Abstract
Many software tools for comparative analysis of genomic sequence data have been released in recent decades. Despite this, it remains challenging to determine evolutionary relationships in gene clusters due to their complex histories involving duplications, deletions, inversions, and conversions. One concept describing these relationships is orthology. Orthologs derive from a common ancestor by speciation, in contrast to paralogs, which derive from duplication. Discriminating orthologs from paralogs is a necessary step in most multispecies sequence analyses, but doing so accurately is impeded by the occurrence of gene conversion events. We propose a refined method of orthology assignment based on two paradigms for interpreting its definition: by genomic context or by sequence content. X-orthology (based on context) traces orthology resulting from speciation and duplication only, while N-orthology (based on content) includes the influence of conversion events. We developed a computational method for automatically mapping both types of orthology on a per-nucleotide basis in gene cluster regions studied by comparative sequencing, and we make this mapping accessible by visualizing the output. All of these steps are incorporated into our newly extended CHAP 2 package. We evaluate our method using both simulated data and real gene clusters (including the well-characterized α-globin and β-globin clusters). We also illustrate use of CHAP 2 by analyzing four more loci: CCL (chemokine ligand), IFN (interferon), CYP2abf (part of cytochrome P450 family 2), and KIR (killer cell immunoglobulin-like receptors). These new methods facilitate and extend our understanding of evolution at these and other loci by adding automated accurate evolutionary inference to the biologist's toolkit. The CHAP 2 package is freely available from http://www.bx.psu.edu/miller_lab.
Keywords: gene clusters, orthology, conversion, evolutionary inference, KIR
Introduction
The release of more and more genomic sequence data has facilitated valuable analyses for reconstructing evolutionary histories and predicting the location of functional elements (Murphy et al. 2001; Siepel et al. 2005; The ENCODE Project Consortium 2007). Most computational methods used for these analyses require accurate multisequence alignments. Although several such methods are reasonably accurate for 95% of the genome (Margulies et al. 2007), we found that current multisequence alignment methods are ineffective for studying gene clusters (Hou 2007; Hsu 2009).
With a correct set of alignments for a gene cluster, we expect that orthologous regions from multiple species are aligned with each other, so identifying orthologs is a key step. The sequence relationships are defined as follows. If similar sequences in the genomes of two species are both descended from the same sequence in their most recent common ancestor species (i.e., their separation was caused by a speciation event), then the regions are defined to be orthologous; whereas, if two genomic regions, from the same or different species, are descended from different copies created by a duplication event, then the regions are paralogous (Fitch 1970). For paralogous sequences in the same genome, if the duplication that created them occurred after a given speciation event in that lineage, then the intervals are said to be in-paralogous (relative to that speciation event); if the duplication occurred before the speciation, they are out-paralogous (Sonnhammer and Koonin 2002).
Although the term orthology has been heavily used, conclusions about orthology have not been consistent because different conceptions of orthology were brought to bear in different fields (Ouzounis 1999; Fitch 2000; Jensen 2001) and different groups refined its definition depending on their computational criteria for predicting orthologs (Dewey 2011; Kristensen et al. 2011). Additionally, the so-called gene conversions complicate the orthology definition for those working in molecular biology and bioinformatics (Fitch 2000). A conversion event (which might not actually involve any genes) overwrites part of one paralog with the corresponding part of another. Although the same effect could be achieved by a coincident duplication and deletion, conversion events are believed to result from a different biological mechanism, namely DNA double-strand breaks or a double Holliday junction dissolution mechanism (Chen et al. 2007). Conversions affect a contiguous run of nucleotides, similar to duplications, deletions, inversions, etc.; however, they do not add or remove base positions (except for the occasional incorporation of small indels) nor disturb the relative location or orientation of genomic structures. They only replace the content of certain intervals with similar but slightly different content, and in that sense are more like large substitution events.
We herein refine the concept of orthology to account for the effects of conversion events, explicitly distinguishing two alternative interpretations, which we define as follows. One, which we call X-orthology (short for context orthology), is based only on duplication and speciation events (i.e., excluding conversions). Thus it tracks the positional origins of relatively large contiguous regions, preserving the genomic context of the genes and other features within the assigned orthologs, and focuses on the history of the intervals comprising the genomic structure rather than the history of the particular nucleotides occupying those intervals. The other version, N-orthology (short for content orthology), tracks the origin of each nucleotide in the sequence contents, including any changes due to conversion events. While conversions also affect contiguous regions, these are typically smaller intervals within the paralogs formed by duplications, so N-orthology tends to produce a finer-grained more fragmented set of ortholog assignments.
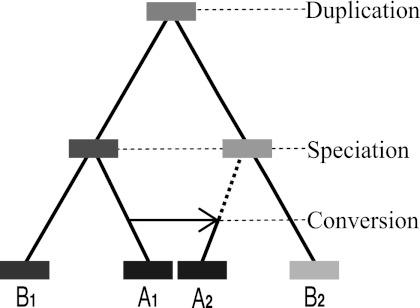
In X-orthology, orthologs are mapped according to where duplication and speciation events occurred—thus the relative positions (order) of intervals assigned as orthologous is typically preserved from the ancestral genome (at least in the absence of subsequent inversions or other rearrangement events). For example, in figure 1, A1 and B1 are orthologous because their intervals originated from the speciation of A and B, and similarly for A2 and B2. However, the assignments may change under the N-orthology interpretation, when orthologous regions are mapped by the origin of their sequence content, since conversion events alter the original X-orthology. For instance, after the conversion event copying A1 over A2, the content origin of A2 is the same as that of A1. Thus according to N-orthology, A2 is orthologous to B1 instead of B2.
FIG. 1.—
Typical scenario for conversion. An ancestral duplication giving rise to two genes 1 and 2 is followed by the split of species A and B. Later, a conversion event may occur between paralogs A1 and A2.
Many studies have aimed to develop orthology-detection methods. Their approaches can generally be classified into two categories. One class attempts to identify orthology relationships by finding the pairs of similar intervals having highest sequence identity, as with COG (Tatusov et al. 2001), TOGA (Lee et al. 2002), OrthoMCL (Li et al. 2003), MBGD (Uchiyama 2007), TOAST (Hou et al. 2009), INPARANOID (Ostlund et al. 2010), eggNOG (Muller et al. 2010), and OrthoDB (Waterhouse et al. 2011). The other is to construct phylogenetic trees, as in HOGENOM (Dufayard et al. 2005), PhyOP (Goodstadt and Ponting 2006), OrthologID (Chiu et al. 2006), TreeFam (Li et al. 2006), LOFT (van der Heijden et al. 2007), SYNERGY (Wapinski et al. 2007), PhylomeDB (Huerta-Cepas et al. 2008), Mestortho (Kim et al. 2008), Evola (Matsuya et al. 2008), PHOG (Datta et al. 2009), EnsemblCompara (Vilella et al. 2009), and PANTHER (Mi et al. 2010).
These methods usually assign orthologs using genomic content as a guide (e.g., Li et al. 2003; Vilella et al. 2009; Muller et al. 2010) and thus are most similar to our N-orthology paradigm, at least in concept. However, many of them use mixed approaches that also depend on context information to varying degrees, sometimes implicitly, making meaningful comparisons difficult. Also, most of them map entire genes to one another, ignoring intergenic regions and making no provision for parts of a gene to have different orthologs on a per-nucleotide basis. And in particular, none of them have considered conversion events complicating the orthology mappings (Chen et al. 2007; Song, Hsu, Riemer, et al. 2011). Although a similarity-based strategy will implicitly account for conversions involving entire regions (in their case genes), the effect of partial conversions on the similarity score can still cause problems, for example, by misleading the assessment of whether duplications occurred before or after speciation. Partial conversions are quite common (Song, Hsu, Riemer, et al. 2011), and since we are interested in the evolutionary history of all DNA in the cluster, we work with the entire duplicated intervals, which are often larger than individual genes and thus more likely to suffer only partial conversion.
