Abstract
Variation among lineages in the mutation process has the potential to impact diverse biological processes ranging from susceptibilities to genetic disease to the mode and tempo of molecular evolution. The combination of high-throughput DNA sequencing (HTS) with mutation-accumulation (MA) experiments has provided a powerful approach to genome-wide mutation analysis, though insights into mutational variation have been limited by the vast evolutionary distances among the few species analyzed. We performed a HTS analysis of MA lines derived from four Caenorhabditis nematode natural genotypes: C. elegans N2 and PB306 and C. briggsae HK104 and PB800. Total mutation rates did not differ among the four sets of MA lines. A mutational bias toward G:C→A:T transitions and G:C→T:A transversions was observed in all four sets of MA lines. Chromosome-specific rates were mostly stable, though there was some evidence for a slightly elevated X chromosome mutation rate in PB306. Rates were homogeneous among functional coding sequence types and across autosomal cores, arms, and tips. Mutation spectra were similar among the four MA line sets but differed significantly when compared with patterns of natural base-substitution polymorphism for 13/14 comparisons performed. Our findings show that base-substitution mutation processes in these closely related animal lineages are mostly stable but differ from natural polymorphism patterns in these two species.
Keywords: base substitution, mutation rate, mutation spectrum, nematode
Introduction
Darwin’s Origin of Species was motivated by his struggle to understand biological variation. That struggle is recapitulated in the age of genomics—we continue to be challenged by the tremendous variability within and among genomes at every scale—within individuals, among individuals within populations and among populations and higher taxa. If the properties of two genomes (or different regions within a single genome) differ in some respect, the most fundamental potential underlying reason for the difference is that mutation differs between the two, that is, the two groups have different mutational biases. However, there are other possibilities—the difference may simply be the result of random genetic drift or, perhaps more interestingly, natural selection may have differentially affected the two groups.
Unambiguously discriminating between the various evolutionary forces as underlying causes of variation in genetic variation is very difficult, for two reasons. First, mutation can never be “turned off,” so any comparison between groups must account for the possibility that mutational biases differ between groups. Mutations are very rare events—a given base in the genome has a probability of mutating on the order of 10−8 to 10−9 per generation—and direct detection of mutations de novo has historically involved extrapolation from a small set of detectable mutations whose properties may not be representative of the genome as a whole (Drake et al. 1998; Sniegowski et al. 2000; Baer et al. 2007; Lynch 2010). Second, the standing genetic variation present in any group has been previously scrutinized by natural selection, so any method employed to infer mutational properties from standing genetic variation must necessarily account for the potential effects of natural selection. The usual method is to identify on logical grounds a fraction of the genome that is putatively evolving neutrally (e.g., 4-fold degenerate silent sites, pseudogenes, intergenic regions, etc.) and compare the features of interest to the putative neutral fraction. However, the history of evolutionary genetics is replete with cases in which putatively neutral features have, upon closer scrutiny, been subsequently identified to bear the signature of nonneutral evolution (Halligan et al. 2011; Kousathanas et al. 2011; Kunstner et al. 2011).
The least assumption-loaded (but not assumption-free) way to discriminate between the effects of mutation and other evolutionary forces in shaping genome evolution is to employ an experimental system in which the effects of selection can be minimized, in which case the observed mutational properties of the genome (rate and spectrum) should be as close to the true values as can be possibly achieved. This is the method of “mutation accumulation” (MA). If mutational properties inferred from MA differ from the standing genetic variation in a group of interest, the most straightforward interpretation is that some evolutionary force other than mutation (i.e., selection and/or drift) has influenced the standing variation. Alternatively, however, it may be that the mutational properties inferred from the MA experiment themselves differ from the natural groups in question. There are now a handful of studies in which genome-wide mutational properties have been inferred from MA experiments combined with high-throughput DNA sequencing (HTS) technologies (Lynch et al. 2008; Denver et al. 2009; Keightley et al. 2009; Ossowski et al. 2010), and those studies have quickly become common references for the relevant mutational properties and are considered more reliable than indirect estimates (Lynch 2010; Appels et al. 2011). However, and importantly, all these studies consider only a single reference genotype, and the taxa in question (fruit flies, roundworms, yeast, plants) constitute a small sample of highly evolutionarily diverged taxa. If, for whatever reason, the mutational properties of a reference genotype (or species) are atypical of the mutational properties of the taxon as a whole, conclusions from that study will be misleading. Thus, it is of considerable importance to establish the generality of the results by investigating the mutational properties of multiple genotypes within multiple related species. An analysis of three genetically distinct sets of Drosophila melanogaster MA lines based on denaturing high-performance liquid chromatography provided evidence for nuclear mutation rate heterogeneity among the three fly genotypes (Haag-Liautard et al. 2007). A recent HTS analysis of human parent-offspring trios suggested considerable mutation rate variation within and between human families (Conrad et al. 2011). Thus, there is evidence that the mutation rate is variable within some animal species.
Here, we report the nuclear genome-wide base-substitution mutational properties of four sets of MA lines derived from two genotypes from each of two species of nematodes in the genus Caenorhabditis. This study extends and generalizes the findings reported in Denver et al. (2009) that focused only on the N2 laboratory strain of Caenorhabditis elegans. We compare our findings to the standing variation present in these two species, relying on HTS data for C. elegans natural isolates published elsewhere (Koboldt et al. 2010; Solorzano et al. 2011). We find several statistically well-supported differences between the mutational and natural standing single nucleotide polymorphism (SNP) spectra in both species.
