Abstract
MicroRNAs (miRNAs) are small RNA molecules involved in the regulation of mammalian gene expression. Together with other transcription regulators, miRNAs modulate the expression of genes and thereby potentially contribute to tissue and species diversity. To identify miRNAs that are differentially expressed between tissues and/or species, and the genes regulated by these, we have quantified expression of miRNAs and messenger RNAs in five tissues from multiple human, chimpanzee, and rhesus macaque individuals using high-throughput sequencing. The breadth of this tissue and species data allows us to show that downregulation of target genes by miRNAs is more pronounced between tissues than between species and that downregulation is more pronounced for genes with fewer binding sites for expressed miRNAs. Intriguingly, we find that tissue- and species-specific miRNAs target transcription factor genes (TFs) significantly more often than expected. Through their regulatory effect on transcription factors, miRNAs may therefore exert an indirect influence on a larger proportion of genes than previously thought.
Keywords: microRNA, transcription factor, gene expression, gene regulation, primates
Introduction
MicroRNAs (miRNAs) are small RNA molecules (∼18 to 24 nt) that regulate gene expression posttranscriptionally via messenger RNA (mRNA) destabilization or translational repression (Lee et al. 1993; Wightman et al. 1993; Lim et al. 2005; Friedman et al. 2009). MiRNAs are characterized by high sequence conservation across highly divergent species (Pasquinelli et al. 2000; Hertel et al. 2006; Chen and Rajewsky 2007; Stark et al. 2007), which reflects the effects of purifying selection due to their evolutionary importance as regulatory molecules. Expression profiling of miRNAs has been performed using cloning and Sanger sequencing (Landgraf et al. 2007), microarrays (Lim et al. 2005), and, most recently, by direct high-throughput sequencing (Creighton et al. 2009). High-throughput sequencing technologies have facilitated both the identification and the large-scale expression profiling of miRNAs in different tissues and cells. MiRNA profiling was also used to study expression across various developmental stages (Somel et al. 2010, 2011) and in comparison between healthy and diseased tissue (Erson and Petty 2008).
The destabilization of target mRNAs is the predominant mechanism of expression regulation by miRNAs (Guo et al. 2010). It is therefore possible to assess the extent to which miRNAs shape gene expression by quantifying both miRNA and mRNA expression in the same samples. The correlation between the expression of single miRNAs and their target genes has previously been studied in cell culture by knocking out/down endogenously expressed miRNAs or by introducing miRNAs into a specific target cell (Baek et al. 2008; Selbach et al. 2008; Guo et al. 2010). Genome-wide patterns of correlation between expression of miRNAs and their target genes in multiple tissues have been less widely explored.
In primates, the main focus has been on discovery and annotation of miRNAs (Berezikov et al. 2005, 2006a, 2006b; Kawaji et al. 2008; Yue et al. 2008; Baev et al. 2009; Brameier 2010), but much less is known about expression variation within and between species, and how these patterns vary in different cell types and tissues. Three studies have correlated the expression of miRNA, mRNA, and proteins in prefrontal cortex comparing humans and rhesus macaques (Somel et al. 2010) and between humans, chimpanzees, and rhesus macaques (Somel et al. 2011; Hu et al. 2011). These studies showed that miRNAs play a role in primate development and aging (Somel et al. 2010, 2011) and that miRNAs with human-specific expression patterns target genes involved in neuronal functions (Hu et al. 2011). However, an understanding of the evolution of gene expression regulation in primates across multiple tissues has not yet emerged. This is of particular interest given that it has been hypothesized that the phenotypic differences between species may be better explained by changes in gene expression than by changes in DNA sequence (King and Wilson 1975). Studies exploring differences in gene expression in multiple primate tissues have yielded lists of genes, which are differentially expressed in closely related species (Khaitovich et al. 2005), but it is less clear which regulatory changes drive these differences.
We have characterized miRNAs from multiple individuals in five different tissues (brain, heart, kidney, liver, and testis) from three primate species (humans, chimpanzees, and rhesus macaques) by Illumina sequencing. We examined the effects of miRNA-mediated regulation by comparing with a gene expression data set generated from the same samples as used for the miRNA expression profiling. Using these data sets, we address the following questions: 1) How big are miRNA expression differences between species and tissues? 2) Is there a particular group of genes regulated by miRNAs, which are differentially expressed between species and/or tissues? 3) Can the functional relationship between miRNAs and their target genes be determined by measuring their expression simultaneously?
Materials and Methods
Samples Library Preparation, Sequencing, and Base Calling
All the individuals used in this study were adult males and suffered sudden death that did not involve the tissues sampled. A description of the samples is available in supplementary tables S1 and S2 (Supplementary Material online).
Total RNA was prepared as described in the Illumina Inc. manual “Small RNA Sample Preparation Guide” (Part # 1004239 Rev. A Illumina Inc., San Diego, CA). Illumina Genome Analyzer I and II sequencing runs were analyzed starting from raw intensities. A detailed summary about the platform a sample was sequenced on, how many cycles and which chemistry were used can be found in supplementary table S2 (Supplementary Material online). Base calling and quality score calculation were performed for all runs using the Improved Base Identification System base caller (Kircher et al. 2009), trained on φX174 control reads of a dedicated control lane in each run. Reads with sequence entropy higher than 0.3 (sequence complexity filtering) and without any bases below a quality score of 10 were kept for downstream analysis.
Composition of Samples and Mapping of Small RNA Reads
We quantified expression of previously annotated miRNAs from miRBase (Griffiths-Jones et al. 2008; www.mirbase.org, release 15) for human, chimpanzee, and rhesus macaque. We mapped the sequenced reads to the official miRNA repository and the corresponding species' annotated miRNAs using PatMaN (Prüfer et al. 2008) allowing zero mismatches. Only reads with a length greater than 18 nt were used. The mature sequences were used as reference sequences for each miRNA. If a read was a substring of a miRNA sequence or vice versa, this read was assigned to this miRNA. If a read mapped to multiple miRNA sequences, the counts of this read were equally distributed to all matched miRNAs.