At the nucleotide level, N-orthology may be regarded as the true orthology of Fitch's definition, though that paper (Fitch 1970) did not explicitly address conversion either. However, many approaches to defining orthology also use genomic context as a guide (reviewed in Dewey 2011). Some of the earliest work on assigning orthology in gene clusters used alignments in flanking DNA sequences as a guide, specifically to avoid confusion introduced by gene conversion events (Hardison 1984; Hardies et al. 1984; Hardison and Gelinas 1986; Hardison and Miller 1993). These and subsequent studies showed that the aligning flanking sequences also harbor gene regulatory modules, such as distal enhancers. Thus the flanking sequences are not simply some connecting sequences that can be ignored in predicting function of genomic regions, but rather they can contain sequences that regulate the expression of the embedded genes. Hence, it is important to know when a converted gene lies in a context (flanking sequences) that is orthologous to a different gene than the source of the conversion. One may expect it to fall under a different regulatory regimen than that of the source gene. This would be the case for gene pairs that are N-orthologous but not X-orthologous. Thus we believe that both N-orthology and X-orthology are informative, complementary, and have their place, as long as the distinction is clearly made and conversion events are accounted for one way or the other (i.e., traced back or explicitly excluded). Our software automatically computes both, so the researcher is free to choose whichever is most appropriate for a particular study.
To infer orthology relationships for both X-orthology and N-orthology, we designed a new approach utilizing the tools from our CHAP package (Song, Hsu, Riemer, et al. 2011) to detect conversion events and using sequence similarity levels for timing evolutionary events. The new software package that includes our previous CHAP tools plus this new orthology-identifying pipeline is called CHAP 2 (freely available from http://www.bx.psu.edu/miller_lab). Whereas the output of our previous CHAP package is primarily conversion calls, the output of the CHAP 2 orthology pipeline is a set of pairwise alignments in MAF format (see http://genome.ucsc.edu/FAQ/FAQformat) that map intervals in one species to the identified orthologous intervals in another, including noncoding and nongenic regions as well as protein-coding genes. We call these orthologous alignments and visualize them using our Gmaj alignment viewer (Song, Hsu, Riemer, et al. 2011), which shows both the orthology calls and the full set of pairwise alignments simultaneously for comparison. In addition to the pairwise relationships, we also visualize a summary of orthology among the genes of multiple species with respect to a given reference species, which is automatically generated using PostScript figures, as in figure 5.
FIG. 5.—
Orthology structure based on pairwise relationships automatically inferred and visualized by our pipeline for human versus other mammals, in the β-globin (A,B) and α-globin (C,D) clusters. The left panels (A,C) show the X-orthology calls, while the right ones (B,D) show the N-orthology results. Colored boxes represent genes; those with dashed borders are pseudogenes. Colors in each nonhuman gene indicate its human orthologs. Genes with multiple human orthologs are split vertically (i.e., with a horizontal line). For instance, in panel B, the fourth and fifth gibbon genes are orthologous to both HBG1 and HBG2. Genes with different partial relationships within their boundaries are partitioned horizontally (e.g., in the second elephant gene in panel A, the left-most part is orthologous to HBB, while the right-most part is orthologous to HBD). Note that the lengths of boxes, spaces, and partitions are not proportional to their actual genomic lengths, but the order is the same as their genomic order. Vertical ordering of colors within a horizontal partition is not significant. A gray color indicates unassigned orthologs.
The CHAP 2 package is designed for Unix/Linux-based systems, including Mac OS X. Users will also need to install the RepeatMasker program (Smit et al. 1996–2010) and a suitable program for preparing gene annotations, such as GeneWise (Birney et al. 2004). For each gene cluster to be analyzed, the user provides 1) genomic sequences from two or more species, 2) a gene annotation file for each of the species, and 3) a Newick-formatted phylogenetic tree for the species. Then, a single command runs the entire pipeline, producing orthologous alignments in MAF format for the reference sequence versus each of the others (for both X- and N-orthology), a list of inferred evolutionary events in the reference species that were used in making the orthology calls, a list of detected gene conversions in all of the species, and ready-to-view PostScript diagrams similar to figure 5. If desired, the orthology calls can then be examined interactively using the included Gmaj viewer.
A major challenge in developing software for detecting orthologs is the lack of gold-standard data for evaluating their correctness. We evaluate our programs using high-quality sequence data for a set of gene clusters (including the well-studied β-globin and α-globin clusters) as well as simulation data produced using the method designed in our study evaluating conversion detectors (Song, Hsu, Riemer, and Miller 2011).
In addition, we illustrate the capability of our method by analyzing a few other gene clusters. To obtain human gene cluster regions, we started by identifying 457 regions containing recent duplications (∼215 Mb; i.e., 7% of the human genome) using self-alignments in the genome (Zhang et al. 2009). We selected 165 clusters that include genes within the duplicated regions (∼111 Mb). From this list, we targeted four clusters that are biomedically interesting due to their association with human genetic diseases, and generated high-quality sequence data for them from seven primate species. Specifically, gene copy number in the chemokine ligand (CCL) cluster (hg19.chr17:34,310,693–34,812,885) correlates with susceptibility to HIV (Degenhardt et al. 2009), the interferon (IFN) cluster (hg19.chr9:21,058,760–21,481,698) is associated with sarcoidosis (Akahoshi et al. 2004), part of the cytochrome P450 family 2 (CYP2abf) cluster (hg19.chr19:41,324,635–41,712,359) is implicated in lung cancer (Wang et al. 2003), and the killer-cell immunoglobulin-like receptors (KIR) cluster (hg19.chr19:55,233,386–55,380,386) is linked to HIV susceptibility (Lopez-Vazquez et al. 2005). Since good-quality primate sequence assemblies for the CCL, IFN, CYP2abf, and KIR clusters were mostly unavailable, accurate Sanger sequences for them were generated by the National Institutes of Health Intramural Sequencing Center (NISC), which we then analyzed using CHAP 2. Such analyses can lead to a better understanding of the evolutionary histories of genes and their functions in complex gene clusters.
Materials and Methods
Adjusting Sequence Similarity for Timing Duplication Events
Similarity of sequence contents is an important signal for timing duplication events; if one match in a set of self-alignments (i.e., when a sequence is aligned to itself) has a higher similarity level than others, then that match is likely to be the most recent duplication (Bailey et al. 2002). Also, if the similarity of one match between two species is higher than any other matches involving the same regions, then it is most likely to be common ancestral—that is, orthologous (Wapinski et al. 2007). Thus we can identify orthologs between two species in a gene cluster using the similarity levels of the intra- and interspecies alignments. These are X-orthologs if we consider only speciation and duplication events (by specifically excluding the effects of conversions); our method identifies the X-orthologs first, followed by the N-orthologs.
Although similarity levels of alignments provide key information for inferring the relative timing of duplication events, they can be misleading due to conversion events (Hsu et al. 2010). When a conversion event occurs, the converted region becomes more similar to the source region than before, which can cause analysis software to mistake prespeciation duplications as being more recent (e.g., postspeciation). We recently developed software for detecting conversion events (Song, Hsu, Riemer, et al. 2011), so we utilize that and then recalculate the similarity level of each alignment by excluding the positions involved in conversion events. If the conversion covers the entire alignment, none of sequence remains to recalculate the similarity. So, we retain the original similarity of this alignment and handle this case as a special one using an additional criterion introduced in Song et al. (2010).
In addition to regions involved in conversion, protein-coding exons may also influence the apparent timing of duplication events when using sequence similarities because they have a tendency to be more conserved than noncoding regions due to their functional constraints (Gish and States 1993). If two regions involved in a duplication include protein-coding regions, their similarity may be higher than later duplications involving only noncoding regions. Therefore, we recalculate the similarity level of alignments that involve protein-coding exons by excluding the protein-coding positions.
