VH. Non-coding RNA in fly dosage compensation. Trends Biochem Sci. 2006;31(9):526–532. doi: 10.1016/j.tibs.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9(5):429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17(1):68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Engström PG, et al. Complex loci in human and mouse genomes. PLoS Genet. 2006;2(4):e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143(2):212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471(7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6(8):R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, et al. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177(3):1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, et al. Conserved introns reveal novel transcripts in Drosophila melanogaster. Genome Res. 2009;19(7):1289–1300. doi: 10.1101/gr.090050.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21(5):289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Inagaki S, et al. Identification and expression analysis of putative mRNA-like non-coding RNA in Drosophila. Genes Cells. 2005;10(12):1163–1173. doi: 10.1111/j.1365-2443.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21(2):93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Jolly C, Lakhotia SC. Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 2006;34(19):5508–5514. doi: 10.1093/nar/gkl711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT-the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm391. (Web Server issue): W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. Detection of intergenic non-coding RNAs expressed in the main developmental stages in Drosophila melanogaster. Nucleic Acids Res. 2009;37(13):4308–4314. doi: 10.1093/nar/gkp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27(13):i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshitz HD, Peattie DA, Hogness DS. Novel transcripts from the ultrabithorax domain of the bithorax complex. Genes Dev. 1987;1(3):307–322. doi: 10.1101/gad.1.3.307. [DOI] [PubMed] [Google Scholar]
- Lunter G, et al. Uncertainty in homology inferences: assessing and improving genomic sequence alignment. Genome Res. 2008;18(2):298–309. doi: 10.1101/gr.6725608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10(11):R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Challenging the dogma: the hidden layer of non–protein-coding RNAs in complex organisms. BioEssays. 2003;25(10):930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351(6328):652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Meader S, Ponting CP, Lunter G. Massive turnover of functional sequence in human and other mammalian genomes. Genome Res. 2010;20(10):1335–1343. doi: 10.1101/gr.108795.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC. Molecular mechanisms that funnel RNA precursors into endogenous small- interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. 2010;16:506–515. doi: 10.1261/rna.1952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Ørom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298(5598):1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5(8):e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17(5):556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. The functional repertoires of metazoan genomes. Nat Rev Genet. 2008;9(9):689. doi: 10.1038/nrg2413. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. 2010;19(R2):R162–R168. doi: 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21(1):11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Bedford T, Lyons AM, Hartl DL. Adaptive impact of the chimeric gene Quetzalcoatl in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2010;107(24):10943–10948. doi: 10.1073/pnas.1006503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17(12):1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila ash1 to Ultrabithorax. Science. 2006;311(5764):1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5(6):e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development, and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupy JL, et al. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2005;102(15):5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkn788. (Database issue):D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8(5):e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Goodhead I, Bähler J. Defining transcribed regions using RNA-seq. Nat Protoc. 2010;5(2):255–266. doi: 10.1038/nprot.2009.229. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14(1):54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2004;3(1):e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yazgan O, Krebs JE. Noncoding but nonexpendable: transcriptional regulation by large noncoding RNA in eukaryotes. Biochem Cell Biol. 2007;85(4):484–496. doi: 10.1139/O07-061. [DOI] [PubMed] [Google Scholar]
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450(7167):304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]