To classify reads that did not map to miRNAs in miRBase, we mapped the remaining reads to multiple databases in order to distinguish alternative read sources. We used the gene database provided by Biomart (ensembl.org/biomart) using all ENSEMBL genes (version 59) with their untranscripted sequence for human. To identify different structural RNAs, we used sequence annotation from UCSC of RNA genes based on the NCBI 36.1 (hg18) human reference genome. We obtained the sequences of piwiRNAs (piRNAs) for human from RNAdb (http://jsm-research.imb.uq.edu.au/rnadb/). Sequences not mapping to any of these databases were aligned to the corresponding species genomes (hg19, panTro2, rheMac2). The reads were distributed to the different categories in the following hierarchical order: miRNA, piRNA, rnaGenes, genes, unknown but mapped to the genome and not mapped to the genome.
mRNA Expression Data
We used mRNA sequence tags quantified using the Illumina NG III Digital Gene Expression approach (Velculescu et al. 1995; Kircher 2011). Samples were obtained from brain (prefrontal cortex), heart, kidney, liver, and testis tissues of male humans, chimpanzees, and rhesus macaques. The samples overlapped extensively with the samples used to generate the miRNA expression set (supplementary table S1, Supplementary Material online).
Normalization of miRNA and mRNA Data
We normalized both the miRNA and the mRNA data using the R package DESeq (Anders and Huber 2010). The package performs a negative binomial fit to the samples. In addition, we log transformed the normalized data.
Expressed miRNAs/Genes and Expression Differences
We defined a miRNA as expressed if more than one transcript was sequenced in one species and one tissue. A gene was considered to be expressed as defined in Kircher et al. (2011), that is, all used samples had to have of at least three mapped reads. Expression differences were calculated using the R library DESeq. The function nbinomTest provides a P value for each compared miRNA. In addition, a correction for multiple testing based on the Benjamini–Hochberg procedure was performed. We defined all miRNAs with an adjusted P value < 0.05 as differentially expressed. The same procedure was performed for the mRNA data.
Expression of Non-annotated miRNAs
A miRNA was defined to be expressed in another species than human if in at least one tissue the expression level was bigger than zero for at least four individuals and a Wilcoxon rank sum test showed no significant (alpha = 0.05) difference between the human samples expression values and the respective species samples expression values.
Species and Tissue Effect
To determine the effect of species and tissues in our data set, we used the set of miRNAs with at least ten transcripts in each species for the analyzed tissue. Additionally, only miRNAs annotated in all three species were included. We used the normalized data set. The fraction of variance explained by tissues and species was calculated by computing the fractions of sum of squares explained by the factor tissue and the factor species in a linear model.
miRNA Sequence Evolution
We calculated miRNA average conservation based on scores obtained from the multiple alignments of 46 vertebrate species using phastCons46way (Siepel et al. 2005). For humans, we calculated miRNA single nucleotide polymorphism (SNP) density using all SNPs from dbSNP (v.135) (Sherry et al. 2001).
Newly Predicted miRNAs
We used miRDeep (Friedländer et al. 2008) to identify potential new miRNAs. For this purpose, we merged reads of all samples in a species. Predictions with positive miRDeep scores and in orthologous regions (UCSC liftOver; Hinrichs et al. 2006) of all species were used for further investigations. Newly predicted miRNAs that were found in orthologous genomic regions of all three species were submitted to miRBase. Accession numbers form miRBase are assigned after publication acceptance.
Target Prediction
We obtained miRNA-binding sites for all mRNA genes from the TargetScanS (Lewis et al. 2003) database. The predictions included both conserved- and nonconserved-binding sites.
Functional Gene Ontology Analysis
We used the Gene Ontology (GO) (Ashburner et al. 2000) and the hypergeometric test from FUNC (Prüfer et al. 2007) to test for enriched GO categories among gene groups. Genes that were regulated by at least one miRNA with different expression were coded “1,” whereas all genes targeted by miRNAs that were expressed and showed no differential expression got the label “0.” FUNC-hyper was run with parameter −c 10, which includes categories with at least ten genes in it. As our significance measure (alpha = 0.05), we used the familywise error rate.
Random Distribution Calculation
Significance levels were computed by calculating a random distribution and comparing the observed values to this distribution. The random distribution is computed by randomly assigning expressed miRNAs to expressed genes from the prediction list. Each miRNA and gene had the same chance of assignment. For the correlation between tissues dependent on the number of binding sites for expressed miRNAs, we randomly permuted the number of binding sites between the miRNA and the mRNA pairs. The random assignments were performed 1,000 times.