Materials and Methods
MA Line Genotypes and Propagation
Four sets of MA lines were initiated, each from a different nematode genotype: C. elegans N2 (Bristol, England, common lab strain), C. elegans PB306 (isolated from an isopod ordered from Connecticut Biological Supply, Inc.), C. briggsae HK104 (Okayama, Japan), and C. briggsae PB800 (Ohio). After eight generations of progenitor strain inbreeding, the MA lines were propagated for 250 generations under single-hermaphrodite bottlenecking across generations in benign laboratory conditions as previously described (Baer et al. 2005). HTS analysis was performed on a randomly chosen subset of the larger set of MA lines (100 MA lines per genotype at the onset of the MA experiment).
HTS Experimentation and Analysis
We analyzed mutations from seven N2 MA lines, five PB306 MA lines, seven HK104 MA lines, and six PB800 MA lines. The genomes of the four progenitor strains used to initiate MA lines were also analyzed. We followed the same basic experimental protocols for Illumina HTS analysis as previously applied (Denver et al. 2009) to the set of seven N2 MA lines reanalyzed here. DNA was extracted by using a Qiagen DNeasy tissue miniprep kit, according to the manufacturer’s protocol, and then prepared according to standard Illumina protocols for genomic DNA samples. A total of 2.5 to 6.0 pmol of prepared DNA sample was loaded into each lane of an Illumina flowcell for analysis. Single-end, 36-cycle (bp) sequencing was done for all experiments on an Illumina GAII system at the Oregon State University Center for Genome Research and Biocomputing (OSU CGRB). Three to seven Illumina lanes were used for each MA line genotype assayed, depending on the sample. After each Illumina run, we applied the standard Illumina data analysis pipeline: Firecrest for tile image analysis, Bustard for base calling, and ELAND for alignment to the reference genome sequence. Reads were aligned to the C. elegans N2 genome (for N2 and PB306 MA line data) and the C. briggsae AF16 reference genome (for HK104 and PB800 data) with ELAND, version 0.2.2.6. To calculate genome-wide coverage and identify SNPs, the first 32 bases of reads from ELAND read categories U0, U1, and U2 were placed on the reference genome by using the coordinates provided by ELAND. U0 reads match unique genomic regions with zero mismatches, U1 reads align to unique regions with one mismatch, and U2 reads align to unique regions with two mismatches. Reads containing missing bases, which appear as ‘‘N’’s in the sequence, were excluded.
Candidate mutations were initially identified at positions that met the following criteria: 1) At least 6-fold coverage, 2) >90% of reads indicated a common nonreference base, 3) there was at least one read from each strand of DNA, 4) the Q scores for all bases contributing to the candidate SNP were 25 or greater, and 5) the coverage was not greater than 25-fold. This heuristic rule set was determined after several rounds of confirmation using conventional polymerase chain reaction (PCR) and ABI (Applied Biosystems) capillary DNA sequencing methods. We reanalyzed the N2 Illumina data, initially analyzed in a previous study (Denver et al. 2009), using the same parameters applied to the other three sets of MA lines for the current study. Identified mutations, sites considered, and other summary data used for our analysis is presented in supplementary table S1 (Supplementary Material online). Supplementary figures S1–S3 (Supplementary Material online) show the chromosomal positions of the mutations detected.
To control for sequence differences between the reference genome sequences (N2 for C. elegans, AF16 for C. briggsae), MA line progenitor strains, and a given MA line, we analyzed the sequence data at the site of a candidate mutation in all other MA lines and the relevant progenitor strain. Candidate mutations were retained only if there was strong evidence for the nonmutant base in all other genomes. As with our initial mutation identification rule set, we imposed conservative criteria in determining whether a given putative mutation was unique to a single MA line. If there was any evidence of the mutant base in the coverage data at that position in another line of the relevant MA line set (of a common progenitor genotype), it was deemed nonunique and was eliminated. Although this approach renders our analysis insensitive to the detection of mutations occurring on more than one MA line (thus, biasing our analysis against the detection of potential mutational hotspots), it was an essential step to disentangle real MA-line-specific mutations from sites confounded by potential cryptic paralogy issues involving the reference and MA line progenitor genomes. Our PCR/capillary DNA sequencing confirmation results (see next section) indicated that cryptic paralogy is not a significant confounder of our analysis.