Homology Graph
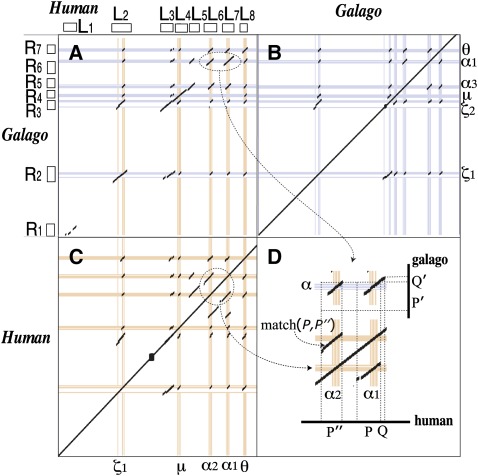
Given two sequences sp1 and sp2 for the same gene cluster in different species, we first construct a graph composed of two disjoint sets of nodes that correspond to the matching regions (i.e., homologous regions) in a set of interspecies alignments between sp1 and sp2. Figure 2 shows example alignments for human and galago in the α-globin cluster obtained from the LASTZ alignment program (Harris 2007).
FIG. 2.—
α-globin cluster in human and galago. (A) Interspecies alignments, (B) galago self-alignments, and (C) human self-alignments are shown. Human and galago both have five genes, but they are not the same five. (D) shows how to determine the parent and child segments in the duplication involving α1 and α2 in human. The hollow boxes next to the “L”s and “R”s are the matching regions represented by nodes in the homology graph of figure 3A.
Let , where such that L and R are sets of nodes representing the matching regions from the interspecies alignments in sp1 and sp2, respectively, and E is a set of edges representing all aligned matches between L and R, weighted by their similarity scores, which we call cross-edges. Self-alignments for each species are also considered in this approach, so G is extended using the self-alignments of sp1 and sp2. Let two matching regions in a self-alignment of sp1 be denoted as P and Q. If they are not included in G from the interspecies alignments yet, the two nodes for P and Q are added in L and connected by a similarity-weighted edge. Similarly, nodes and edges for the sp2 self-alignments are added to R. These self-alignment edges are called in-edges, and they represent candidates for duplications. We call G a homology graph. The α-globin cluster in figure 2 corresponds to the homology graph shown in figure 3A.
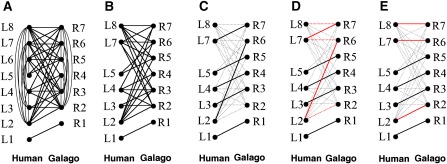
FIG. 3.—
Illustration of getting one-to-one ancestral alignments. (A) Bipartite graph of eight human nodes in L and seven galago nodes in R. The nodes are connected with in-edges based on self-alignments in figure 2 (B,C) and cross-edges based on interspecies alignments in figure 2A. (B) Bipartite graph after removing all postspeciation duplicated regions (accordingly, L6 was removed). Because the in-edges are not used after this step, they are also removed. (C) Example of an initial matching. Nodes that belong to this matching still remain in black lines, but the others are dimmed in gray dotted lines. (D) Augmenting path of which both end-nodes are unmatched in C. The path in red that starts from unmatched node L8 and ends in node R2 demonstrates an augmenting path of the graph and matching in C. For each augmenting path, an incremental weight (i.e., the sum of weights of the edges in dotted red lines) is calculated. The augmenting path having the maximum increment is selected. (E) The matching modified by adding red dotted edges and excluding red bold edges in D. Steps D and E are repeated until there are no more augmenting paths. E is the maximum-weight bipartite matching that corresponds to the one-to-one ancestral alignments. The algorithm used in these steps is proved to construct the maximum-weight bipartite matching in Johnson and McGeoch (1993).
Removing Recent Duplications
G is reduced to a less complicated graph by removing regions formed by duplication events that occurred after speciation. The postspeciation duplications are identified as follows. The candidates for duplication events are the in-edges in G. Note that a duplication event involves a single local alignment because alignments split by insertion of repeats are chained right after obtaining LASTZ alignments by a preprocessing step of the CAGE pipeline (Song et al. 2010). If an in-edge has a higher weight (adjusted similarity) than any cross-edges entering or leaving the two nodes that it connects, it is considered to represent a postspeciation event except for in-edges involved in conversions covering the entire local self-alignment. When the entire self-alignment is covered by conversion, it is determined by the CAGE criterion based on the overlap relationships of the matching regions from the alignments whether it is a pre- or postspeciation event as well as comparing edges' weight (Song et al. 2010). First, an in-edge with the highest similarity of all the postspeciation ones is selected as the latest duplication. In this case, in-edges entirely covered by conversion may be inferred as later events than their actual time, but they do not influence orthology results if self-alignments do not overlap any other alignments, and the CAGE criterion adjusts the duplication order otherwise (see details in Song et al. 2010). Then, its parent (original copy) and child (inserted copy) regions are identified, assuming the parent segment will keep a longer conserved synteny with the other species than the child (Han et al. 2009). Unlike the method of Han et al. (2009), which relies on gene order information in a syntenic region, we use the similarity of syntenic regions, including nongenic parts as well as genes (Wapinski et al. 2007).
Figure 2 shows interspecies alignments and self-alignments for human and galago in the α-globin cluster. After constructing the homology graph based on these alignments, one postspeciation duplication was inferred. Match (P,P″) involving the human α1 and α2 genes was entirely covered by a conversion. So, we checked additional CAGE criterion to determine if it is either pre- or postevent, although the match keeps 98% identity. As a result, it is inferred as a postspeciation event. To decide the parent–daughter relationship between two regions, we observe their flanking regions. The match between P in human and P′ in galago has a contiguous 600-bp flanking match (Q,Q′), as shown in figure 2D, so we infer that P including α1 is the parent segment and P″ including α2 is the child. For a tandem duplication, the flanking region may be the duplicate itself. Some deletion events that occur in the boundary area of a duplication may cause loss of the conserved syntenic information as well. If the parent–daughter relationship is not identified by the flanking regions, it is marked as an “undetermined” state.
Once the parent–daughter relationship is identified, the child duplicate segment is removed from the alignments (i.e., the duplication event is “rewound”). For instance, in figure 2, P″ (including α2) is removed. In case of undetermined ones, either one is removed.
Finally, G is reduced by removing all nodes contained in representing alignments of the removed child duplicate segments. These steps are iterated until there are no remaining postspeciation duplications. As a result of this procedure, G in figure 3A is reduced to figure 3B by removing nodes L6. Since in-edges are not necessary to keep in G any longer after all postspeciation duplication events are dealt with, the remaining in-edges are also removed.
Reconstructing One-to-One Common Ancestral Orthologous Alignments
Now, we have only orthologous and out-paralogous alignments of the interspecies alignments between sp1 and sp2 after removing regions determined to have been inserted by duplication events after the species split (i.e., all remaining edges in G represent one-to-one orthologous and out-paralogous mappings). The task of constructing the common ancestral orthologous alignments for sp1 and sp2 is accomplished by obtaining one-to-one mappings in G based on best reciprocal hits. This problem can be stated as follows.
Problem 1. Suppose L has nl match regions and R has nr match regions. Let denote nodes in L and nodes in R. The weight of an edge between li and rj, denoted as wij, is recomputed by multiplying the alignment similarity of li and rj (denoted as sij) by the length of the match. Building the ancestral orthologous alignments of sp1 and sp2 is formulated as a maximum-weight bipartite matching problem, namely: given G, find a matching M, which maximizes the sum of the weights of the edges that belong to M.
We use an efficient algorithm for solving the maximum-weight bipartite matching problem (Johnson and McGeoch 1993). Figure 3 illustrates how the algorithm works to construct the one-to-one ancestral mappings. Final mappings in figure 3E correspond to ancestral orthologous alignments in figure 4A.