Results
Small RNA Composition and Annotation
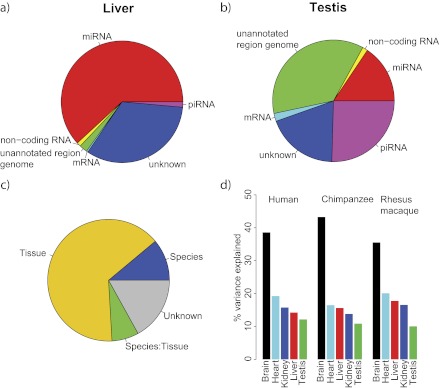
We sequenced 73 small RNA libraries (25 from humans, 24 from chimpanzees, and 24 from rhesus macaques) derived from 5 individuals of each of the species (human, chimpanzee, and rhesus macaque) for the tissues brain, liver, and heart. Five individuals of each species were sequenced for kidney (except chimpanzee with four individuals) and testis (with four rhesus macaques). When we mapped the reads to miRBase (www.mirbase.org, release 15; Griffiths-Jones et al. 2008; supplementary table S1, Supplementary Material online), the majority of reads matched known miRNAs (median 59%). However, testis yielded a consistently smaller fraction of matches for all species (median 9%). A considerable proportion of reads in testis were assigned to piRNAs, which are a different class of small RNAs that are mainly found expressed in the germ line (Girard et al. 2006; Aravin et al. 2007) (fig. 1a and b). An excess of 5′ uridine (U) residues is a signature of the piRNA family (Malone and Hannon 2009). This 5′ uridine (U) enrichment in piRNAs is clearly visible in testis-derived molecules (55% U in first position) compared with molecules from other tissues (supplementary fig. S1, Supplementary Material online). In addition, we found that in testis a higher fraction of reads failed to be assigned to small RNAs, structural RNAs, or mRNAs. However, these reads could be mapped to the respective genome and showed an even higher fraction of uridine in the first position than observed for the reads mapping to annotated piRNAs suggesting that they represent unannotated piRNAs (supplementary fig. S1, Supplementary Material online).
FIG. 1.—
Small RNA libraries composition and variance distribution for miRNA expression. (a and b) Small RNA composition for a sample of human brain and human testis, respectively. (c) Percentage of miRNA expression variance explained by factors tissue and species and their interaction. (d) Percentage of miRNA expression variance explained by different factors in tissues.
Combining the data of all five tissues, we identified 585 of the 718 (81%) known miRNAs in human, 431 of the 530 (81%) known miRNAs in chimpanzee, and 399 of the 502 (79%) known miRNAs in rhesus macaque (table 1). Although the number of different miRNAs detected is similar between tissues, brain and testis showed consistently more expressed miRNAs than other tissues (table 1).
Table 1.
Number of miRNAs Expressed in Each Tissue and in Each Species
| Cortex | Liver | Testis | Kidney | Heart | Total | Annotated | |
| Human | 522 | 412 | 497 | 477 | 445 | 585 | 718 |
| Chimpanzee | 375 | 298 | 357 | 341 | 306 | 431 | 530 |
| Rhesus macaque | 330 | 297 | 352 | 335 | 307 | 399 | 502 |
When comparing the repertoire of miRNAs between species, we found some instances in which a particular miRNA was annotated in human but not in one or both of the other species. Using our rhesus and chimpanzee expression data, we were able to detect 16 of these unannotated miRNAs in chimpanzee and 27 miRNAs in rhesus macaque. The expression level of these miRNAs in these other species was generally comparable with the expression level in human.
In order to identify miRNAs independently of their miRBase annotation, we applied the miRDeep algorithm (Friedländer et al. 2008) to our data set. miRDeep identifies miRNAs from deep sequencing of small RNA libraries by comparing the position and frequency of reads with the secondary structure of the predicted pre-miRNA. The algorithm provides a combined score indicating the reliability of the prediction; the more positive the score the more reliable the prediction. To reduce the false positive prediction rate, we applied a score cutoff of zero. That is, we required a higher likelihood for a true miRNA than random background. This resulted in the prediction of 649 miRNAs in human (2,993 total predictions, 331 known), 377 (3,063 total, 239 known) in chimpanzee, and 859 (3,538 total, 249 known) in rhesus macaque. Of these, 17 miRNAs were located in orthologous genomic regions in all three species and had a positive miRDeep score. One miRNA was predicted to be functional for both strands of the mature/star duplex in human. Seven miRNAs were independently described by others and were added to the new miRBase releases (version 17) during the preparation of this manuscript (Berezikov et al. 2006b; Goff et al. 2009; Creighton et al. 2010; Jima et al. 2010; Liao et al. 2010; Stark et al. 2010; Persson et al. 2011; Schotte et al. 2011; Dannemann et al. 2012). Twelve of the newly predicted miRNA candidates were tissue specific and eight of them were brain specific (supplementary fig. S2, Supplementary Material online).
For the analyses presented in this paper, we used only the set of all miRBase-annotated miRNAs detected in our sequencing data. For the between-species comparisons, we used miRNAs expressed in all three species and for the between-tissues comparisons, we used miRNAs expressed in all compared tissues in a given species. This is a rather stringent requirement, but it ensures that we compare bona fide miRNAs.
miRNA Expression Differences
We quantified miRNA expression and compared expression levels between species and tissues using the R package DESeq (Anders and Huber 2010). We found an approximately six times higher divergence in expression between tissues than between species. Divergence between tissues explained 65% of the miRNA expression variance, whereas differences between species explained around 11% (fig. 1c). Expression differences in brain explained the highest fraction of the tissue variance (41%) followed by differences in heart (20%), liver (16%), kidney (14%,) and testis (9%) (fig. 1d).
We compared the number of miRNAs with a significant expression difference between any two species (supplementary fig. S3, Supplementary Material online). When comparing the fraction of miRNAs with significant difference in expression, we found that brain and heart show consistently fewer differences between species than other tissues. Between humans and chimpanzees, 4% of all detected miRNAs differ significantly in expression in brain and 11% in heart. In contrast, liver and testis had the most differentially expressed miRNAs (52% for both tissues). When comparing the fractions of differentially expressed miRNAs between 1) human and rhesus macaque and 2) chimpanzee and rhesus macaque, we found a significant difference between the two proportions for testis (45% and 25% for human–macaque and chimpanzee–macaque, respectively; P = 0.006, Fisher's exact test) but no other tissue (P > 0.05).
When we summed the number of differently expressed miRNAs over all three pairwise species comparisons, we observed a higher number of differently expressed miRNAs between human–macaque and between chimpanzee–macaque as compared with human–chimpanzee. The same pattern was consistently observed in all individual tissue comparisons except in testis, which departed significantly from this trend. In testis, 38 miRNAs showed different expression between human and chimpanzee, 39 between chimpanzee and macaque, and 74 between human and macaque.