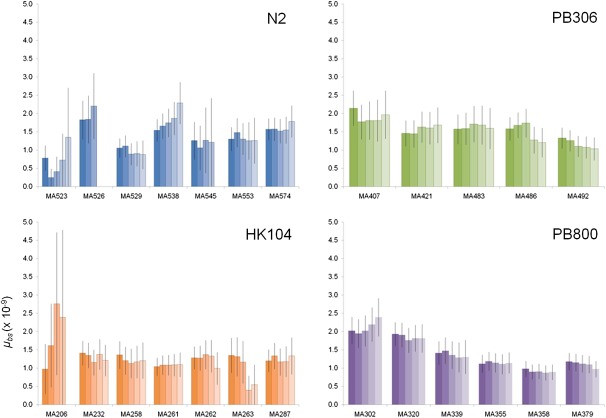
Heterozygosity at nucleotide sites in progenitor strain genomes constitutes another potential confounder of our analysis. An initially heterozygous site in a progenitor strain could quickly (in the first few generations) undergo differential segregation and fixation in different MA line lineages, leading to false positives that appear as de novo line-specific mutation events. We have four lines of evidence to argue that heterozygosity does not impact our findings: 1) C. elegans and C. briggsae natural strains reproduce primarily through self-fertilizing hermaphroditism in nature and their genomes are expected and observed in C. elegans (Cutter et al. 2009), to be highly homozygous as a consequence; 2) the MA line progenitor strains were inbred in the laboratory for eight generations prior to initiating MA experiments—thus, >99.9% heterozygous sites present prior to inbreeding (expected to be very few, see previous point) are expected to be homozygous by the end of inbreeding; 3) we confirmed 30/30 mutations using PCR and capillary sequencing (see below)—none of the progenitor strain sites were observed to be heterozygous; 4) we analyzed mutation rates across increasing n-fold coverage thresholds (6X–10X) and observed strong stability across cutoffs (fig. 1)—if heterozygosity was a confounder at lower (e.g., 6X) cutoffs, we would expect inflated rate estimates at these lower thresholds instead of the observed uniformity.
Fig. 1.—
Mutation rate estimates across varying n-fold coverage thresholds. For each of the 25 MA lines, the darkest far left bar shows the μbs estimate for ≥6X coverage; increasingly lighter shading shows rate estimates of increasingly higher n-fold coverage thresholds, up to ≥10X on the far right. Error bars show standard error of the mean approximations.
Our original experimental plan was to sequence seven MA lines from each of the four genotypes, but we report mutation data from only five PB306 MA lines and six PB800 MA lines. We obtained sequence data for two additional lines thought to be PB306 MA lines and one thought to be a PB800 MA line, though downstream analysis revealed DNA sequences identical to that of the respective MA line progenitor strain in each of the three cases: zero mutations were detected at ≥6X coverage using our criteria (see above). A small number (2–7, depending on the line) of likely false-positive mutations (3X–5X coverage) were detected in these data sets; two of these low-covered sites were targeted in our first PCR/capillary sequencing confirmation screen (see below) and found to be false positives. The most parsimonious explanation for this observation is technical error—either the wrong DNA sample was loaded onto the Illumina system or the progenitor nematode strain contaminated and overtook the targeted MA line laboratory populations near or after the end of the MA experiment or while the targeted nematode strains were being expanded for Illumina sequencing. We cannot rule out the distant possibility that these three lines each actually accumulated zero mutations during the experiment, though this possibility is extremely unlikely given the uniformity in mutation processes observed in the other 25 MA lines analyzed and the fact that nonzero mutation rates are observed in all biological systems analyzed (Baer et al. 2007). We thus concluded that these three data sets constituted cases of technical error and eliminated them from further analysis.
The coding context of each identified mutation was determined with custom Perl scripts that parsed the General Feature Format (gff) files for C. elegans build WS170 and C. briggsae build WS212. If a position was not found within the boundaries of a curated gene, it was deemed intergenic. For positions within genes, the site was categorized by its position relative to exon or intron; we did not consider untranslated regions because they are not annotated in the C. briggsae build. Furthermore, although majority of the C. briggsae reference genome is composed of sequences with well-defined chromosome positions, there also remain sequences that have been assigned to contigs of known chromosome source but unknown precise position within the chromosome (ChrN_random in supplementary table S1, Supplementary Material online), as well as sequences assigned to contigs of unknown chromosome source (ChrUn in supplementary table S1, Supplementary Material online). Mutations mapped to these two positionally uncertain C. briggsae sequence types were used for total mutation rate calculations but omitted from analyses involving chromosome domain and coding region analyses. The C. elegans and C. briggsae intrachromosomal recombination domain boundaries (tip, arm, core) used in our analyses were taken from a recent analysis of recombination rate variation in these two species (Ross et al. 2011). For exon positions, the relative coding regions were translated by using both the reference base and the mutant base, and the type of resulting amino acid change was determined. These were categorized into synonymous and nonsynonymous groups. Premature termination codons were treated as nonsynonymous changes. For calculations of expected numbers of synonymous and nonsynonymous mutations, initial null expected values based on the universal genetic code alone were first adjusted to account for patterns of codon usage in the C. elegans and C. briggsae genomes (Stein et al. 2003). We also extended the approach developed by Moran et al. (2009), also previously applied by us to the N2 MA line data (Denver et al. 2009), to accounting for the effects of patterns of mutational bias, observed in the MA lines analyzed here, in determining expected numbers of nonsynonymous and synonymous substitutions.