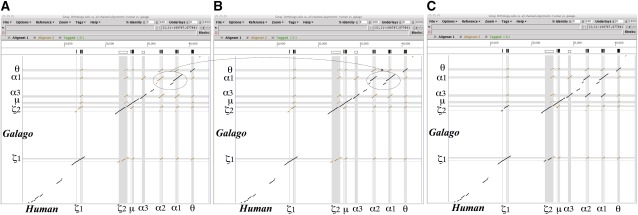
FIG. 4.—
Illustration of orthologous alignments between human and galago for (A) ancestral orthologous alignments, (B) X-orthology, and (C) N-orthology in the α-globin gene cluster using the Gmaj viewer. The ancestral orthologous alignments are one-to-one orthologous alignments, which map all orthologs immediately after human and galago split (i.e., right before all postspeciation events). Our method determines these ancestral orthologous alignments first, and then obtains X-orthology and N-orthology by adding orthologs formed by postspeciation events, such as those indicated with ovals in (A,B). For details, see text. Local alignments in brown represent all interspecies homologous regions between human and galago, and those in black, the orthologous regions as a subset of the brown alignments.
Obtaining X-Orthology
After the one-to-one ancestral alignments are obtained, a set of many-to-many X-orthologs is mapped by repeating the postspeciation duplications that were removed in the previous step, in the order of their event time. When a duplication event is reapplied to the ancestral alignments, which map all orthologs between two species before this duplication event happened, all orthologs of the parental copy are orthologous to the daughter one of that duplication. So, we add interspecies alignments that align the daughter copy to the orthologs of the parental one to the ancestral alignments. For example, a duplication between the α2 and α1 genes in human is reapplied, as the dotted circles and arc in figure 4A and B show. This step is repeated until all postspeciation duplication events are restored. Finally, we have many-to-many X-orthologous alignments between the two species.
Because these steps are pairwise based, duplication events inferred in our pipeline occasionally may not be consistent in all species. For instance, parent and daughter copies of a duplication may not be consistent when they are not determined by flanking regions, such as tandem duplication. Duplication time may sometimes be inconsistent when the sequence similarity of a self-alignment from a duplication event is equal to orthologous interspecies alignments involving the parent and daughter copies of the duplication. Note that the CAGE criterion determines whether a duplication is a pre- or postspeciation event when similarity comparison can not determine the event time. In order to adjust and refine orthologs having conflicts caused by the inference of inconsistent time or parent–daughter relationship of duplications, we check each event from the most recent one in a bottom-up approach based on the species tree. If a duplication event in species sp1 occurs after the split of sp1 and another species sp2, the duplication should be inferred in sp1 versus all out-group species of sp1 and sp2 (note the pairwise steps of sp1 and the out-group species infer older events as well as this recent one). If all out-group species agree with the recent duplication, it stays in the events inferred by all pairwise comparisons of sequences. If not, it is removed in the event results in sp1 and sp2 and treated as an older one that happened before the split of sp1 and sp2. While the duplication time is adjusted, the consistency of its parent–daughter relationship is also checked. We choose the majority of all the cases including that event. Minor cases are adjusted to the parent–daughter relationship of the majority in the pairwise results containing that duplication. This step ends when we reach the root of the species tree.
Obtaining N-Orthology
In order to obtain N-orthologs, we consider conversion events as well as duplications. Conversion time is estimated by similarity comparison. First, sequence similarity is calculated between two regions involved in each conversion event. If their similarity is higher than any other orthologous alignments involving those regions, the conversion is called as a postspeciation event. Their similarity is also compared with that of self-alignments corresponding to postspeciation duplication events to determine its relative event order among the duplications. Once all postspeciation conversion events are placed in the results of postspeciation events in their time order in each pairwise comparison, all the postspeciation conversion and duplication events are repeated in the order of their event time, similar to X-orthology. The consistency of conversion events is also handled in the same way as the X-orthology case. As a result, our pipeline generates all N-orthologs between human and galago in the α-globin cluster, as shown in figure 4C.
Results
Orthologous Relationships of Genes in the β-Globin and α-Globin Clusters
Using CHAP 2, we obtained X- and N-orthology mappings of human versus 13 other species for the β-globin cluster, and human versus 14 other species for the α-globin cluster. The DNA sequence data for these clusters are available at the ENSEMBL (http://www.ensembl.org) and GenBank (http://www.ncbi.nlm.nih.gov/genbank) Web sites. Human gene annotations [HBB(β), HBD(δ), HBH(η), HBG1(γ1), HBG2(γ2), and HBE(ε) for the β-globin cluster (listed 3′ → 5′) and HBZ-T1(ζ1), HBZ-T2(ζ2), HBK(μ), HBA-T1(α3), HBA-T2(α2), HBA-T3(α1), and HBQ(θ) for the α-globin cluster (5′ → 3′)] were downloaded from the University of California–Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu). Annotation information for nonhuman species was obtained using GeneWise (Birney et al. 2004) for coding genes (followed by manual curation) and using CHAP 2's pseudogene detector based on LASTZ sequence alignments (Harris 2007) for pseudogenes. The panels in figure 5 (generated automatically by our software) summarize the orthologous relationships among genes in the β-globin and α-globin clusters (for further explanation, see Supplementary Material online).
Our inference for X-orthology (fig. 5A) in the β-globin cluster is consistent with other studies (Fitch et al. 1991; Opazo et al. 2008), likewise for the α-globin cluster (fig. 5C; e.g., Hoffmann et al. 2008). In particular, our inference of a fusion event for the second elephant gene in figure 5A agrees with published results (Opazo et al. 2009). Figure 5B and D show that in both globin clusters, the results for N-orthology are somewhat different (for additional details, see Supplementary Material online).
In addition to validating our X-orthologs in the β-globin and α-globin clusters against existing studies using context-based methods (e.g., Opazo et al. 2008, 2009), we wanted to compare our N-orthologs to those from other methods based on sequence content. Although many existing methods have used approaches that are primarily content based, most of them are limited to calling orthologs on a per-gene basis, that is, for assigning which entire gene is orthologous to each given entire gene. Because our method often identifies different orthologs for different parts of a gene, it is difficult to make a meaningful comparison between our results and others on a per-gene basis. In addition, our method needs only DNA sequences to determine orthology, whereas others require protein sequences and/or gene annotation information which can be difficult to obtain, as protein-coding annotations for nonhuman species in gene clusters are usually either unavailable or not as accurate as for human. (Our method uses gene annotations for visualization and if provided will take advantage of them to slightly improve its sequence similarity calculations, but it does not require them to determine orthology.)
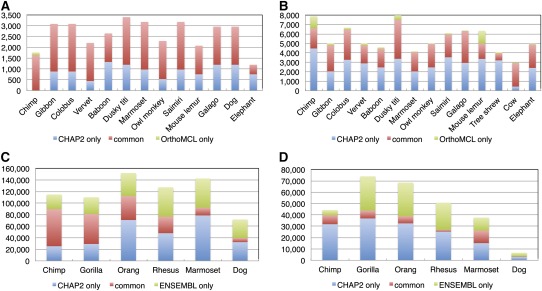
Nevertheless, we have attempted to compare our N-orthology results to those from OrthoMCL (Li et al. 2003), for which software is available (many methods are available only as precomputed output in databases). We ran OrthoMCL with the same sequences and gene annotations used for the analyses of the β-globin and α-globin clusters in figure 5, and compared its results with the N-orthologs from CHAP 2 (note that OrthoMCL requires protein sequences, so this comparison also depends upon the accuracy of GeneWise for determining protein sequences in the nonhuman species). Because OrthoMCL's output consists of groups of orthologous genes rather than per-nucleotide calls, we performed the comparison in coding regions only, treating 100% of the coding bases of each human gene as being mapped to the nonhuman genes placed in the same orthologous group. Figure 6A and B shows the differences between the two programs. Out of all pairs of coding nucleotides in the β-globin cluster that are called as orthologous between human and any of the 13 other species by either or both methods (based on human bases), 67.0% are called in common by both programs, 32.6% by CHAP 2 only and 0.4% by OrthoMCL but not by CHAP 2. For the α-globin cluster, 48.6% are called in common, 49.5% by CHAP 2 only, and 1.9% by OrthoMCL only. Of course, we expect substantial differences here, since CHAP 2 allows parts of a gene to have different orthologs and OrthoMCL does not.