Interestingly, miRNAs with expression differences between tissues show often tissue-specific patterns, that is, these miRNAs differ in expression in only one tissue while expression is unchanged in the other four tissues. In humans, 50% of these tissue-specific differentially expressed miRNAs were found in brain (chimpanzee 73%, rhesus macaque 43%). In total, we found that nine of the ten miRNAs with a consistent tissue-specific expression in all three species were brain specific.
Correlation between miRNA Expression and Sequence Evolution
To calculate the correlation between miRNA expression and sequence evolution, we used two measures: sequence conservation among vertebrates and sequence variation within humans. We found positive Spearman correlations between average miRNA hairpin conservation score and miRNA expression for all species and tissues (supplementary fig. S4, Supplementary Material online). All but one (chimpanzee testis) were statistically significant (alpha = 0.05; supplementary table S3, Supplementary Material online). Conversely, we found negative Spearman correlations between the miRNA hairpin SNP number and miRNA expression though only in brain was the correlation statistically significant (supplementary fig. S4 and table S3, Supplementary Material online).
Functional Analysis of Differentially Expressed miRNAs
To evaluate the effect of miRNA expression on the transcriptome, we used mRNA sequence tags quantified using sequencing using the Illumina NG III Digital Gene Expression approach (Velculescu et al. 1995; Kircher et al. 2011). Of the 73 samples, 64 were common to both the mRNA and the miRNA studies (supplementary table S2, Supplementary Material online). mRNA targets of our expressed miRNAs were identified using TargetScanS (Lewis et al. 2003).
We tested for functional enrichment of target genes in two of the GO (Ashburner et al. 2000) domains (molecular function and biological process) using FUNC (Prüfer et al. 2007). The multitissue/species comparisons allow us to distinguish two groups of differentially expressed miRNAs. The first group is the tissue-specific miRNAs, which were identified in the within-species comparisons (a total of 15 pairwise tests: 3 species × 5 tissues). For a given species, we required that all miRNAs were expressed in all five tissues and filtered for miRNAs that were differentially expressed in one tissue compared with all others. The second group is the species-specific miRNAs, which were identified in the between-species comparisons (a total of 15 pairwise tests: 3 pairwise species comparisons × 5 tissues). For a given tissue, we required all miRNAs to be expressed in all species and filtered for those miRNAs that are differentially expressed in a species comparison.
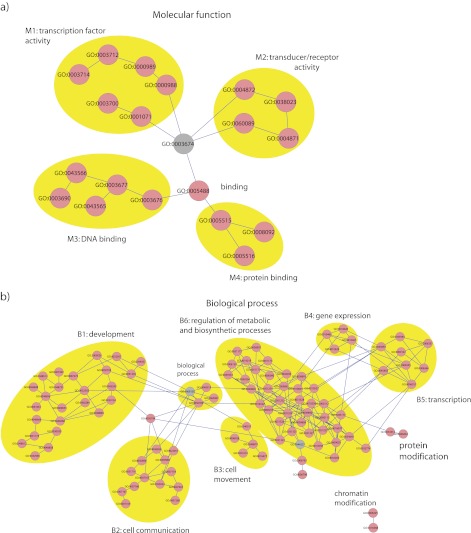
We found 93 GO categories to be significantly enriched in at least one test for tissue-specific miRNA targets and 103 categories in at least one test for species-specific miRNA targets. The significantly enriched categories overlapped substantially (62 categories) between the species- and the tissue-specific tests (supplementary table S4, Supplementary Material online). We identified several highly connected clusters in the directed acyclic graph representation of the GO, which were related to development, cell communication, gene and transcript expression, cell movement, regulation of metabolic, and biosynthetic processes and DNA/protein/transcription binding (figs. 2a, 2b, and 3; supplementary figure S5, Supplementary Material online).
FIG. 2.—
Enriched GO categories in the GO domains molecular function (a) and biological process (b). For each domain, a subgraph from the GO is depicted. GO categories with related functions (clusters M1–M4 and B1–B6) are highlighted in yellow ellipses. Significantly enriched categories are graph nodes shown in pink and categories linking the clusters are graph nodes shown in gray.
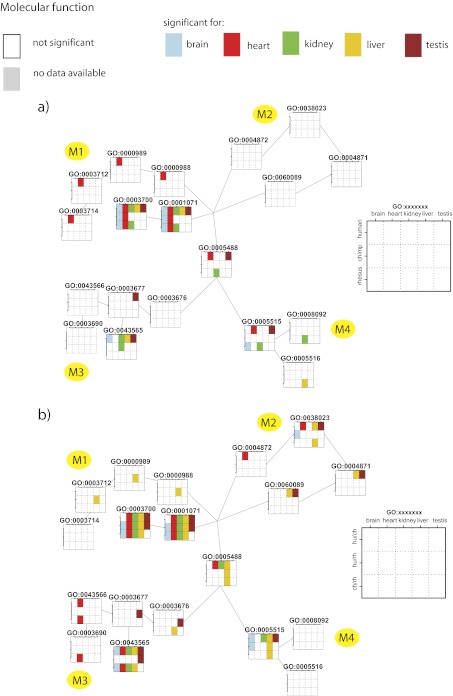
FIG. 3.—
Enriched GO categories for the tissue-specific (a) and species-specific (b) tests in the GO domain molecular function. The same subgraph as in figure 2b is depicted. Each node (GO category) is represented by a grid, and the clusters are named from M1–M4 as in figure 2b. Test results are color encoded: squares in white are not significantly enriched for a given category, squares in gray depict tests not carried out due to insufficient data, and other colors depict significant tests (P < 0.05). Colors were used to better visualization of tissues distribution of significant tests. Color code explained in the inset.