Mutation Confirmation
Upon collection of the raw Illumina data for the four sets of MA lines analyzed here, we initially applied an analytical pipeline identical to that originally used for the N2 MA lines (3X or better coverage, otherwise same rules described above). In our previous analysis of the N2 MA line Illumina data, 51/52 mutations identified using this approach were confirmed using PCR and ABI capillary sequencing (Denver et al. 2009). After identifying candidate mutations for the current study, we randomly selected 15 sites from the resultant PB306, HK104, and PB800 candidate mutation lists for confirmation analysis using PCR and ABI capillary sequencing. PCR primers were designed in the ∼800 bp flanking each candidate mutant site and then used to amplify target regions in the corresponding MA line and the MA line progenitor. PCR products were then directly sequenced using an ABI3730 capillary sequencing system at the OSU CGRB. Confirmation required the detection of the mutant base in the MA line sample and the ancestral wild-type base in the progenitor sample. However, only 7/15 candidate mutations evaluated in this fashion were confirmed. Upon examination of the coverage patterns of the 15 candidate mutations evaluated, it was found that all eight mutations not confirmed were originally supported by five or fewer Illumina reads. Among the seven candidate mutations that were confirmed, six were covered by six or more Illumina reads; one candidate mutation covered by five reads was confirmed. We thus initiated a second PCR/capillary sequencing confirmation effort involving 24 candidate mutation sites, all of which were covered by six or more Illumina reads. 24/24 of these candidate mutations were confirmed. We thus decided upon 6X or greater mutant site coverage as the threshold for calling mutant sites since 30/30 candidate mutations evaluated at this threshold were confirmed. The higher false positive rate at mutant sites covered 3X–5X in the PB306, HK104, and PB800 MA lines (compared with N2) is most likely related to the fact that the reference genome sequence required for mutation mapping differed from strains used as MA line progenitors. The sequence differences between reference genomes (N2, AF16) and MA line progenitor strains without references (PB306, HK104, PB800) are expected to lead to cryptic paralogy confounders in our analyses when sequence coverage is low.
Mutation Rate and Statistical Analyses
Individual MA-line-specific mutation rates were calculated with the equation μbs = m/(LnT), where μbs is the base substitution mutation rate (per nucleotide site per generation), L is the number of MA lines, m is the number of observed mutations, n is the number of nucleotide sites, and T is the time in generations, as previously described (Denver et al. 2009). We approximated standard errors for individual mutation rates as [μbs /(nT)]1/2, as previously described (Denver et al. 2009). Values used for n reflect the total number of base pairs surveyed that met our criteria for consideration of a possible mutation site.
To evaluate the significance of mutation rate differences across different species, strains, chromosomes, chromosome regions, and coding regions, we employed X2 goodness-of-fit tests. Our measured numbers of mutation “hits” were sufficiently low that was is preferable to treat the data as categorical rather than continuous. We evaluated the observed numbers of mutant versus nonmutant sites across comparisons against null expectations calculated based on null expectations of discrete uniform mutation distributions. For example, the null distributions were calculated based on the null expectation that the total summed number of mutations observed across a group of MA lines would be uniformly distributed across those MA lines in accordance with the numbers of sites considered in each line. This same basic approach was extended to all of our X2 tests.
Results
Experimental Overview
We analyzed the nuclear genomes of four sets of 250-generation nematode MA lines, each derived from a different progenitor genotype: C. elegans N2 (laboratory strain also analyzed in Denver et al. 2009), C. elegans isolate PB306, C. briggsae isolate HK104, and C. briggsae isolate PB800. Details about the propagation and maintenance of these MA lines were previously described (Baer et al. 2005). This group of four Caenorhabditis nematode MA line sets has previously been analyzed in terms of the deleterious genomic mutation rate for fitness (Baer et al. 2005), nuclear microsatellite mutation rates (Phillips et al. 2009), and mitochondrial genome mutation rates (Howe et al. 2010). For this study, we analyzed the genomes of seven C. elegans N2 MA lines (the same lines analyzed in Denver et al. 2009, though under more conservative analysis parameters), five C. elegans PB306 lines, seven C. briggsae HK104 lines, and six C. briggsae PB800 lines. We also analyzed the genomes of the four progenitor strains used to initiate the MA experiments.
Nuclear genomes were analyzed using Illumina HTS technology. We followed the same basic sample preparation protocols and sequencing approach (36-bp single-end reads) as was previously applied to the N2 MA lines (Denver et al. 2009). The same HTS analysis parameters were also applied, with one key distinction. A more stringent n-fold coverage threshold (≥6X here vs. ≥3X in the previous analysis of N2) was required for effective mutation identification in the current study. As detailed in the Materials and Methods, when we applied a ≥3X cutoff to identify putative mutations in the four MA lines sets analyzed here, evaluation of putative mutant sites using PCR and capillary sequencing revealed very high false positive rates (∼50%) in the PB306, HK104, and PB800 MA lines. When the more stringent ≥6X threshold was applied, all (24/24) putative mutant sites evaluated by PCR and capillary sequencing were confirmed. The higher incidence of false positives at the ≥3X cutoff in the PB306, HK104, and PB800 MA lines as compared with the low rate at this threshold previously reported by us for the N2 MA lines only (51/52 supported) is most likely associated with the fact that we were able to use the N2 genome sequence for mutation mapping in the N2 MA lines, whereas the other three sets relied on reference genomes of different genotypes (N2 for PB306, C. briggsae AF16 for HK104 and PB800). In particular, the necessary use of reference sequences that differ from MA line progenitor genotypes in these cases is expected to lead to cryptic paralogy confounders at lower sequence coverage levels. All mutations reported here, including in N2, conformed to the ≥6X coverage cutoff as well as the other analysis parameters required for mutation identification (see Materials and Methods). Our analysis approach resulted in the effective survey of large amounts of nonrepetitive nuclear DNA sequence in each MA line, ranging from 8.2 to 85.9 Mb (table 1).
Table 1.