FIG. 6.—
Comparison of CHAP 2's N-orthology calls with results from OrthoMCL (A,B) and ENSEMBL Compara's “EPO” alignments (C,D), for human versus other mammalian species in the β-globin (A,C) and α-globin (B,D) clusters. In A and B the red bars show how many pairs of coding nucleotides are assigned as orthologous by both CHAP 2 and OrthoMCL, while blue and green indicate those called by CHAP 2 only or by OrthoMCL only, respectively. In C and D, the color meanings are similar, except that Compara enables us to include noncoding bases as well. For the comparison of CHAP 2 with Compara, we reran the CHAP 2 pipeline using the same sequence assemblies that Compara used, which are slightly different from those we used for our main analysis of the globin clusters in figure 5 and the comparison in A and B.
In addition, the ENSEMBL Compara database (http://www.ensembl.org) provides content-based orthology calls on a per-nucleotide basis similar to ours, for parts of genes and even in nongenic regions. Thus we were able to use that for comparison, at least in the β-globin and α-globin clusters where Compara has orthology data available. Figure 6C and D shows that the results are quite different. Out of all pairs of nucleotides in the β-globin cluster that are called as orthologous between human and any of the six other species by either or both methods (based on human bases), 76.4% are called in common by both methods, 17.6% are called only by CHAP 2, and 5.9% are called only by Compara. For the α-globin cluster, 56.4% are called in common, 32.3% are called only by CHAP 2, and 11.4% are called only by Compara. In addition, we counted the number of human protein-coding nucleotides for which orthologs in one or more of the six other species are assigned by CHAP 2 and by Compara. CHAP 2 assigns orthologs for 95.0% of the human β-globin coding bases and 66.2% for α-globin (77.4% if dog is excluded; the dog α-globin coding exons do not align with the human coding exons at all in the initial LASTZ alignment step), while Compara assigns 94.7% and 52.7% (63.2%), respectively.
Patterns of Homology and Evidence for Gene Conversion in the KIR Locus
The KIR locus is a highly polymorphic locus found only in simian primates and encoding receptors used by Natural Killer (NK) cells (and certain T cells) to recognize MHC Class I ligands. Much of the gene content variation at the locus in humans is captured by haplotypes A and B (Martin et al. 2004). These haplotypes share the genes KIR3DL3, KIR3DP1, KIR2DL4, and KIR3DL2, collectively described as framework loci (Wilson et al. 2000). Other genes at the locus are variably present and subject to linkage disequilibrium (LD) that is strongest on either side of KIR2DL4 (Abi-Rached et al. 2010); KIR2DL has relatively weak LD and is also present in both sequences analyzed here. The notably high level of polymorphism at this locus is thought to be caused by high levels of gene duplication and asymmetric recombination resulting in duplications/deletions, while patterns of LD may reflect reciprocal recombination on either side of KIR2DL4 and extensive gene conversion or exon shuffling (Rajalingam et al. 2004). The maintenance of diversity, in turn, is linked to balancing selection relating to the dual role of NK cells in immune and reproductive functions (Parham 2005).
Given the inferred role of recombination and conversion at the KIR locus, complex orthology mappings are expected, yet previous efforts to establish phylogenetic relationships among primate KIR genes using full protein sequences (Guethlein et al. 2002; Sambrook et al. 2006) are effectively grounded in the X-orthology concept. However, corrections for recombination events have been attempted in other studies in which coding sequences were decomposed into distinct functional domains (Rajalingam et al. 2004; Cadavid and Lun 2009), and some specific instances of gene conversion have been noted (Shilling et al. 1998; Graef et al. 2009; Abi-Rached et al. 2010; summarized in the Supplementary Material online). A central role for gene duplication is also invoked in models of the evolution of the locus in humans (Martin et al. 2004) and across species (Guethlein et al. 2007), indicating that a systematic application of the N-orthology concept to this locus would have utility for evolutionary studies.
The presence of contiguous sequence for the A and B haplotypes presents an opportunity for direct and unbiased observation of duplications leading to in-paralogy and of gene conversion occurring within human lineages and between all parts of the locus: exonic, intronic, and intergenic. Sequences from other great apes (while they will only sample diversity therein) permit the assignment of ancestral states and differentiation of sources and targets of gene conversion. GenBank annotations for both human haplotypes were propagated into the package for this analysis, while genes and pseudogenes in other species were inferred.
Figure 7A and B shows the orthology relations detected. CHAP 2 was able to recapitulate previously described orthologies between genes and to do so in both concepts, suggesting that some areas are relatively unperturbed by gene conversion. For example, in humans, the orthology between KIR3DL1 (A haplotype) and KIR3DS1 (B haplotype) (Sambrook et al. 2006) was well supported by the X-orthology analysis (fig. 7A), and the N-orthology analysis suggests that this relationship has been free of conversions within the sampled human lineages. Similar conclusions can be made between species. For example, orthology established by both concepts was confirmed between human and chimp KIR3DL3 (cf. Abi-Rached et al. 2010).
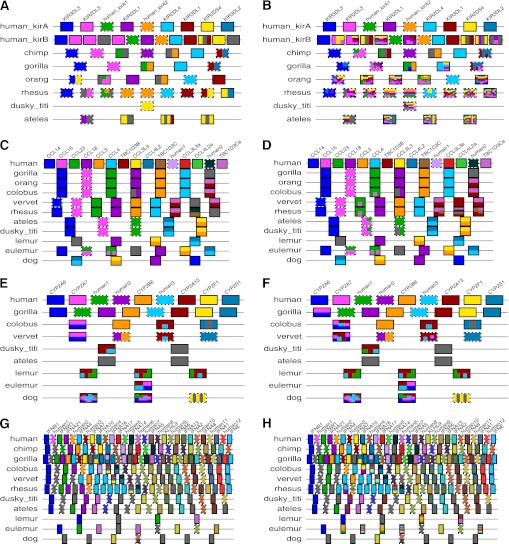
FIG. 7.—
Orthology structure automatically inferred and visualized by our pipeline for human versus other mammals in four additional clusters: KIR (A,B), CCL (C,D), CYP2abf (E,F), and IFN (G,H). As in figure 5, the left panels (A,C,E,G) show the X-orthology calls, while the right ones (B,D,F,H) show the N-orthology results.
Within the human lineage 2 and 14, gene conversion events were identified in haplotypes A and B, respectively, listed in supplementary table S1 (Supplementary Material online). Two events detected in haplotype B with high confidence affected exons. The last four exons of KIR2DS5 are superposed on KIR2DS2, and the first exon of KIR2DS5 transferred to KIR2DL5B. These events, revealed here by our CHAP 2 conversion detector, expand on the generally described pattern of exon shuffling (Rajalingam et al. 2004); however, events affecting intronic sequence can also be described within humans. For example, in haplotype A, conversions occur between the KIR2DS4/KIR3DL2 and KIR2DL3/KIR2DL1 gene pairs, with both sets involving solely intronic sequences—a pattern only detectable when contiguous noncoding segments are analyzed systematically for N-orthology. The application of this concept, made possible by CHAP 2, therefore permits an exploration of the broader pattern of gene conversion within the human lineage and beyond (for trends noted in the New World monkeys, see Supplementary Material online).
Summary of Orthologous Relationships in the CCL, CYP2abf, and IFN Clusters
In addition to the two globin clusters and the KIR cluster, we used CHAP 2 to obtain X- and N-orthologous alignments for three more gene clusters: CCL, CYP2abf, and IFN (fig. 7C–H).