We found that transcription factor–binding categories were significantly enriched—23 of the total 30 GO tests performed. The signal was even more pronounced in the species-specific tests, where 13 of 15 tests were significant. Additionally, all tissue-specific tests in human were significant. In categories related to development, most of the significant enrichments were found in brain for both tissue-specific tests (16 of 25 significant tests in these categories were brain specific) and species-specific comparisons (15 of 21 significant tests in these categories were brain specific). Another cluster that showed enrichment for brain-related categories was cell communication in the species-specific test (of 22 significant tests, 13 were brain specific and 8 were testis specific). Additionally, we found that categories related to regulation of metabolic and biosynthetic processes (a total of 101 significant tests) were preferentially enriched in kidney (35) and liver (32) compared with brain (8), heart (6), and testis (10).
Correlation between Transcription Factor Expression and miRNA Regulation
Based on the functional enrichment for transcription factor activity, we sought to further investigate the regulatory relationship between miRNAs and transcription factors for the tissue comparisons. As a proxy for this functional relation, we used transcription factor expression and the number of miRNA-binding sites in each transcription factor. To obtain an annotation of transcriptions factors that is independent of GO categories, we designated a gene as a transcription factor if it was annotated in TRANSFAC (Kel et al. 2003). Using mRNA expression data and TRANSFAC transcription factor annotation, we identified 94 transcription factors that were expressed in all five tissues. We identified differentially expressed transcription factors by performing pairwise tissue comparisons. We defined a transcription factor as tissue specific if it was differentially expressed between at least one tissue compared with the four others, that is, a transcription factor was allowed to show specific tissue expression patterns for one or more tissues. For each transcription factor, we obtained the number of 3′ untranslated region (UTR)-binding sites for expressed miRNAs. We calculated the Spearman correlation (rho) between the number of times a transcription factor is tissue-specific and the number of binding sites per expressed mRNA for an expressed miRNA and obtained a negative correlation (rho = −0.15, P = 0.16). We compared the distributions of the number of binding sites for expressed miRNAs between tissue-specific and nontissue-specific transcription factors and found a nonstatistically significant shift between the distributions (supplementary fig. S6, Supplementary Material online). We repeated the two analyses using only binding sites for miRNAs that are highly differentially expressed between tissues (tissue specific for more than two tissues) and obtained a negative correlation (rho = −0.18, P = 0.09) and a significant difference between the number of miRNA-binding sites for the two groups of transcription factors (Wilcoxon rank sum test, P = 0.03). Transcription factors with fewer binding sites for expressed miRNAs tend to show a higher variability in their expression between tissues than those with more binding sites.
Correlation of Expression between miRNAs and Their Predicted Target Genes
To determine the functional relationship between miRNAs and their predicted target genes, we correlated the expression values of miRNAs and genes. For each miRNA–mRNA target pair, we required that both the miRNA and the mRNA were expressed in all species or tissues. We then carried out two tests to determine the level of correlation at different levels of granularity. Each test calculates the Pearson correlation coefficient (r) for each pair.
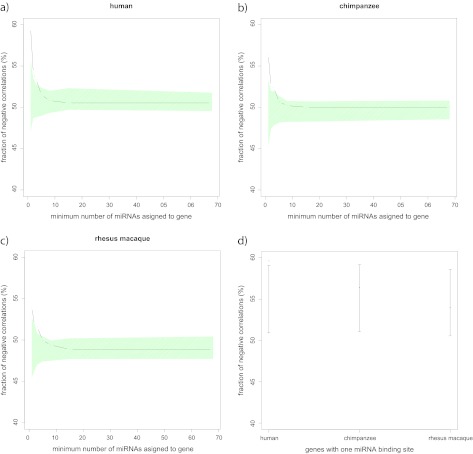
For our first test, we correlated miRNA and mRNA expression between tissues in each species. We averaged the miRNA and mRNA expression values over all individuals per tissue and species. We then calculated the Pearson correlation coefficient between miRNA expression and mRNA expression levels for the five tissues. The fraction of negative correlations observed was not different from the fraction obtained by randomly assigning miRNAs and mRNAs. When comparing genes with one miRNA-binding site to genes with more than one, we found a significant signal in all species (to obtain significance, we randomly permuted the number of binding sites in genes, p_hu < 0.01, p_ch < 0.01, p_rh < 0.01; fig. 4). However, when we restricted our analysis to genes with only one miRNA-binding site, we found that the fraction of negative correlations was significantly higher than by randomly assigning miRNAs and mRNAs in human but none of the other species (p_hu = 0.02, p_ch = 0.26, p_rh = 0.65; fig. 4).
FIG. 4.—
Relation amid the fraction of negative correlations between the expression of miRNA–mRNA pairs (in the between-tissue correlation) and the number of miRNA-binding sites in 3′ UTRs of genes for human (a), chimpanzee (b), and rhesus macaque (c). The black points and lines show the observed fraction of negative correlations. The green-shaded area shows the 95% confidence intervals (CIs) obtained by randomly permuting miRNA-binding sites across genes. Fraction of negative correlations between the expression of miRNA–mRNA pairs (in the between-tissue correlation) for genes with only one 3′ UTR miRNA-binding site in human, chimpanzee, and rhesus macaque. (d) The red points show the observed fraction of negative correlations and in black the 95% CIs obtained by randomly assigning miRNAs and mRNAs from the original miRNA–mRNA set.