Summary of HTS Mutation Data for MA Lines
| Species | Strain | Line | Sites Cons. | No. Mut. | Rate (x10−9) | SEM (x10−9) |
| Cb | HK104 | MA206 | 8,220,587 | 2 | 0.97 | 0.69 |
| Cb | HK104 | MA232 | 51,028,392 | 18 | 1.41 | 0.33 |
| Cb | HK104 | MA258 | 38,268,317 | 13 | 1.36 | 0.38 |
| Cb | HK104 | MA261 | 69,378,157 | 18 | 1.04 | 0.24 |
| Cb | HK104 | MA262 | 53,078,426 | 17 | 1.28 | 0.31 |
| Cb | HK104 | MA263 | 23,732,939 | 8 | 1.35 | 0.48 |
| Cb | HK104 | MA287 | 50,102,000 | 15 | 1.20 | 0.31 |
| Cb | PB800 | MA302 | 57,260,340 | 29 | 2.03 | 0.38 |
| Cb | PB800 | MA320 | 72,458,123 | 35 | 1.93 | 0.33 |
| Cb | PB800 | MA339 | 53,805,568 | 19 | 1.41 | 0.32 |
| Cb | PB800 | MA355 | 74,998,209 | 21 | 1.12 | 0.24 |
| Cb | PB800 | MA358 | 85,891,433 | 21 | 0.98 | 0.21 |
| Cb | PB800 | MA379 | 84,789,081 | 25 | 1.18 | 0.24 |
| Ce | N2 | MA523 | 25,738,392 | 5 | 0.78 | 0.35 |
| Ce | N2 | MA526 | 26,302,925 | 12 | 1.82 | 0.53 |
| Ce | N2 | MA529 | 60,805,045 | 16 | 1.05 | 0.26 |
| Ce | N2 | MA538 | 64,944,055 | 25 | 1.54 | 0.31 |
| Ce | N2 | MA545 | 19,117,865 | 6 | 1.26 | 0.51 |
| Ce | N2 | MA553 | 49,167,471 | 16 | 1.30 | 0.33 |
| Ce | N2 | MA574 | 71,382,969 | 28 | 1.57 | 0.30 |
| Ce | PB306 | MA407 | 37,320,025 | 20 | 2.14 | 0.48 |
| Ce | PB306 | MA421 | 46,660,508 | 17 | 1.46 | 0.35 |
| Ce | PB306 | MA483 | 38,202,177 | 15 | 1.57 | 0.41 |
| Ce | PB306 | MA486 | 63,270,543 | 25 | 1.58 | 0.32 |
| Ce | PB306 | MA492 | 66,338,507 | 22 | 1.33 | 0.28 |
Note.—Sites Cons. indicates the total numbers of sites analyzed that fit the parameters required for potential identification of a mutation. No. Mut. shows the numbers of mutations detected for a given MA line. SEM indicates the approximate standard error of the mean.
Genome-Wide Rates
Our analysis identified 448 total mutations: 108 in seven N2 MA lines, 99 in five PB306 MA lines, 91 in seven HK104 MA lines, and 150 in six PB800 MA lines (table 1, supplementary table S1, Supplementary Material online). We calculated the per-generation, base-substitution mutation rate (μbs) in each MA line by dividing the number of observed line-specific mutations by the product of the number of sites considered (total sites sequenced that conformed to the analysis parameters required to identify a mutation) and the number of generations. Among the 25 MA lines analyzed, line-specific total μbs estimates varied nonsignificantly (P = 0.47, X2 test), from 0.8 to 2.1 × 10−9 mutations per site per generation (table 1). Pooling mutations by progenitor strain, we observed no significant mutation rate variation among the four nematode genotypes (P = 0.42, X2 test); pooling by species, we also observed no significant variation (P = 0.38, X2 test). We evaluated mutation rates under increasing n-fold stringencies for mutation identification and site consideration (≥6X to ≥10X) and observed stability in rate estimates (fig. 1). These findings indicate that there is little variation in total μbs among the nuclear genomes of the four Caenorhabditis strains analyzed.
We next analyzed conditional mutation-rates specific to the six nonstrand-specific base substitution types, expressed as the type-specific rate conditioned on the underlying number of sites that fit our criteria for evaluation (i.e., numbers of considered G:C and A:T sites for each respective set of associated mutation types). The bias toward G:C→A:T transitions and G:C→T:A transversions previously observed in the N2 MA lines (Denver et al. 2009) was observed here in all four sets of lines (fig. 2); no significant variation in type-specific mutation rates was observed among strains (P = 0.64, X2 test). No significant variation was observed among strains when pooling mutations types into transitions and transversions (P = 0.07, X2 test).
Fig. 2.—
Conditional rate estimates for the six base substitution types. Error bars show standard error of the mean approximations.