The CCL gene family encodes chemokines, small proteins regulating the migration of lymphocytes. This role of chemokines is important to control the immune response to bacterial and viral infections, inflammation, and cancer. Among the CCL genes, CCL3 and CCL4 have been studied extensively since their association with HIV susceptibility was reported (Gonzalez et al. 2005). The CCL3 and CCL4 genes are in a duplication unit along with one copy of TBC1D3, and three copies of this unit were identified in the human reference genome (fig. 7C and D). The human CCL4 and CCL4-like genes have only one amino acid difference, and the CCL4-like genes such as CCL4L2 and CCL4L2a have no difference in their coding sequences. However, one nucleotide substitution (AG → GG) at the acceptor splice site of intron 2 of CCL4L2 generates nine alternative transcripts (Colobran et al. 2005). The alternative transcripts produced by the GG site were predicted to lack five amino acids encoded by the third exon of CCL4L2. Decreased expression of CCL4L2 may have functional implications. Interestingly, CHAP 2 detected conversion events (CCL4 → CCL4L2) involving the entire region of the two genes. For the substitution of A (CCL4 and CCL4L2a) for G (CCL4L2) in the acceptor splice site of the second intron of CCL4L2, the recent gene conversion converted G back again to A. This event may generate polymorphism at the site (rs4796195) and contribute to recovering gene function.
The CYP2abf cluster contains four subfamilies of the CYP2 family: CYP2A, CYP2B, CYP2F, and CYP2S in primates (Hu et al. 2008). Genomic rearrangements in each lineage have altered the copy number of CYP2 genes among species (fig. 7E and F). For example, human and lemur have the most CYP2A subfamily gene copies. However, the origin of the three CYP2A copies is different between these species. As shown in figure 7E, all three copies in lemur showed X-orthology with CYP2A13 and the two human pseudogenes. This supports independent duplications generating each copy of the lemur and human CYP2A genes. Moreover, our CHAP 2 package detected conversion events in these regions (fig. 7F). The burgundy color in the first gene of colobus monkey and vervet represent conversion events between CYP2A13 and the ancestor of CYP2A6 and CYP2A7; the sequence of the CYP2A6/CYP2A7 ancestor was converted by the content of CYP2A13. The function of the genes could be affected by these events. The CYP2A6 enzyme metabolizes nicotine, the primary compound in tobacco (Pianezza et al. 1998). CYP2A13 is known to play important roles in metabolism of a major tobacco-specific carcinogen, 4(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) (Su et al. 2000). The function of CYP2A7 is not yet known, but it is expressed in the human liver, as is CYP2A6. Therefore, all three genes may have significant roles responding to chemicals from the external environment. The dynamic evolution of these genes in the cluster could help organisms to adapt to rapid environmental changes.
The IFN cluster has very complicated genomic structures due to gene copy number variations and high similarity among these gene copies (fig. 7G and H). This cluster includes mainly the interferon alpha (IFNA) family, which plays an important role in the innate immune response (Levy and Farcia-Santre 2001). This gene family shows species-specific gene duplications and frequent gene conversion events (Miyata et al. 1985; Woelk et al. 2007). This is corroborated by our results for the species-specific gene compositions using both X- and N-orthology. All species have different numbers of gene copies (fig. 7G), indicating gene gains and losses by frequent rearrangements. Moreover, our package detected frequent conversion events in the IFN cluster (fig. 7H), many of which occurred in coding regions. For example, a gene conversion in the human lineage occurred from IFNA4 to IFNA7. Also, the entire genic region of IFNA1 was converted by IFNA13. These events correspond with species-specific gene groupings on the phylogenetic tree identified in a previous study (Woelk et al. 2007).
Summary of Orthologous Relationships in Six Gene Clusters
The nonhuman species used for analyzing the six gene clusters are listed in supplementary tables S2–S7 (Supplementary Material online), which include the average sequence similarity of the human regions and their orthologs in each species. Sequences for gorilla, colobus, vervet monkey, dusky titi, ateles, lemur, and eulemur (black lemur) were newly generated by NISC; those for the other species (including the human KIR haplotype B sequence) were downloaded from the ENSEMBL and GenBank Web sites. We summarize the orthology results in Table 1.
Table 1.
Comparison of Orthologous Alignments between Human and Other Species according to X- and N-Orthology in Six Gene Clusters
| β-globin | α-globin | CCL | IFN | CYP2abf | KIR | |
| Number of all interspecies homologous aligning pairs of segments | 551 | 616 | 1,441 | 8,487 | 2,711 | 1,515 |
| Number of pairs called in common as both X- and N-orthologous | 143 (26.0%) | 143 (23.2%) | 305 (21.2%) | 286 (3.4%) | 329 (12.1%) | 66 (4.4%) |
| Number of pairs called only as N-orthologous | 24 (4.4%) | 66 (10.7%) | 35 (2.4%) | 115 (1.4%) | 64 (2.4%) | 291 (19.2%) |
| Number of pairs called only as X-orthologous | 0 (0.0%) | 2 (0.3%) | 0 (0.0%) | 1 (0.0%) | 1 (0.0%) | 1 (0.1%) |
| Total length of interspecies homologous alignments based on human bases | 1,165,759 | 720,851 | 3,404,892 | 12,470,575 | 2,727,002 | 4,719,583 |
| Total length of alignments called in common as both X- and N-orthologous based on human bases | 757,212 (65.0%) | 326,801 (45.3%) | 1,577,090 (46.3%) | 2,123,807 (17.0%) | 1,110,567 (40.7%) | 597,357 (12.7%) |
| Total length of alignments called only as N-orthologous based on human bases | 81,950 (7.0%) | 119,451 (16.6%) | 149,822 (4.4%) | 214,097 (1.7%) | 139,497 (5.1%) | 1,126,059 (23.9%) |
| Total length of alignments called only as X-orthologous based on human bases | 463 (0.0%) | 5,987 (0.8%) | 991 (0.0%) | 5,001 (0.0%) | 12,543 (0.5%) | 21,488 (0.5%) |
First, we counted all homologous aligning pairs of segments between human and the other species. These numbers show the evolutionary complexity of each gene cluster. Next, we counted the number of homologous pairs called only as X-orthologs, only as N-orthologs, and in common by both methods. We found that 26.0%, 23.2%, 21.2%, 3.4%, 12.1%, and 4.4% of the interspecies homologous pairs were called as orthologous according to both paradigms for the β-globin, α-globin, CCL, IFN, CYP2abf, and KIR clusters, respectively. Because homologous pairs vary in their length, we calculated the fraction of orthologous sequence in terms of the alignment length based on human bases. The portions called in common by both paradigms were 65.0%, 45.3%, 46.3%, 17.0%, 40.7%, and 12.7% of the interspecies homologous base pairs. Next, we computed the portions of orthologous pairs that fell in only one orthology category. 7.0%, 16.6%, 4.4%, 1.7%, 5.1%, and 23.9% of the interspecies homologous base pairs were called only as N-orthologous in the six clusters, respectively, while the portions called only as X-orthologous were less than 1% in all six clusters. On average, the orthologous portions of the IFN and KIR clusters are quite low compared with other clusters. This means that many homologs in the IFN and KIR clusters are out-paralogous and implies that many evolutionary events occurred in these clusters before the split of human and each other species—that is, the IFN and KIR clusters have been very active in terms of large-scale genomic changes compared with the other clusters. Interestingly, the portion of KIR orthologs increased markedly under N-orthology; this suggests that many recent conversion events have occurred in the KIR cluster.