For our second test, we sought to test the correlation of miRNA to target mRNA between species. For each tissue and miRNA–mRNA pair, we arrived at three datapoints that are used as input for the individual correlations. The correlations were calculated for each pair, and the resulting fraction of negative correlations was tested against random target assignments. We also tested the fraction of negative correlations dependent on the number of miRNA-binding sites in the genes. None of the tissues gave a significant excess of negative correlations, and the amount of negative correlation was not different from the amount obtained from random assignments of genes to miRNAs (all P values > 0.05). The excess of negative correlation also did not differ depending on the number of miRNA-binding sites in the genes.
It has been proposed that miRNAs exert their effect by setting the mean and reducing the variance of the genes that they regulate, and in this way, they stabilize phenotypes a process called canalization (Wu et al. 2009). Our results are consistent with miRNAs setting the mean expression of the genes they regulate. For technical reasons, we are not able to test where there is also a reduction in the variance of mRNA expression. We found a correlation between the mean expression level of the genes and the relative variance to the mean in the mRNA data set, that is, miRNAs tend to target highly expressed genes, and these genes also show a higher variance due to the fact that their expression level and their variance are not independent. Unfortunately, we found no general normalization that eliminated the correlation between the variance estimate and the expression of a given gene.
Discussion
In this study, we have characterized small RNA libraries in five tissues of humans, chimpanzees, and rhesus macaques using high-throughput sequencing. For four of the five tissues, the majority of the sequencing reads mapped to known miRNAs. In contrast, testis showed only a small percentage (9%) of reads matching known miRNAs. A larger fraction mapped to piRNAs (17%) while about one-third mapped to the genome but did not overlap any known RNA annotation. PiRNAs are known to be expressed specifically in germ line and gonadal somatic cells (Siomi et al. 2011) and are associated with the PIWI proteins, which are indispensable proteins for germ line development (Siomi et al. 2011) in many animals (Ghildiyal and Zamore 2009). They are divided in two classes: the first class is involved in silencing transposons, whereas the function of the second class remains unknown (Aravin et al. 2007). PiRNAs have a length distribution of between 25 and 35 nt (Aravin et al. 2001). Our library preparation protocol aims at extracting molecules of up to 25 nt in length, implying that the set of piRNAs discovered in our study is biased toward smaller sizes. Despite this limitation, we were able to detect a bias for 5′ uridine (U) residue (Malone and Hannon 2009) and an excess of adenosine at position 10 (Friedländer et al. 2009) that characterize piRNAs. Interestingly, testis reads that mapped to each species' corresponding genome but did not overlap any known RNA annotation showed the same patterns, suggesting that a large fraction of piRNAs remain to be characterized, as they are not among the ∼32,000 human piRNAs reported in the database we used for mapping (Pang et al. 2007).
Summing over all tissues, we were able to detect approximately 80% of all known miRNAs for each of the three species. When counting the number of miRNAs expressed, we observed a difference between tissues. In all three species, brain and testis show the most diverse miRNA repertoire. For testis, this result is surprising given the reduced power that we have to detect lowly expressed miRNAs because of the large fraction of reads representing other small RNAs that may or may not be transcribed from unannotated parts of the genome. In contrast, the miRNA repertoires of heart and liver are less diverse (table 1). Given that miRNAs have been shown to regulate mRNA abundance it is to be expected that the diversity of expressed miRNAs is linked to the number of different mRNAs expressed in each tissue. MRNA expression diversity was shown in previous studies to be highest in testis (Khaitovich et al. 2005; Kircher et al. 2011). However, gene expression diversity in brain was similar to the diversity seen in liver, kidney, and heart, and not more diverse, as observed for miRNA expression in the brain. This discrepancy between mRNA and miRNA diversity is further supported by our observation that a large fraction of the newly predicted miRNAs were brain specific (supplementary fig. S2, Supplementary Material online). Our data thus suggest that a larger fraction of genes may be regulated by miRNAs in brain as compared with other tissues. If true, this would lend further support to the central role attributed to regulatory RNAs in brain function (Mattick 2011). However, we also note that brain is an intensively studied tissue and that miRNAs annotated in miRBase could be potentially biased toward brain-specific miRNAs. A bias in annotation cannot, however, explain why newly discovered miRNAs are often brain specific. MiRNAs show high sequence conservation between species (Pasquinelli et al. 2000; Hertel et al. 2006; Chen and Rajewsky 2007; Stark et al. 2007). In this study, we found a similar high conservation in their expression between primate species. In agreement with previous gene expression studies, the divergence between tissues is higher than the divergence between species. This is reflected in the high amount of variance (65%) explained by the factor “tissue” in the linear model that we applied (fig. 1c). The high conservation of miRNA expression levels and tissue-specific expression patterns were previously observed in a comparative study of 26 tissues in humans and rodents (Landgraf et al. 2007). In our study, miRNAs expressed in brain explained the highest fraction of tissue variance (fig. 1d). It has been shown that brain has the smallest mRNA expression differences between species, and it was concluded that purifying selection is extremely efficient at eliminating mutations that modify gene expression in this tissue (Khaitovich et al. 2005, 2006). We found that many miRNAs are specifically expressed in brain. We therefore hypothesize that miRNAs, in addition to purifying selection, may explain the highly conserved mRNA expression patterns for this tissue.
Although the expression differences in miRNAs between species are small, they are sufficient to pinpoint miRNAs with a difference in expression between the primate species studied. Humans and chimpanzees show more similarity than either shows to rhesus macaques. The differences between species are particularly small in brain and heart and much bigger in liver and testis. It has been previously shown that mRNA expression is subjected to different levels of constraint in different tissues (Khaitovich et al. 2005, 2006). Using rhesus macaque as outgroup to assign differences, we found no excess of differential expression on the human or chimpanzee lineage for the tissues brain, kidney, liver, and heart. In testis, however, we observed that chimpanzees and rhesus macaques are much closer to one another than either is to humans (supplementary fig. S3, Supplementary Material online). Reproductive pressures such as sperm competition may be similar for chimpanzees and rhesus macaques (Dixson 1998), and it has been hypothesized that selection may have shaped both the protein sequence evolution and the mRNA expression of male reproductive genes (Wyckoff et al. 2002; Khaitovich et al. 2005, 2006). Our data suggest that adaptive changes may have shaped the expression patterns of miRNAs in testis on the human lineage.