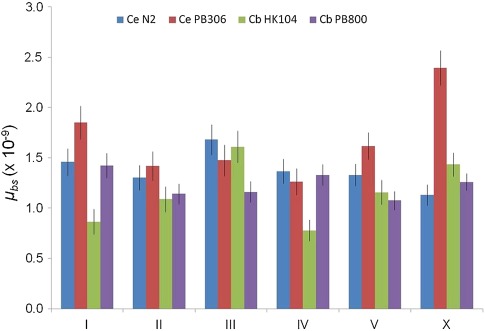
Chromosomal Rates
We next analyzed patterns of mutational variation among and within chromosomes. The chromosomal positions of detected mutations are depicted in supplementary figures S1–S3 (Supplementary Material online). First, we compared chromosome-specific mutation rates within and among MA line sets. The highest rate was observed for the C. elegans PB306 X chromosome (fig. 3). Pooling autosomal mutations to compare against X mutations revealed a marginally significant difference in the autosomal versus X rate in PB306 (P = 0.04, X2 test) but not in the other three MA lines sets (0.30 < P < 0.86, X2 tests). However, when the five autosomal rates were considered individually along with the X chromosome (rather than pooling autosomal data as in previous analysis), no significant rate variation was detected among the six PB306 chromosomes (P = 0.21, X2 test). Aside from the marginally significant evidence for an elevated X-specific rate in PB306, chromosome-specific rates were otherwise mostly uniform (fig. 3). The chromosomes of C. elegans and C. briggsae are subdivided into three major intrachromosomal domains: tip, arm, and core (Consortium, CeS 1998; Ross et al. 2011). The core domains have high gene densities and low recombination rates; the arm domains have low gene densities and high recombination rates; the tip domains have low gene densities and low recombination rates. There was no significant variation in the distribution of mutations across the autosomal tip, arm, and core intrachromosomal recombination domains in any strain (0.25 < P < 0.99, X2 test).
Fig. 3.—
Chromosome-specific μbs estimates. Error bars show standard error of the mean approximations.
Rates in Functional Sequence Categories
We analyzed mutation rates across exon, intron, and intergenic functional sequence categories (supplementary fig. S4, Supplementary Material online) and observed no significant variation among these three categories in any of the four strains analyzed (0.19 < P < 0.85, X2 tests). The numbers of mutations detected at nonsynonymous and synonymous codon positions in protein-coding sequence were not significantly different than the null expectation of equal distributions across nonsynonymous and synonymous codon positions (0.07 < P < 0.13, X2 test), though observed mutation numbers at nonsynonymous sites were smaller than the expectations in all four cases.
Discussion
Mutation Rates
Our analysis of base substitution mutation processes in two strains of C. briggsae and two strains of C. elegans revealed extensive mutational uniformity in many contexts suggesting that, for the most part, nuclear base-substitution mutation processes have been stable over the evolutionary history of this nematode group. Direct mutation rate estimates derived from MA studies are often extrapolated to other species for evolutionary analysis; for example, MA line–derived rates from C. elegans and D. melanogaster have been used for internally calibrated molecular clock–based approaches to estimate divergence times among species in these two animal genera (Cutter 2008). The base-substitution mutational stability reported here indicates that MA line–based rate estimates for a given species can be extended to related species with confidence. However, although our comparative analysis expanded beyond a single species and strain, it is still limited in that only four genotypes were analyzed. A larger scale analysis of many more species and strains of Caenorhabditis nematodes might reveal more mutational heterogeneity. Variation in genome-wide patterns of SNP types in two other C. elegans natural isolates (CB4856, CB4858) suggest that base-substitution mutation processes might vary between these two strains (Solorzano et al. 2011), though selective differences on base substitution processes between the strains might also be responsible for the different SNP patterns between the strains.
The uniformity in nuclear genome base-substitution mutation processes reported here is inconsistent with patterns of mutational fitness decay in this set MA lines that suggest higher mutation rates in the C. briggsae strains relative to the C. elegans strains (Baer et al. 2005). Mitochondrial μbs estimates for the two sets of C. briggsae MA lines (Howe et al. 2010) were also highly similar to the C. elegans N2 mitochondrial μbs (Denver et al. 2000). However, nuclear microsatellites displayed higher rates of insertion–deletion mutation in the C. briggsae MA lines relative to the C. elegans lines (Phillips et al. 2009) and a higher rate of large mitochondrial DNA deletions was observed for the C. briggsae MA lines (Howe et al. 2010). Thus, differences in insertion–deletion mutation processes, rather than base-substitution events, are potentially responsible for the observed variation in the mutational decay of fitness between these two species.
Mutation Spectra
The mutation spectrum, as measured by the conditional μbs estimates for each of the six base substitution types (fig. 2), was found to not significantly vary among the four nematode genotypes. A strong bias toward G:C → A:T transitions and G:C → T:A transversions was observed in each set of lines, as was previously observed for the C. elegans N2 MA lines (Denver et al. 2009). The previous analysis of N2 MA lines also reported a significant difference between the MA line mutation spectrum and patterns of natural polymorphism in C. elegans at sites commonly presumed to be neutral (e.g., pseudogenes, introns, and intergenic DNA). In particular, the ratio of transition to transversion variants (Ts/Tv) in such presumably neutral sequences in C. elegans natural populations is observed to be 1.2 to 3.0 (depending on the analysis), whereas the average Ts/Tv observed in the C. elegans N2 MA lines was 0.45 with line-specific values ranging from 0.19 to 0.79 (Denver et al. 2009). This discrepancy might reflect stronger genome-wide purifying selection against transversions as compared with transitions. Alternatively, the Ts/Tv differences might result if mutation processes differ between laboratory-reared nematodes and those evolving in nature. The previous study could not rule out the possibility that N2 might have an unusual mutation spectrum associated with its multidecade evolution in the laboratory environment.