Evaluation by Simulation
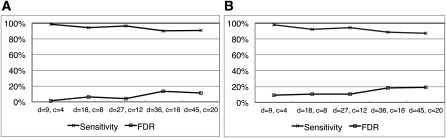
We evaluated the performance of our orthology pipeline using simulation data sets. These data were generated by a simulator for gene cluster evolution (Song, Hsu, Riemer, and Miller 2011). The simulation starts with a 200-kb duplication-free sequence treated as an ancestral cluster. Large-scale events, such as duplications, deletions, and conversions, as well as small-scale mutations were simulated, and finally three sequences mimicking human, Old World monkeys, and New World monkeys were generated (note that our simulator also applies purifying selection; see details in Song, Hsu, Riemer, and Miller 2011). Since the actual scenario for each data set is already known from the simulation, we have true orthologs that can be used to compare our results and to evaluate the performance of our methods. With each simulation data set, we ran CHAP 2 to infer both types of orthologs. Figure 8 shows the accuracy of our results with simulation data sets. Note that to the best of our knowledge, no other studies have developed software for discriminating between the concepts of X- and N-orthology that we could use for comparison. Although there are existing methods that infer content-based orthology, most of them are limited to calling orthologs on a per-gene basis. Moreover, they require protein sequences as input, but our simulated data sets are DNA only and do not necessarily correspond to realistic proteins, since CHAP 2's orthology calls are not gene based (the automatic figures use genes as convenient illustrative groupings that are likely to be meaningful and interesting to see, but the orthology algorithms make very little use of gene information). ENSEMBL Compara provides orthology results on a per-nucleotide basis similar to ours, but unfortunately, we could not run their program for our simulation study because it is not publicly available.
FIG. 8.—
Sensitivity and FDR of (A) X-orthology and (B) N-orthology results from our pipeline using simulation data sets reflecting d duplications and c conversions. Sensitivity is obtained by calculating the fraction of orthologous pairs of nucleotides that were detected correctly and FDR by computing the fraction of called pairs that were incorrect.
Conclusion
Our methods for accurately and automatically detecting orthologs should accelerate the biomedical analysis of complex gene clusters. We believe that the combination of outputs, including a visual overview, facilitates accurate identification of conversion events and the impact these have on inferences about orthology. This, in turn, should help correct misapprehensions regarding the evolution of gene clusters subject to frequent conversion events and encourage the use of conversion detection prior to phylogenetic inference (Hsu et al. 2010) or the estimation of purifying or positive selection (Edwards et al. 2006; Wilson and McVean 2006). Another benefit is the conceptual clarity brought by refining the concept of orthology while still respecting its traditional definition (Fitch 1970).
A major motivation for this investigation was our desire to ultimately supply the community with better whole-genome sequence alignments. We feel that multispecies alignments of entire mammalian genome sequences currently provide reasonable accuracy for single-copy regions of the genome, but often perform inadequately and/or inconsistently for gene clusters. A major use of interspecies alignments is to transfer functional data from one species to another, making an alignment most useful if aligned functional regions have the same or analogous function in the two species. When a gene has one X-ortholog and a different N-ortholog, which should it be aligned to? One could reason that the structure of a protein, and by implication its function, is determined by its gene sequence, so the N-ortholog is to be preferred. On the other hand, the regulatory signals lying outside of the coding region may influence function more than the coding region does, suggesting that at least in some cases the X-ortholog may be preferable.
Strategies for producing whole-genome sequence alignments also need to determine how genes (or more generally genomic intervals) are handled when they have no ortholog in a second species. For instance, according to fig. 5A and B, HBG1 and HBG2 in the human β-globin cluster have no ortholog in the dog genome (for either kind of orthology). Indeed, alignments available at the ENSEMBL Web site (ensembl.org) leave them unaligned to dog, whereas those at the UCSC Genome Browser (genome.ucsc.edu) align them to the most similar dog paralog. It is currently unclear to us, which approach is the correct one.
Figure 7 shows that gene clusters can have evolutionary histories that are much more complex than those of the globins, and unambiguous assignment of gene orthologs is frequently impossible (e.g., when an evolutionary operation affects only part of a gene), although multiway comparisons might help to resolve some inconsistent or ambiguous mappings. Moreover, a strict determination of orthology is confounded by typical genome-sequencing strategies, which cannot accurately assemble complex gene clusters. While any analysis of high-level genomic structure and evolutionary history necessarily depends on the quality of the input sequences and of fundamental lower-level analyses, such as assembly and local-alignment construction, the method reported here provides a rational framework for creating sequence alignments of human gene clusters to the corresponding clusters in other mammals under such conditions. For example, the conversion detector used by CHAP 2 takes precautions to minimize false positives that may be caused by alignment errors (Song, Hsu, Riemer, et al. 2011) and by purifying selection (Hsu et al. 2010).
While the amount of color assigned to a nonhuman gene in figure 7 is not informative, the underlying analysis can quantify the “amount of orthology,” which could be used to decide which gene in that species to align to a particular human gene. Alternatively, this analysis could be performed on a per-exon or even per-nucleotide basis. Although the general approach seems relatively straightforward, many details remain to be resolved in this ongoing project, including how best to splice the resulting alignments of gene clusters into a whole-genome sequence alignment so as to retain the existing alignments outside of gene clusters. In this and many other endeavors, the two concepts of orthology that are defined and explored here should be kept in mind.
Supplementary Material
Supplementary material and tables S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Federico Hoffmann for helpful discussion. This work was supported by grant HG02238 to W.M. from the National Human Genome Research Institute and National Science Foundation award DEB 0733029.
References
- Abi-Rached L, Moesta A, Rajalingam R, Guethlein L, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010;6(11):e1001192. doi: 10.1371/journal.pgen.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akahoshi M, et al. Association between IFNA genotype and the risk of sarcoidosis. Hum Genet. 2004;114(5):503–509. doi: 10.1007/s00439-004-1099-5. [DOI] [PubMed] [Google Scholar]
- Bailey J, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadavid L, Lun C. Lineage-specific diversification of killer cell Ig-like receptors in the owl monkey, a New World primate. Immunogenetics. 2009;61(1):27–41. doi: 10.1007/s00251-008-0342-y. [DOI] [PubMed] [Google Scholar]
- Chen J, Cooper D, Chuzhanova N, Ferec C, Patrinos G. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- Chiu J, et al. OrthologID: automation of genome-scale ortholog identification within a parsimony framework. Bioinformatics. 2006;22:699–707. doi: 10.1093/bioinformatics/btk040. [DOI] [PubMed] [Google Scholar]
- Colobran R, et al. Multiple products derived from two CCL4 loci: high incidence of a new polymorphism in HIV+ patients. J Immunol. 2005;174(9):5655–5664. doi: 10.4049/jimmunol.174.9.5655. [DOI] [PubMed] [Google Scholar]
- Datta R, Meacham C, Samad B, Neyer C, Sjlander K. Berkeley PHOG: phyloFacts orthology group prediction web server. Nucleic Acids Res. 2009;37:W84–W89. doi: 10.1093/nar/gkp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, et al. Copy number variation of CCL3-like genes affects rate of progression to simian-AIDS in Rhesus Macaques (Macaca mulatta) PLoS Genet. 2009;5:e1000346. doi: 10.1371/journal.pgen.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C. Positional orthology: putting genomic evolutionary relationships into context. Brief Bioinform. 2011;12(5):401–412. doi: 10.1093/bib/bbr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufayard J, et al. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- Edwards C, et al. Evolution of the human immunodeficiency virus envelope gene is dominated by purifying selection. Genetics. 2006;174(3):1441–1453. doi: 10.1534/genetics.105.052019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch D, et al. Duplication of the gamma-globin gene mediated by L1 long interspersed repetitive elements in an early ancestor of simian primates. Proc Natl Acad Sci U S A. 1991;88(16):7396–7400. doi: 10.1073/pnas.88.16.7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- Fitch W. Homology: a personal view on some of the problems. Trends Genet. 2000;16(5):227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Gish W, States D. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307(5714):1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Goodstadt L, Ponting C. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput Biol. 2006;2:e133. doi: 10.1371/journal.pcbi.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef T, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206(11):2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guethlein L, Aguilar AO, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J Immunol. 2007;179(1):491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- Guethlein L, Flodin L, Adams E, Parham P. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J Immunol. 2002;169(1):220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- Han M, Demuth J, McGrath C, Casola C, Hahn M. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardies S, Edgell M, Hutchison C. Evolution of the mammalian beta-globin gene cluster. J Biol Chem. 1984;259:3748–3756. [PubMed] [Google Scholar]