We sought to study the relation between expression and sequence evolution. We found a positive correlation between miRNA sequence conservation and expression level. It has previously been shown in humans and Drosophila that purifying selection is weaker in lowly expressed miRNAs and that they therefore have less constraint in sequence evolution than highly expressed miRNAs (Lu et al. 2008; Liang and Li 2009). Highly expressed miRNAs are important regulators of gene expression, and their sequence conservation is therefore accordingly higher. The levels of purifying selection can be also assessed by the within species diversity. We took advantage of the extensive human diversity databases and found negative correlations between miRNA expression and the number of miRNA polymorphisms. A similar effect has been reported in humans (Liang and Li 2009). However, the effect was not as strong as the correlation between expression and conservation. The lack of polymorphism data available for other primate species meant that we were not able to perform the same analysis on these.
In addition to known miRNAs, we predicted 17 new miRNAs in the three species studied. It has been debated whether newly predicted miRNAs are functional or not, especially because they tend to have lower expression and more sequence divergence than known miRNAs (Lu et al. 2008; Liang and Li 2009). While this paper was under review, 7 of our 17 newly predicted miRNAs were reported by other studies and add to miRBase (supplementary fig. S2, Supplementary Material online). The fact that these miRNAs were independently predicted by other groups, together with their orthologous location in the three primate species and their tissue-specific expression patterns support that our predictions are reliable (supplementary table S5, Supplementary Material online). Our comparison of the expression levels of newly predicted miRNAs with known miRNAs showed that the former group tended to have lower expression values (supplementary fig. S7, Supplementary Material online). This is in agreement with previous studies and supports the hypothesis that newly emerged miRNAs are raw material for evolution that only have weak or negligible impact on gene expression after their emergence (Liang and Li 2009).
Mammalian miRNAs exert their regulatory action by decreasing target mRNA levels (Guo et al. 2010). We therefore expected to find a negative correlation between miRNAs and their target genes. We used miRNA targets that were computed based on genome-wide prediction algorithms that take sequence complementarity into account. For these predicted targets, we performed the analysis at tissue and species level. We did not find an excess of negative correlation between miRNA and mRNA expression in the between tissues and between species comparisons. However, when we restricted our set of targets to genes with only one binding site for expressed miRNAs, we did find an excess of negative correlation in the between tissues comparison, whereas the between species comparison gave no signal. This difference in results may be due to a difference in power; tissue divergence exceeds species divergence, with tissue differences explaining the majority of variance in miRNA expression (fig. 1c). It is possible that technical variation exceeds the effect exerted by miRNAs at the gene level in the between species comparisons, whereas the comparison between tissues yields significant results due to more pronounced differences in expression. The lack of power in the between-species comparison may be further exacerbated by the use of human-based target prediction databases that could potentially obscure species-specific effects. However, a substantial source of error may lie in the high false positive rate of computational miRNA target prediction, which poses a challenge for understanding the impact of miRNAs on gene expression regulation (Alexiou et al. 2009). Another source of noise for finding the regulatory relation between the expression of miRNA–mRNA pairs is the involvement of other regulatory molecules that hinder a clear measurement of the effect of miRNAs alone. Transcription factors, in particular, can regulate transcription positively and negatively (Hobert 2008) and their effect strength has been reported to be larger than that of miRNAs: in Caenorhabditis elegans individual deletion of miRNA loci only caused developmental and morphological defects in <10% of the cases (Miska et al. 2007), whereas RNA interference experiments for transcription factors resulted in observable effects in ∼30% of the cases reviewed in Hobert (2008). The action of transcription factors may therefore lead to false signals of apparent positive correlation between miRNA–mRNA pairs, and mRNA expression differences have to be seen as the result of the sum of many regulating factors impacting the expression level.
The pronounced excess of negative correlations, we observe when using only genes with fewer binding sites for expressed miRNAs indicates that the power to measure the functional relation between miRNA–mRNA pairs is higher for this group of genes. Genes regulated by multiple expressed miRNAs introduce more noise and hinder our ability to measure the impact of each miRNA on mRNA expression values. Two studies of development and aging in humans and macaque brains (Somel et al. 2010), and humans, chimpanzee, and macaque brains (Somel et al. 2011) have shown that age-related miRNA expression profiles are more negatively correlated with their targets than random expectations. A similar effect has been reported by comparing mRNAs, miRNAs, and proteins from human and chimpanzees using two different brain regions (Hu et al. 2011).
By comparing multiple species and tissues, we found that genes targeted by differentially expressed miRNAs were enriched in some GO categories, whereas genes targeted by miRNAs with uniform expression showed no signal of enrichment. The significant categories formed functionally related clusters in the GO graphs for the biological process and molecular function domains (fig. 2a and b). A significant enrichment for genes with transcription factor–binding activity was found consistently in the majority of tests both between species and between tissues (fig. 3). This suggests that miRNAs, as downregulators of gene expression, preferentially modulate transcription factors, which are themselves transacting regulatory molecules. It has been shown that transcription factors that are differentially expressed between human and chimpanzee brains preferentially regulate genes involved metabolism and transcription among others (Nowick et al. 2009). Additionally, in plants, miRNAs preferentially regulate transcription factors that are involved in development (Rhoades et al. 2002; Jones-Rhoades and Bartel 2004). We speculate that miRNAs may therefore also have an indirect influence on the expression of these types of genes.