To gain a broader understanding of Ts/Tv variation among MA lines and natural populations of Caenorhabditis nematodes, we analyzed Ts/Tv ratios observed in each of the four sets of MA lines, comparing them to each other and to Ts/Tv ratios observed in recent HTS analyses of C. elegans and C. briggsae natural isolates. We relied on data resulting from an analysis of SNPs between N2 and two C. elegans natural isolates, CB4856 from Hawaii and CB4858 from California (Solorzano et al. 2011). For C. briggsae, data resulting from a recent analysis of SNPs occurring between reference strain AF16 (India) and two natural isolates, HK104 from Japan and VT847 from Hawaii, was used (Koboldt et al. 2010). We also included the SNP polymorphisms detected in our analysis between natural isolate progenitors of MA lines and reference genome sequences (C. elegans N2 ↔ PB306, C. briggsae AF16 ↔ HK104, C. briggsae AF16 ↔ PB800). This data resulted as a consequence of our broader MA line mutational analysis; we conservatively identified natural isolate SNPs as those sites in unique genomic regions with ≥6X unanimous HTS data reporting the progenitor SNP in the MA line natural isolate (PB306, HK104, PB800) and derivative MA line HTS data.
In the MA lines, Ts/Tv ranged from 0.64 to 1.14 (table 2), though the variation was outside of usual significance thresholds (P = 0.07, X2 test). In seven sets of natural isolate total SNP comparisons (includes two independent analyses of C. briggsae AF16 ↔ HK104), Ts/Tv had a narrower range from 1.01 to 1.42 though the variation was highly significant (P = 3.3 × 10−24, X2 test), primarily due to the unusually low ratio in the N2 ↔ CB4856 SNPs. A significant Ts/Tv difference between the two C. elegans SNP data sets was previously noted (Solorzano et al. 2011). We compared observed Ts/Tv in the seven SNP data sets to expectations based on Ts/Tv observed in the MA lines (table 2) and found that the patterns of natural isolate total SNPs deviated from expectations based on MA line Ts/Tv in 14/14 comparisons (two sets of expected values calculated for each SNP data set, one from each MA line of the same species), though just marginally so when the C. elegans PB306 MA line data was used to predict N2↔CB4856 SNP patterns. When limiting the analysis to intron and intergenic positions, sequences commonly presumed to be neutral, significant differences were observed in 13/14 cases. The single exception was when the C. elegans PB306 MA line Ts/Tv (1.06) was used to calculate expected values for the N2-CB4856 SNP data, when limited to intron and intergenic sites (Ts/Tv = 1.08). Higher Ts/Tv ratios were also observed in an analysis of 22 autosomal intron loci across 16 genetically diverse C. briggsae natural isolates where Ts/Tv = 1.26 (Cutter and Choi 2010). Likewise, a genome-scale HTS analysis of polymorphism in 200 C. elegans natural isolates showed an overall Ts/Tv = 1.27 (Andersen et al. 2012). These observations support the general conclusion that Ts/Tv ratios differ between mutation spectra observed in MA lines and natural SNP patterns in C. elegans and C. briggsae. The Ts/Tv similarity between the PB306 MA lines and the N2-CB4856 SNP data, however, shows that parallels in MA line base substitution mutation processes and natural polymorphisms in noncoding DNA do occur, albeit in only 1/14 cases analyzed here. Further analysis is required to understand the complex differences between MA line mutation processes and natural patterns of polymorphism. The present study suggests that the previously reported Ts/Tv dissimilarity between N2 MA line mutation spectra and patterns of presumably neutral polymorphism in C. elegans was not an artifact of unusual mutation in the laboratory-domesticated N2 strain. The Ts/Tv dissimilarities between the MA lines and natural isolates might reflect weak but efficient purifying selection against transversion mutations. One intriguing hypothesis recently put forth suggests that transversions might be under stronger genome-wide selection due to their disruptive effects on chromatin organization (Babbitt and Cotter 2011). However, it also cannot be ruled out that these dissimilarities result from underlying mutational differences in laboratory versus natural environments.
Table 2.
Transitions and Transversions in MA Lines and Natural Isolates
| MA Line Mutations | Natural Isolate SNPs | ||||||||||
| Ce a N2 | Ce a PB306 | Cb a HK104 | Cb a PB800 | Ce a N2↔PB306 | Cb a AF16↔HK104 | Cb a AF16↔PB800 | Ce b N2↔CB4856 | Ce b N2↔CB4858 | Cb c AF16↔HK104 | Cb c AF16↔VT847 | |
| Ts | 42 | 51 | 38 | 80 | 45,224 | 68,118 | 59,249 | 2,727 | 21,145 | 13,801 | 5,766 |
| Tv | 66 | 48 | 53 | 70 | 33,556 | 49,555 | 41,861 | 2,709 | 15,993 | 10,029 | 4,245 |
| Ts/Tv, tot | 0.64 | 1.06 | 0.72 | 1.14 | 1.35 | 1.37 | 1.42 | 1.01 | 1.32 | 1.38 | 1.36 |
| P-value 1, tot | N2, ≈0 | HK104, ≈0 | HK104, ≈0 | N2, 3.2 × 10-65 | N2, ≈0 | HK104, ≈0 | HK104, 1.5 × 10-226 | ||||
| P-value 2, tot | PB306, ≈0 | PB800, ≈0 | PB800, ≈0 | PB306, 0.046 | PB306, 4.3 × 10-97 | PB800, 1.3 × 10-45 | PB800, 1.2 × 10-17 | ||||
| Ts, IN + IG | 31,938 | 41,399 | 35,764 | 1,698 | 17,838 | 10,022 | 4,875 | ||||
| Tv, IN + IG | 22,922 | 28,909 | 24,555 | 1,578 | 13,333 | 7,265 | 3,522 | ||||
| Ts/Tv, IN + IG | 1.39 | 1.43 | 1.46 | 1.08 | 1.33 | 1.44 | 1.38 | ||||
| P-value 1, IN + IG | N2, ≈0 | HK104, ≈0 | HK104, ≈0 | N2, 3.8 × 10-52 | N2, ≈0 | HK104, ≈0 | HK104, 1.3 × 10-162 | ||||
| P-value 2, IN + IG | PB306, ≈0 | PB800, ≈0 | PB800, ≈0 | PB306, 0.71 | PB306, 1.6 × 10-90 | PB800, 3.8 × 10-19 | PB800, 2.3 × 10-10 | ||||
Note.—Ts indicates transitions and Tv indicates transversions; tot indicates numbers observed across all genomic regions; IN + IG indicates numbers observed in intron and intergenic regions. For each natural isolate SNP data set, two sets of X2 tests were performed to evaluate how observed Ts and Tv numbers fit predictions based on Ts and Tv numbers observed in MA lines from the same species. For a given P-value row, the MA line genotype data set used to calculate expected values is shown on top and the corresponding P-value is indicated below.