- Hardison R. Comparison of the beta-like globin gene families of rabbits and humans indicates that the gene cluster 5'-epsilon-gamma-delta-beta-3' predates the mammalian radiation. Mol Biol Evol. 1984;1:390–410. doi: 10.1093/oxfordjournals.molbev.a040326. [DOI] [PubMed] [Google Scholar]
- Hardison R, Gelinas R. Assignment of orthologous relationships among mammalian alpha-like globin genes by examining flanking regions reveals a rapid rate of evolution. Mol Biol Evol. 1986;3:243–261. doi: 10.1093/oxfordjournals.molbev.a040392. [DOI] [PubMed] [Google Scholar]
- Hardison R, Miller W. Use of long sequence alignments to study the evolution and regulation of mammalian globin gene clusters. Mol Biol Evol. 1993;10:73–102. doi: 10.1093/oxfordjournals.molbev.a039991. [DOI] [PubMed] [Google Scholar]
- Harris R. Improved pairwise alignment of genomic DNA [PhD thesis] [University Park (PA)]: Pennsylvania State University; 2007. [Google Scholar]
- Hoffmann F, Opazo J, Storz J. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol Biol Evol. 2008;25(3):591–602. doi: 10.1093/molbev/msn004. [DOI] [PubMed] [Google Scholar]
- Hou M. Algorithms for aligning and clustering genomic sequences that contain duplications [PhD thesis] [University Park (PA)]: Pennsylvania State University; 2007. [Google Scholar]
- Hou M, Riemer C, Berman P, Hardison R, Miller W. Aligning two genomic sequences that contain duplications. In: Ciccarelli F, Miklos I, editors. Comparative genomics: International Workshop (RECOMB-CG 2009), volume 5817 of Lecture Notes in Bioinformatics. Budapest, Hungary: Springer; 2009. pp. 98–110. [Google Scholar]
- Hsu C. Inference of orthologs, while considering gene conversion, to evaluate whole-genome multiple sequence alignments [PhD thesis] [University Park (PA)]: Pennsylvania State University; 2009. [Google Scholar]
- Hsu C, et al. An effective method for detecting gene conversion events in whole genomes. J Comput Biol. 2010;17(9):1281–1297. doi: 10.1089/cmb.2010.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, et al. Evolution of the CYP2ABFGST gene cluster in rat, and a fine-scale comparison among rodent and primate species. Genetica. 2008;133:215–226. doi: 10.1007/s10709-007-9206-x. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Bueno A, Dopazo J, Gabaldn T. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008;36:D491–D496. doi: 10.1093/nar/gkm899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. Orthologs and paralogs—we need to get it right. Genome Biol. 2001;2(8):1002.1–1002.3. doi: 10.1186/gb-2001-2-8-interactions1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, McGeoch C. Network flows and matching: first DIMACS implementation challenge. American Mathematical Society, Providence, RI; 1993. [Google Scholar]
- Kim K, Sung S, Caetano-Anolles G, Han J, Kim H. An approach of orthology detection from homologous sequences under minimum evolution. Nucleic Acids Res. 2008;36:e110. doi: 10.1093/nar/gkn485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen D, Wolf Y, Mushegian A, Koonin E. Computational methods for gene orthology inference. Brief Bioinform. 2011;12(5):379–391. doi: 10.1093/bib/bbr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, et al. Cross-referencing eukaryotic genomes: tIGR orthologous gene alignments (TOGA) Genome Res. 2002;12:493–502. doi: 10.1101/gr.212002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Farcia-Santre A. The virus battles: iFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001;12:143–156. doi: 10.1016/s1359-6101(00)00027-7. [DOI] [PubMed] [Google Scholar]
- Li H, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:572–580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert C, Roos D. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vazquez A, et al. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum Immunol. 2005;66(3):285–289. doi: 10.1016/j.humimm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Margulies E, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17(6):760–764. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, et al. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene. 2004;335:121–131. doi: 10.1016/j.gene.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Matsuya A, et al. Evola: ortholog database of all human genes in H-InvDB with manual curation of phylogenetic trees. Nucleic Acids Res. 2008;36:787–792. doi: 10.1093/nar/gkm878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, et al. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Hayashida H, Kikuno R, Toh H, Kawada Y. Evolution of interferon genes. Interferon. 1985;6:1–30. [PubMed] [Google Scholar]
- Muller J, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Opazo J, Hoffmann F, Storz J. Differential loss of embryonic globin genes during the radiation of placental mammals. PNAS. 2008;105(35):12950–12955. doi: 10.1073/pnas.0804392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo J, Sloon A, Campbell K, Storz J. Origin and ascendancy of a chimeric fusion gene: the β/δ-globin gene of paenungulate mammals. Mol Biol Evol. 2009;26(7):1469–1478. doi: 10.1093/molbev/msp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund G, et al. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounis C. Orthology: another terminology muddle. Trends Genet. 1999;15(11):445. doi: 10.1016/s0168-9525(99)01845-4. [DOI] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5(3):201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Pianezza M, Sellers E, Tyndale R. Nicotine metabolism defect reduces smoking. Nature. 1998;393:750. doi: 10.1038/31623. [DOI] [PubMed] [Google Scholar]
- Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172(1):356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- Sambrook J, et al. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;7:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilling H, Lienert-Weidenbach K, Valiante N, Uhrberg M, Parham P. Evidence for recombination as a mechanism for KIR diversification. Immunogenetics. 1998;48:413–416. doi: 10.1007/s002510050453. [DOI] [PubMed] [Google Scholar]
- Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2010. [cited 2012 Mar 22] Available from: http://www.repeatmasker.org. [Google Scholar]
- Song G, Hsu C, Riemer C, Miller W. Evaluation of methods for detecting conversion events in gene clusters. BMC Bioinfomatics. 2011;12(Suppl 1):S45. doi: 10.1186/1471-2105-12-S1-S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, et al. Conversion events in gene clusters. BMC Evol Biol. 2011;11:226. doi: 10.1186/1471-2148-11-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Zhang L, Vinar T, Miller W. CAGE: combinatorial analysis of gene-cluster evolution. J Comput Biol. 2010;17:1227–1242. doi: 10.1089/cmb.2010.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E, Koonin E. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- Su T, et al. Human cytochrome P450 CYP2A13 predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 2000;60:5074–5079. [PubMed] [Google Scholar]
- Tatusov R, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007;35:D343–D346. doi: 10.1093/nar/gkl978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden R, Snel B, van Noort V, Huynen M. Orthology prediction at scalable resolution by phylogenetic tree analysis. Nucleic Acids Res. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella A, et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19(2):327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 2003;63(22):8057–8061. [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Waterhouse R, Zdobnov E, Tegenfeldt F, Li J, Kriventseva E. OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011. Nucleic Acids Res. 2011;36:D271–D275. doi: 10.1093/nar/gkm845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, McVean G. Estimating diversifying selection and functional constraint in the presence of recombination. Genetics. 2006;172:1411–1425. doi: 10.1534/genetics.105.044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, et al. Plasticity in the organization and sequences of human KIRILT gene families. Proc Natl Acad Sci U S A. 2000;97(9):4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk C, Frost S, Richman D, Higley P, Pond S. Evolution of the interferon alpha gene family in eutherian mammals. Gene. 2007;397(1–2):38–50. doi: 10.1016/j.gene.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Evolutionary history reconstruction for mammalian complex gene clusters. J Comput Biol. 2009;16(8):1051–1070. doi: 10.1089/cmb.2009.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]