As a second line of evidence for the connection between miRNAs and transcription factors, we measured the functional relationship between transcription factor expression differences and miRNA regulation between tissues. To do that we used as a proxy the number of binding sites for expressed miRNA present in transcription factors 3′ UTRs. We found that differentially expressed transcription factors contain fewer miRNA-binding sites than transcription factors that are not differentially expressed. MiRNAs have been speculated to have the function of stabilizing the expression of their targets (Kircher et al. 2008; Wu et al. 2009). In our between-tissue comparison, this would mean that the miRNA expression level is adjusted to maintain homogeneous mRNA expression. Transcription factors that are differentially expressed show a depletion in miRNA-binding sites. We hypothesize that these transcription factors achieve differential expression by reducing the number of miRNA-binding sites and therefore avoiding the control by expressed miRNAs.
Further studies integrating miRNA–mRNA profiles with data sets produced by shotgun proteomics and ribosome profiling will further improve the understanding of gene expression and gene expression regulation in primates and help to disentangle the relative contributions of different gene expression regulatory machinery and their impact on phenotype in primate evolution.
Supplementary Material
Supplementary figures S1–S7 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Thomas Giger for performing the frozen tissue dissections; Ines Drinnenberg, Matthias Meyer, and the Sequencing Group of the Max Planck Institute for Evolutionary Anthropology for providing sequencing runs; Martin Kircher for technical assistance with sequence processing; Marike Schreiber for assistance with the figure preparation; Cesare de Filippo, Martin Kircher, Michael Lachmann, Mehmet Somel, and Martin Surbeck for useful discussions during the project and for comments on the manuscript. The project was funded by a grant of the Max Planck Society.
References
- Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baev V, Daskalova E, Minkov I. Computational identification of novel microRNA homologs in the chimpanzee genome. Comput Biol Chem. 2009;33:62–70. doi: 10.1016/j.compbiolchem.2008.07.024. [DOI] [PubMed] [Google Scholar]
- Berezikov E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Berezikov E, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006a;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Berezikov E, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006b;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameier M. Genome-wide comparative analysis of microRNAs in three non-human primates. BMC Res Notes. 2010;3:64. doi: 10.1186/1756-0500-3-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief Bioinformatics. 2009;10:490–497. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, et al. Discovery of novel microRNAs in female reproductive tract using next generation sequencing. PLoS One. 2010;5:e9637. doi: 10.1371/journal.pone.0009637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M, Nickel B, Lizano E, Burbano HA, Kelso J. Annotation of primate miRNAs by high-throughput sequencing of small RNA libraries. BMC Genomics 13:116. 2012 doi: 10.1186/1471-2164-13-116. Advance Access publication March 27, 2012. doi:10.1186/1471-2164-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixson AF. Primate sexuality. Oxford: Oxford University Press; 1998. [Google Scholar]
- Erson AE, Petty EM. MicroRNAs in development and disease. Clin Genet. 2008;74:296–306. doi: 10.1111/j.1399-0004.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, et al. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci U S A. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Goff LA, et al. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS One. 2009;4:e7192. doi: 10.1371/journal.pone.0007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, et al. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- Hu HY, et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 2011;7:e1002327. doi: 10.1371/journal.pgen.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jima DD, et al. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifies hundreds of novel microRNAs. Blood. 2010;116:e118–e127. doi: 10.1182/blood-2010-05-285403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Kawaji H, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel AE, et al. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, et al. Positive selection on gene expression in the human brain. Curr Biol. 2006;16:R356–R358. doi: 10.1016/j.cub.2006.03.082. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kircher M. 2011. Understanding and improving high-throughput sequencing data production and analysis [dissertation]. [Leipzig (Germany)]: Fakultät für Mathematik und Informatik. [Google Scholar]
- Kircher M, Bock C, Paulsen M. Structural conservation versus functional divergence of maternally expressed microRNAs in the Dlk1/Gtl2 imprinting region. BMC Genomics. 2008;9:346. doi: 10.1186/1471-2164-9-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 2009;10:R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Liang H, Li WH. Lowly human expressed microRNA genes evolve rapidly. Mol Biol Evol. 2009;26:1195–1198. doi: 10.1093/molbev/msp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JY, et al. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lu J, et al. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol Biol Evol. 2008;25:929–938. doi: 10.1093/molbev/msn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. The central role of RNA in human development and cognition. FEBS Lett. 2011;585:1600–1616. doi: 10.1016/j.febslet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci U S A. 2009;106:22358–22363. doi: 10.1073/pnas.0911376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, et al. RNAdb 2.0—an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–D182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Persson H, et al. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011;71:78–86. doi: 10.1158/0008-5472.CAN-10-1869. [DOI] [PubMed] [Google Scholar]
- Prüfer K, et al. FUNC: a package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, et al. PatMaN: rapid alignment of short sequences to large databases. Bioinformatics. 2008;24:1530–1531. doi: 10.1093/bioinformatics/btn223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Schotte D, et al. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011;25:1389–1399. doi: 10.1038/leu.2011.105. [DOI] [PubMed] [Google Scholar]
- Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Somel M, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–1218. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MS, et al. Characterization of the melanoma miRNAome by deep sequencing. PLoS One. 2010;5:e9685. doi: 10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu CI, Shen Y, Tang T. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome Res. 2009;19:734–743. doi: 10.1101/gr.084640.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff GJ, Li J, Wu CI. Molecular evolution of functional genes on the mammalian Y chromosome. Mol Biol Evol. 2002;19:1633–1636. doi: 10.1093/oxfordjournals.molbev.a004226. [DOI] [PubMed] [Google Scholar]
- Yue J, Sheng Y, Orwig KE. Identification of novel homologous microRNA genes in the rhesus macaque genome. BMC Genomics. 2008;9:8. doi: 10.1186/1471-2164-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]