indicates results from this study.
indicates results from Solorzono et al. 2011.
indicates results from Koboldt et al. 2010.
Conclusion
This study provides important insights into the extent of mutational variation among related animal lineages, and the interrelationships between underlying mutation spectra and patterns of natural polymorphism at loci widely presumed to be neutral. Although our study suggested substantial mutational uniformity in most regards, mutation processes might be more variable than indicated by our findings. Variation among the four sets of MA lines in terms of mutation spectra was just outside of common significance thresholds (P = 0.07). Although our study provided the largest mutation data set for MA line–based mutational analysis to date, a much larger survey of mutational variation that includes MA lines derived from many dozens to hundreds of progenitor genotypes will be required to more broadly and effectively address the extent of mutational variation within and between related animal species.
Supplementary Material
Supplementary figures S1–S4 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank M. Dasenko and C. Sullivan at the OSU Center for Genome Research and Biocomputing for assistance with DNA sequencing and computing. We thank M. Salomon for helpful discussions and explorations of HTS analysis approaches. We thank J. Shapiro and E. C. Andersen for helpful discussions. This work was supported by NIH grant GM072639 to C.F.B. and D.R.D. and NIH grant GM087628 to D.R.D.
References
- Andersen EC, et al. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44:285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appels R, et al. Genome studies at the PAG 2011 conference. Funct Integr Genomics. 2011;11:1–11. doi: 10.1007/s10142-011-0215-6. [DOI] [PubMed] [Google Scholar]
- Babbitt GA, Cotter CR. Functional conservation of nucleosome formation selectively biases presumably neutral molecular variation in yeast genomes. Genome Biol Evol. 2011;3:15–22. doi: 10.1093/gbe/evq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- Baer CF, et al. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc Natl Acad Sci U S A. 2005;102:5785–5790. doi: 10.1073/pnas.0406056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Choi JY. Natural selection shapes nucleotide polymorphism across the genome of the nematode Caenorhabditis briggsae. Genome Res. 2010;20:1103–1111. doi: 10.1101/gr.104331.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Dey A, Murray RL. Evolution of the Caenorhabditis elegans genome. Mol Biol Evol. 2009;26:1199–1234. doi: 10.1093/molbev/msp048. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- Denver DR, et al. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci U S A. 2009;106:16310–16314. doi: 10.1073/pnas.0904895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C, et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445:82–85. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- Halligan DL, et al. Positive and negative selection in murine ultraconserved noncoding elements. Mol Biol Evol. 2011;28:2651–2660. doi: 10.1093/molbev/msr093. [DOI] [PubMed] [Google Scholar]
- Howe DK, Baer CF, Denver DR. High rate of large deletions in Caenorhabditis briggsae mitochondrial genome mutation processes. Genome Biol Evol. 2010;2:29–38. doi: 10.1093/gbe/evp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, et al. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19:1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, et al. A toolkit for rapid gene mapping in the nematode Caenorhabditis briggsae. BMC Genomics. 2010;11:236. doi: 10.1186/1471-2164-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A, Oliver F, Halligan DL, Keightley PD. Positive and negative selection on noncoding DNA close to protein-coding genes in wild house mice. Mol Biol Evol. 2011;28:1183–1191. doi: 10.1093/molbev/msq299. [DOI] [PubMed] [Google Scholar]
- Kunstner A, Nabholz B, Ellegren H. Significant selective constraint at 4-fold degenerate sites in the avian genome and its consequence for detection of positive selection. Genome Biol Evol. 2011;3:1381–1389. doi: 10.1093/gbe/evr112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips N, Salomon M, Custer A, Ostrow D, Baer CF. Spontaneous mutational and standing genetic (co)variation at dinucleotide microsatellites in Caenorhabditis briggsae and Caenorhabditis elegans. Mol Biol Evol. 2009;26:659–669. doi: 10.1093/molbev/msn287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, et al. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 2011;7:e1002174. doi: 10.1371/journal.pgen.1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. Bioessays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Solorzano E, et al. Shifting patterns of natural variation in the nuclear genome of caenorhabditis elegans. BMC Evol Biol. 2011;11:168. doi: 10.1186/1471-2148-11-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]